Abstract
The yjbEFGH operon is implicated in the production of an exopolysaccharide of an unknown function and is induced by osmotic stress and negatively regulated by the general stress response sigma factor RpoS. Despite the obvious importance of RpoS, negative selection for rpoS has been reported to take place in starved cultures, suggesting an adaptive occurrence allowing the overexpression of RpoD-dependent uptake and nutrient-scavenging systems. The trade-off of the RpoS-dependent functions for improved nutrient utilization abilities makes the bacterium more sensitive to environmental stressors, e.g., osmotic stress. In this work, we addressed the hypothesis that overinduction of genes in rpoS-deficient strains indicates their essentiality. Using DNA microarrays, real-time PCR, and transcriptional fusions, we show that genes of the wca operon, implicated in the production of the colanic acid exopolysaccharide, previously shown to be induced by osmotic stress, are also negatively controlled by RpoS. Both exopolysaccharides in the synthesis of which yjb and wca are involved are overproduced in an rpoS mutant during osmotic stress. We also show that both operons are essential in an rpoS-deficient strain but not in the wild type; promoters of both operons are constitutively active in yjb rpoS mutants; this strain produces extremely mucoid colonies, forms long filaments, and exhibits a reduced growth capability. In addition, the wca rpoS mutant's growth is inhibited by osmotic stress. These results indicate that although induced in the wild type, both operons are much more valuable for an rpoS-deficient strain, suggesting that the overproduction of both exopolysaccharides is an adaptive action.
The role of the general stress response sigma factor RpoS (σ38) in Escherichia coli's physiology has been extensively studied. While most studies have focused on the characterization of genes positively controlled by RpoS and on the physiological implications of the underexpression of these genes in rpoS-deficient strains (22, 61), only a few refer to genes negatively regulated by RpoS and to the consequences of their overexpression.
Inactivation of rpoS is common; mutations in the rpoS gene have been detected among laboratory bacterial stocks (25, 52), as well as in environmental and clinical isolates of pathogenic and commensal enteric bacteria (1, 43), suggesting that under certain conditions, the loss or attenuation of RpoS activity may be of adaptive value. Other reports indicated that the rpoS gene tends to undergo frequent mutations that lead to loss of activity and that the mutated forms appear to spread and become dominant in glucose-limited chemostat cultures (39) or during incubation in stationary phase (5, 66). It was also shown that the rate of rpoS mutations spreading throughout the population decreased when osmotic stress was applied to the system (14, 28). The selection pressure on the rpoS gene is thought to be largely due to a competition between the sigma factors RpoS and RpoD for the limited number of RNA polymerase core subunits (13, 32). This suggests that rpoS inactivation may be beneficial to a starved cell due to the overexpression of RpoD-dependent nutrient-sensing and uptake systems.
It was previously shown (24, 44) that among E. coli genes shown to be significantly induced by osmotic stress, one gene, yjbF, stood out in that the promoter that drives its induction appeared to be negatively regulated by RpoS. This gene, a member of a four-gene operon (yjbEFGH), was listed (15) among genes that are positively dependent upon RcsC, the inner membrane sensor kinase of the Rcs phosphorelay system. The Rcs system controls a variety of physiological functions in prokaryotes, such as extracellular polysaccharide (EPS) synthesis (27, 42, 51, 58), biofilm formation (10, 15, 41), cell division (3), and motility (17, 53). It has been shown (16) that the four genes of the yjbEFGH operon are transcribed together, that this operon is induced during growth on solid surfaces, and that it is involved in the production of an unknown EPS.
E. coli K-12 possesses five known sets of genes promoting EPS production: (i) genes involved in O-antigen synthesis (46); (ii) the wca operon, responsible for colanic acid (CA)-EPS synthesis (19); (iii) the pgaABCD operon which encodes genes involved in the production of poly-β-1,6-N-acetyl-d-glucosamine (PGA), a polysaccharide shown to be crucial for biofilm formation (59); (iv) the dfc pseudo-operon (comprised of gfcABCDE, etp, and etk), responsible for the production of type IV capsule in E. coli O127:H6 (this operon is nonfunctional in E. coli K-12 due to the presence of an IS1 element in its promoter region [40]); and (v) the yjbEFGH operon, a paralogue of dfcABCD (15).
We propose that overinduction of the yjb operon and, possibly, that of other genes involved in EPS production may be an adaptive response to osmotic stress in an rpoS-deficient background and that these overinduced genes are of special value for an rpoS-deficient strain. Our results indicate that alternative stress response strategies may come into play in the absence of RpoS or when its activity is diminished, allowing the cells to survive and proliferate even without its general stress protection.
MATERIALS AND METHODS
Strains and plasmids.
The strains used in this study are listed in Table 1. All strains were grown in Luria-Bertani medium (LB broth; 5 g liter−1 yeast extract, 10 g liter−1 Bacto-tryptone, and 5 g liter−1 NaCl) supplemented with the appropriate antibiotics (100 μg ml−1 ampicillin, 10 μg ml−1 tetracycline, 50 μg ml−1 kanamycin) to an optical density at 600 nm (OD600) of 1 to 2 (Spekol 1200; Analytikjene, Germany), mixed with 50% glycerol, and kept at −80°C. Cells were recovered from freezing by overnight growth on LB agar plates. Plates were kept for up to 1 month at 4°C.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype | Source or reference(s) |
|---|---|---|
| Strains | ||
| MG1655 | F− λ− | Laboratory stock |
| QC2410 | MG1655 rpoS::Tn10 | 31 |
| MK1399 | MG1655 ΔyjbEFGH(kanR) | This work |
| MK1310 | MG1655 ΔyjbE(kanR) | This work |
| MK1351 | MG1655 ΔPwza(kanR) (allele introduced from MS1651 [40]) | This work |
| MK2499 | QC2410 ΔyjbEFGH(kanR) | This work |
| MK2431 | QC2410 ΔyjbE(yjbE::kanR) | This study |
| MK2411 | QC2410 ΔPwza(kanR) | This work |
| CF6343 | MG1655 ΔlacIZ(MluI) | 8 |
| SK1150 | CF6343 rcsA::kanR | 17 |
| SK1158 | CF6343 rcsB::Tn10 | 8 |
| SK1154 | CF6343 rcsC::Tn10 | 8 |
| MS1651 | EPEC O126:H6 ΔPwza(kanR) | 40 |
| DY378 | W3110 λcI857Δ(cro-bioA) (permissive strain for gene inactivation) | 64 |
| Plasmids | ||
| pDEW201 | Promoterless; luxCDABE ori pMB1(pBR322) rop ampR | 56, 57 |
| pDEW/yjbF (pDEW609) | pDEW201; yjbF′::luxCDABE (pgi-yjbF) | 56, 57 |
| pDEW/wza | pDEW201; wza′::luxCDABE | This study |
| pDEW/osmY | pDEW201; osmY′::luxCDABE | 56, 57 |
| pDEW/lon | pDEW201; lon′::luxCDABE | 56, 57 |
| pKD13 | Template plasmid for kanR amplification | 11 |
| pES2 | recA′::gfpUV ori pUC ampR | 45 |
| pATC400 | pBR322 rcsA+ ampR | 54 |
Detection of rpoS and wca mutants.
RpoS deficiency was routinely checked qualitatively by a catalase activity assay. A drop of 5 μl of 32% H2O2 was placed on a colony; immediate vigorous bubbling indicated wild-type RpoS activity (39), and lack of bubbling was interpreted as RpoS deficiency. Deficiency of wca (ΔPwza) was tested by transforming the strain with plasmid pATC400 (rcsA+) (54). Wild-type strains became extremely mucoid (CA-EPS overproduction), and ΔPwza strains did not.
Construction of new inactive alleles.
Nonpolar gene deletions were carried out as described previously (11, 64), using the primers listed in Table 2. Briefly, pKD13 (11) was used as a template DNA for PCR with primer pairs 60 bases long designed to amplify the kanamycin resistance gene from the plasmid (20 bases at the 3′ end of each primer) and to undergo a recombination process with the edges of the chromosomal target site (40 bases at the 5′ side of each primer identical to the recombination sites in the chromosome). The ΔPwza allele was directly amplified from the MS1651 genome (40), which was extracted with a DNeasy plant mini kit (Qiagen) according to the manufacturer's instructions. PCR (TGradient; Biometra, United States) was carried out with proofreading Bio-X-Act DNA polymerase (Bioline) in the presence of 200 nM template and primers using the manufacturer's reagents and instructions. The ∼1,500-bp product was gel purified using a QIAquick gel extraction kit (Qiagen), digested with DpnI to eliminate template plasmid, and then desalted using the same kit. The PCR product was then transformed into DY378 (64), a recombination-permissive strain harboring a lysogenically defective lambda phage. An overnight culture grown at 30°C was regrown to an OD600 of 0.6, heat shocked for 15 min to induce the lambda PL promoter (controlling lambda recombination promoting factors), cooled on ice, washed four times in cold double-distilled water (DDW), and finally resuspended in 1 ml DDW. Aliquots (100 μl) were mixed on ice with 50 μl of the purified PCR product. Electrical DNA transformation was carried out at 2.5 mV (Electro Cell manipulator ECM 935; BTX, United States), and the cells were then grown for 1 h in 37°C prewarmed LB broth. Positive recombinants were selected on LB agar plates supplemented with kanamycin and verified by PCR and sequencing.
TABLE 2.
Primers used in this study
| Primer name, purpose | Sequence (5′ → 3′)a | Use |
|---|---|---|
| Gene inactivation | ||
| Pwza-F | CGTTGGTGATCGCCTGTTGACCGG | Transfer of Pwza allele from MS1651 to MG1655 |
| Pwza-R | GCCGTTCGTTCAGCTCCATCGTGG | Transfer of Pwza allele from MS1651 to MG1655 |
| yjbE-F | ATTTTTGCCATATCTGCGCTTGCGGCGACTTCTGCGT GGGCGTGTAGGCTGGAGCTGCTTC | Inactivation of yjbEFGH and yjbE |
| yjbE-R | GTCGTGGTTGTGGTGGTCCCGGTATTAGAACCA TCACCGCATTCCGGGGATCCGTCGACC | Inactivation of yjbE |
| yjbH-R | CGCACGGCTGCGTGTCGGGCCAGACGAGAAGAGAT CCAACGGTACATTCCGGGGATCCGTCGACC | Inactivation of yjbEFGH |
| Cloning (pDEW201 based) | ||
| wza-F | TATACATGGAATTCCGATTAACCCGGCCCAGATAGACG | EcoRI-mediated ligation |
| wza-R | ATCAGTGGGGTACCCCCCCGCCCTCGCCTTTCAGCG | KpnI-mediated ligation |
| rRT-PCR analysis | ||
| yjbE-F | TTTGCCATATCTGCGCTTG | |
| yjbE-R | TGGTGGTCCCGGTATTAGAA | |
| yjbF-F | AAGCGACCTGCACTCATTCT | |
| yjbF-R | AGGCCAGCACCACAAATAAC | |
| yjbG-F | TATTGTCGCGTTGCTTTTGA | |
| yjbG-R | CCAGCTCTTCGCTAATCACC | |
| yjbH-F | GCTTGCTACGGCGAAACATA | |
| yjbH-R | GGGAAGAGTTGCACTGAAGC | |
| wzc-F | CCTCGATATTGCAGTGAGCA | |
| wzc-R | CGCTGCTCAGGGTGTAGTTT | |
| wcaD-F | CCCATCACCATCGTCACTTT | |
| wcaD-R | CCACACCATGCCAATAATGA | |
| gmd-F | ATGGCGACCTGAGTGATACC | |
| gmd-R | TTTCTTTTTCCAGACCGAGGA | |
| wcaH-F | AGGAAGACTTTGCCACGGTA | |
| wcaH-R | CTTCCAGCGTTTCGTCTTTC | |
| manC-F | ATGAGCAGCACCGCTTTATT | |
| manC-R | CCGCCAATACCAGCATTAAC | |
| rcsA-F | CGACATTGAAACCGTTGATG | |
| rcsA-R | GCAATTGCCATAAAAACGAT | |
| osmY-F | TGCTGGCTGTAATGTTGACC | |
| osmY-R | TCGGTGCTCTTGATGTTGTC | |
| rpmA-F | TAACGGTCGCGATTCAGAAG | |
| rpmA-R | GCAAACAGAGTGTGGTCACG |
Underlined letters indicate restriction enzyme digestion sites.
Allele transduction by P1 kc coliphage.
The new inactive alleles were transduced into MG1655 by P1 kc transduction as described previously (48). The rpoS::Tn10 allele from QC2410 was also transduced into MK1399, MK1310, and MK1351 to produce the double mutants MK2499, MK2431, and MK2411. Briefly, an overnight donor bacterial strain culture (carrying a defective allele) was regrown in 10 ml LB supplemented with 5 mM CaCl2 for 40 min before the addition of 100 μl P1 kc coliphage (produced on MG1655) and then incubated at 37°C with shaking (200 rpm) for 3 h until the culture was completely lysed by the phage. Cells that survived the lysis were killed by chloroform, and cell debris was removed by centrifugation. The supernatant containing the new phages was collected, and various volumes were incubated at 37°C for 40 min with an overnight MG1655 culture suspended in 10 mM MgSO4, 5 mM CaCl2. P1 infection was stopped by adding 100 μl 0.1 M Na3 citrate to each sample, and positive transductants were selected on LB agar plates supplemented with the appropriate antibiotics.
Cloning of the wza promoter.
MG1655 genomic DNA (extracted with a DNeasy plant mini kit [Qiagen]) was used as a template for amplifying the wza promoter with primers designed to have a calculated annealing temperature of 72°C to its target site and a tail containing the restriction enzyme digestion site followed by 8 mismatched nucleotides at its far 5′ end. pDEW201 (56, 57) was used as an acceptor for the new DNA fragment. DNAs (plasmid and PCR product) were digested with EcoRI and KpnI and desalted by using a QIAquick gel extraction kit (Qiagen). Ligation was carried out with T4 ligase (Roche) according to the manufacturer's instructions after treating the linear vector with shrimp alkaline phosphatase (Fermentas). Chemical transformation into AG1688 competent cells was carried out. Positive plasmids were selected by colony PCR and verified by sequencing.
DNA microarray and rRT-PCR analysis.
Single colonies of strains MG1655 and QC2410 (rpoS::Tn10) grown (37°C) overnight on LB broth were regrown in fresh LB broth containing 0.09 or 0.7 M NaCl for 90 and 180 min, respectively, until the wild-type strain reached an OD600 of 0.3. The cultures were then diluted to the density obtained by the rpoS mutant (OD600 = 0.15), and 50 ml of each strain was subjected to total RNA extraction using an RNeasy midi kit (Qiagen) and DNase I (Qiagen) treatment. The RNA concentration was determined by using an ND-1000 spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE). Twenty micrograms of total RNA was reverse transcribed with a poly(T18) primer using a RevertAid first-strand cDNA synthesis kit (Fermentas), and mRNA expression was assayed by using an Affymetrix GeneChip E. coli antisense genome array (Weizmann Institute of Science [Rehovot, Israel] MicroArray Unit) according to the manufacturer's instructions. The DNA microarray results were confirmed by relative real-time PCR (rRT-PCR) analysis (30). Twenty-five nanograms of cDNA was mixed with Sybr green reaction mixture (ABI) and assayed with an ABI Prism 7000 sequence detection system (Applied Biosystems) for the relative quantification of the abundance of each of the following genes: yjbE, yjbF, yjbG, yjbH, wzc, gmd, manC, wcaH, wcaD, and rcsA. Two additional genes that were similarly amplified were the RpoS-dependent osmY that served as a negative control and the RpoS-independent housekeeping gene rpmA that served as a reference. The latter gene yielded average threshold cycle (CT) values (n = 3) of 17.8 ± 0.7 (mean ± standard deviation) and 17.5 ± 0.6 for MG1655 and QC2410, respectively. The reaction conditions were as follows: 50°C for 2 min, hot start at 95°C for 10 min, 40 cycles of 95°C for 15 s, and 60°C for 1 min. A dissociation test (12) was performed at the end of each run to ensure a single PCR product in each reaction mixture. Results were obtained with the SDS analysis program (ABI) using a relative quantification algorithm with automatic CT calculation. A ΔCT was calculated for each gene as ΔCT = CT(studied gene) − CT(rpmA). ΔΔCT was calculated in two different ways. (i) RpoS dependency was calculated with the equation ΔΔCT = ΔCT(rpoS::Tn10) − ΔCT(MG1655). (ii) The relative expression of wca genes compared to yjb genes was calculated with the equation ΔΔCT = [averaged ΔCT(wzc, gmd, manC, wcaH, wcaD)] − [averaged ΔCT(yjbE, yjbF, yjbG, yjbH)]. RQ (relative RNA quantification) was calculated as RQ = 2−ΔΔCT for the results of each experiment. Results are presented as averaged RQ values (n = 3).
Bioluminescence monitoring of promoter induction.
Induction of promoters was routinely monitored by following the luminescence of E. coli strains bearing pDEW201-derived plasmids (56, 57) (Table 1) harboring promoter::luxCDABE (Photorhabdus luminescens luxCDABE bioluminescence genes) transcriptional fusions in 96-well microtiter plates. All experiments were carried out in LB broth supplemented with 0.09, 0.3, 0.5, or 0.7 M NaCl. To ensure similar plasmid copy numbers (24), the lag phase was eliminated by initiating the experiments with a growing culture. A single colony was grown to an OD600 of 0.5 on LB broth supplemented with the appropriate antibiotic at 37°C, diluted 1:10 in fresh medium, and then regrown to an OD600 of 0.2. Fifty microliters of bacterial culture was added to the wells of an opaque, white, clear-bottom 96-well microtiter plate already containing 50 μl fresh LB broth supplemented with either twice the required NaCl concentration or 0.09 M NaCl (basal growth medium concentration). The plate was then sealed with a transparent cover and incubated at 37°C for 300 min in a microtiter plate reader (Victor2; Wallac, Finland). Bioluminescence (reported in the instrument's arbitrary relative light units) and OD600 were measured at intervals of 15 min, each following a 5-s shaking. For several hours following exposure to NaCl in the microtiter plate, the OD600 of the NaCl-exposed cells did not change. Bioluminescence data were normalized to a uniform cell density by dividing the measured light intensity (relative light units) by the OD600 value measured in the same well at the same time point. Maximum bioluminescence refers to the peak of activity obtained in the course of 300 min of measurements.
EPS-related phenotype characterization.
Single colonies were spread on LB agar plates supplemented with either 0.09 or 0.7 M NaCl with or without 150 μg ml−1 Congo red (16) and allowed to grow for 24 h at 37°C. Plates were photographed over a black background (without Congo red) or a light screen (with Congo red) with an Olympus Stylus-770 SW digital camera. A mucoid appearance indicated overproduction of CA-EPS by the wca operon; red-stained cells indicated the presence of the EPS driven by the yjb operon.
Light/fluorescence microscopy.
Five microliters of culture was mounted on a microscopic slide or in a Neubauer counting chamber (400-μm2 by 10-μm-deep Clay Adams; Becton Dickinson, NJ), covered with a coverslip, and analyzed with an epifluorescence microscope (Axivert 135TV; Zeiss, Germany) or a light microscope (Eclipse N100; Nikon, Japan). Photographs were acquired with a mounted Canon PowerShot A95 digital camera.
Total carbohydrate determination.
Overnight colonies grown on LB agar plates supplemented with 0.7 or 0.09 M NaCl were collected and suspended in 2% Na2SO4, vigorously mixed for 10 min, and calibrated to a uniform cell concentration of 1010 ml−1. The following analytical protocol was used (65): 500 μl of cells was extracted by the addition of 800 μl chloroform, vigorous mixing, and phase separation by centrifugation (14,000 rpm for 10 min). A 300-μl amount of the upper (aqueous) phase was mixed with 200 μl DDW and with 1 ml anthrone reagent (Sigma) stock solution (1 mg ml−1 in concentrated H2SO4 [95 to 98%, wt/vol]). The mixture was incubated at 90°C for 10 min, and the OD630 of the sample was determined. Total carbohydrate content was calculated from a trehalose (Sigma) calibration curve.
Characterization of growth capability.
Single colonies were grown on LB broth supplemented with the appropriate antibiotics to an OD600 of 0.5, diluted 1:10 into fresh LB broth supplemented with a final concentration of either 0.7 or 0.09 M NaCl, and incubated at 37°C with shaking (200 rpm) for 6 h. The OD600 was monitored at 2-h intervals. To verify the correlation of OD measurements to the real cell concentration, a determination of growth was also performed at some time points by direct microscopic counts (Eclipse N100; Nikon, Japan).
RESULTS
Genes of the wca operon are induced by osmotic stress and are repressed by RpoS.
As previously reported (24, 44), the promoter regulating the induction of the yjbEFGH operon is activated by osmotic stress and is repressed by RpoS. In an attempt to identify additional EPS synthesis-related genes that may be coregulated with yjbEFGH, we have screened for genes that share these two regulatory features: induction by NaCl and overinduction in an rpoS::Tn10 mutant.
For this purpose, the MG1655 wild-type strain and its rpoS::Tn10 derivative were both grown in LB supplemented with either 0.09 or 0.7 M NaCl until the wild-type cultures reached an OD600 of 0.3. All samples were then diluted to the same OD and subjected to total RNA extraction, followed by whole-cell DNA microarray analysis. Sampling times were chosen based on the activity of the yjbF′::luxCDABE transcriptional fusion in each strain in each NaCl dose; after 180 min, it was an order of magnitude higher in the rpoS mutant than in the wild type with both grown in LB broth supplemented with 0.7 M NaCl.
Three members of the yjb operon, yjbE, yjbF, and yjbH, displayed osmotic-stress response ratios of 33 (result of a very low background), 3.9, and 2.2 (Table 3). In the few previous reports of DNA microarray studies that have investigated the response of E. coli to osmotic shock (e.g., reference 60), members of the yjb operon have not been listed among the osmotically regulated genes. This may be attributed to the lower NaCl doses (<0.5 M) applied in these studies, as well as to the short duration of exposure (<30 min). In the present study, based on the observed induction characteristics of the yjbF′::luxCDABE transcriptional fusion, the experimental conditions were harsher. In our DNA microarray, yjbE, yjbF, and yjbH displayed RpoS dependency ratios of 1.7, 1.6, and 1.7, respectively; the RpoS dependency threshold ratio was therefore set at 1.5.
TABLE 3.
Expression ratios obtained by DNA microarray analysis and rRT-PCR of gene members of various EPS production systemsa
| Gene | Microarray analysis ratio
|
rRT-PCR RpoS dependency ratioc | Comment | |
|---|---|---|---|---|
| Osmotic- stress responseb | RpoS dependencyc | |||
| yjbE | 33.2 | 1.7 | 7.1 ± 0.4 | yjb operon |
| yjbF | 3.9 | 1.6 | 5.3 ± 0.4 | |
| yjbG | 1.9 | 1.8 | 4.1 ± 0.4 | |
| yjbH | 2.2 | 1.7 | 4.1 ± 0.3 | |
| wza | 5 | 2.7 | ND | wca operon |
| wzb | 3.8 | 1.7 | ND | |
| wzc | 4.4 | 2 | 5 ± 0.3 | |
| wcaA | 3.1 | 2.1 | ND | |
| wcaB | 9 | 2.7 | ND | |
| wcaC | 8.8 | 1.9 | ND | |
| wcaD | 27 | 2.6 | 4.4 ± 0.7 | |
| wcaG | 4 | 1.8 | ND | |
| gmd | 5.1 | 1.6 | 7.9 ± 0.3 | |
| wcaH | 5.7 | 2.2 | 8.4 ± 0.4 | |
| wcaI | 2.6 | 1.7 | ND | |
| manC | 18 | 1.4 | 6.1 ± 0.5 | |
| wzxC | 4.3 | 1.8 | ND | |
| wcaK | 4 | 1.5 | ND | |
| wcaM | 3.2 | 1.6 | ND | |
| rcsA | 5.9 | 1.9 | 4.1 ± 0.7 | Transcription activator of wca/yjboperons |
| osmY | 6.3 | 0.3 | 0.3 ± 0.4 | RpoS dependent |
Expression ratios for the wild type and its rpoS mutant were obtained following growth in LB supplemented with either 0.7 or 0.09 M NaCl.
MG1655 (0.7 M NaCl)/MG1655 (0.09 M NaCl).
rpoS::Tn10 (0.7 M NaCl)/MG1655 (0.7 M NaCl). ND, not determined.
Among 774 genes that demonstrated ≥2-fold overexpression both in the wild type and in the rpoS mutant in the presence of 0.7 M NaCl, only 187 also displayed a ≥1.5-fold-increased level of expression in the rpoS mutant in comparison to their level of expression in the wild type (see Table S1 in the supplemental material). Also selected by this double criterion were 14 out of the 21 known genes of the wca operon, displaying osmotic-shock response ratios of 3.2 to 27 and RpoS dependency ratios of 1.5 to 2.8 (Table 3). The wca operon was previously reported (49) to be induced by osmotic stress, but its negative dependence on rpoS was not demonstrated. A few additional RcsC-dependent genes, as defined previously (15), behaved similarly, including osmB, rcsA, ugd, ykfE, ygaC, ymgD, yhaL, and yhaK (see Table S1 in the supplemental material). As previously reported (18), another functional EPS production operon, pgaABCD, is also induced by osmotic stress. In our experiment, its members displayed osmotic-stress response ratios of 1.6 to 6.1 but exhibited neither positive nor negative dependency upon RpoS.
The apparently similar induction characteristics (osmotic stress and RpoS dependency) of yjb and wca were confirmed by relative real-time PCR analysis, applying the same sample preparation procedure employed for the DNA microarray analysis. The RpoS-dependent gene osmY served here as a negative control. Indeed, as also shown in Table 3, all genes tested (four members of the yjb operon, five members of the wca operon, and their common regulator rcsA) displayed RpoS dependency ratios ranging from 4 to 8, confirming the general trend obtained by the DNA microarray analysis. As expected, the RpoS-dependent gene osmY was induced by NaCl but was repressed in the rpoS mutant.
The yjb and wca promoters display similar induction patterns.
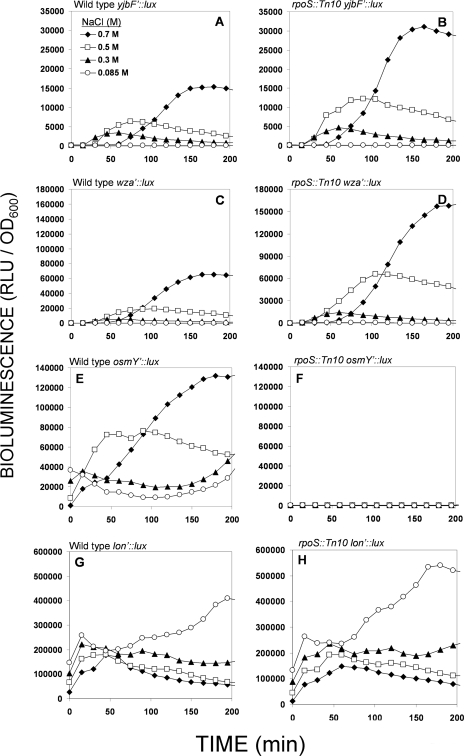
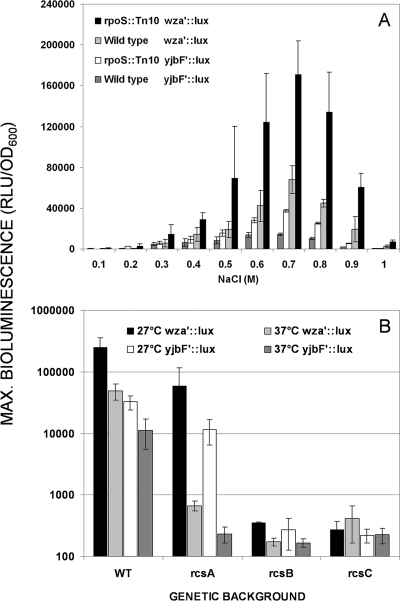
To study the induction characteristics of the two EPS biosynthesis operons, wca and yjb, we have cloned the promoter of wza, the first gene of the wca operon (50), to produce a wza′::luxCDABE transcriptional fusion. The activity of this construct was compared to that of the previously described (24) yjbF′::luxCDABE in the presence of different NaCl concentrations, both in the wild type and in the rpoS mutant (Fig. 1). As in the rRT-PCR analysis, the RpoS-dependent promoter of osmY (osmY′::luxCDABE) (56, 57) served as the control. As is clearly evident from the first four panels (A to D) of Fig. 1, the yjb and wca promoters exhibit similar patterns of induction in response to three different NaCl concentrations, both in the wild type (Fig. 1A and C) and in the rpoS::Tn10 mutant (B and D); furthermore, the activation of both promoters was similarly enhanced by the rpoS mutation and peaked at the same NaCl concentration (0.7 M) (Fig. 2A). The only difference observed in the induction of the two promoters was in the intensity; the bioluminescence exhibited by the wza′ fusion was approximately fivefold higher in both the wild type and the rpoS mutant. The same was true for the rRT-PCR analysis; the averaged RQ values (see Materials and Methods) for wca gene expression compared to the expression of yjb genes were 4.7 ± 2.2 and 5.1 ± 1.1 for the wild type and the rpoS mutant, respectively. As expected, the osmY promoter exhibited a strong positive response to osmotic stress, but its activity was strongly inhibited in the rpoS::Tn10 mutant (Fig. 1E and F). An RpoS-independent gene promoter (lon, an RpoH-dependent gene) exhibited similar activity patterns and intensities in both strains (Fig. 1G and H).
FIG. 1.
Activities of yjbF′::luxCDABE (A, B), wza′::luxCDABE (C, D), osmY′::luxCDABE (E, F), and lon′::luxCDABE (G, H) transcriptional fusions in response to different NaCl concentrations in the wild-type strain (A, C, E, and G) and in the rpoS::Tn10 mutant (B, D, F, and H). RLU, relative light units.
FIG. 2.
(A) NaCl dose dependency of yjbF′::luxCDABE and wza′::luxCDABE induction in the wild-type strain and the rpoS::Tn10 mutant. (B) Induction of yjbF′::luxCDABE and wza′::luxCDABE transcriptional fusions in response to 0.7 M NaCl in rcsA, rcsB, and rcsC mutants and their ancestral wild type at 27°C and 37°C. Error bars show standard deviations. Max, maximum; RLU, relative light units.
The yjb operon was listed (14) among genes controlled by the Rcs phosphorelay system in an RcsA-dependent manner (16). The same authors also pointed out that a sequence highly similar to the RcsAB box (62) is located in the yjbE promoter region. We confirmed these findings by assessing the osmotic (0.7 M NaCl) induction and activity of the yjbF′::luxCDABE and wza′::luxCDABE transcriptional fusions in rcsA-, rcsB-, and rcsC-inactive strains (Fig. 2B). The experiments were conducted at two temperatures, 27°C and 37°C, as it was previously shown (49) that the wca operon is more active at the lower temperature. As before, the induction of both promoters exhibited very similar patterns but different intensities. Both were almost completely inhibited in the rcsC and rcsB mutants, and both displayed a higher activity at the lower temperature in the wild type. In the rcsA mutant, the inhibition of activity was temperature dependent: it was significantly inhibited (74-fold for wca and 49-fold for yjb) at 37°C and only moderately reduced (4.2-fold for wca and 2.8-fold for yjb) at 27°C.
Overproduction of EPS in rpoS mutants.
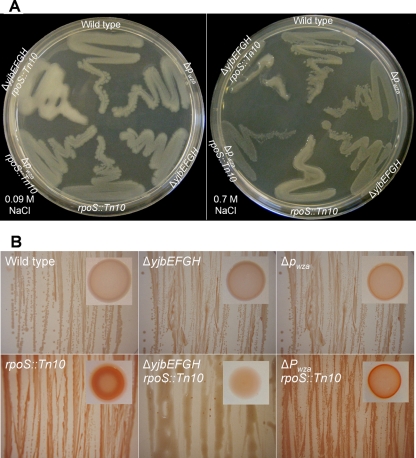
CA-EPS production (mucoid appearance) was unnoticeable in wild-type colonies grown at 37°C on LB agar plates supplemented with either 0.09 or 0.7 M NaCl (Fig. 3A); however, in 0.7 M NaCl, the rpoS::Tn10 mutant produced moderate mucosity. Inactivation of the wca promoter on top of rpoS mutation (ΔPwza rpoS::Tn10) appeared to limit growth somewhat but clearly abolished mucosity (Fig. 3A). In a parallel manner, the same was true for the yjb-dependent EPS (Fig. 3B) characterized by Congo red staining: it was not observed in the wild type but was apparent in the rpoS mutant. Inactivating the yjb operon in the rpoS mutant (ΔyjbEFGH rpoS::Tn10) suppressed the overproduction of the EPS stained with Congo red and, in addition, gave rise to extreme mucosity. The carbohydrate contents of these cultures (Table 4) support this visual observation. The carbohydrate concentrations of all strains were higher when strains were grown on LB agar plates supplemented with 0.7 M NaCl. The increase in carbohydrate in the wild-type strain may be due to enhanced activity of several EPS production systems known to be induced by osmotic stress: PGA, yjb-dependent EPS, and CA-EPS. In the rpoS mutant, only the latter two are expected to be overproduced; from the data in Table 4 it may be roughly estimated that the mixture is dominated by the CA-EPS.
FIG. 3.
(A) Appearance of culture after 24 h of growth on LB agar plates supplemented with 0.09 M or 0.7 M NaCl. (B) Congo red staining of cultures grown on LB agar plates supplemented with 0.7 M NaCl. Representative colonies are shown in the insets.
TABLE 4.
Carbohydrate content of cells grown on LB agar plates supplemented with different concentrations of NaCl
| Genotype | Carbohydrate content ± SD (μg/109 cells)
|
|
|---|---|---|
| 0.09 M NaCl | 0.7 M NaCl | |
| Wild type | 3.4 ± 2.6 | 11.6 ± 6 |
| ΔPwza | 2.7 ± 2.4 | 9.1 ± 4.5 |
| ΔyjbEFGH | 2.4 ± 1.4 | 16.1 ± 9.5 |
| rpoS::Tn10 | 0.9 ± 0.3 | 393 ± 124a,b |
| ΔPwza rpoS::Tn10 | 1.7 ± 1.1 | 136 ± 16b |
| ΔyjbEFGH rpoS::Tn10 | 79 ± 39a | 806 ± 136a |
Mucoid.
Congo red positive.
Δyjb rpoS::Tn10 double mutants are filamentous.
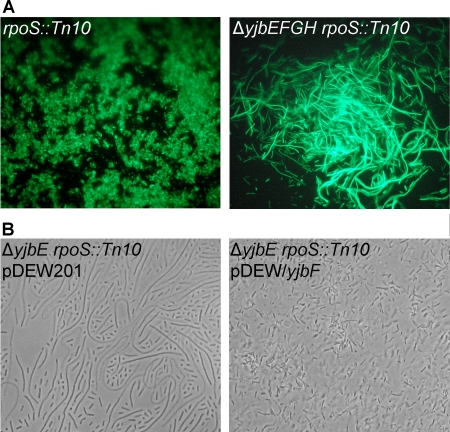
Upon deletion of yjbEFGH, the rpoS::Tn10 strain not only lost the red staining described above but also became extremely mucoid (Fig. 3A and B). In addition, the cells elongated into filaments up to 200 μm long (Fig. 4A). These phenotypes were no longer dependent upon osmotic stress and were also observed during growth on regular LB agar plates (0.09 M NaCl). To better visualize this phenomenon, the cells were transformed with a plasmid containing a green fluorescent protein gene (pES2, recA′::gfpUV) (45). Furthermore, a ΔyjbE rpoS::Tn10 double mutant was found to exhibit identical colony (not shown) and cellular (Fig. 4B) appearance. Both phenotypes were negated when this strain was transformed with plasmid pDEW/yjbF, which harbors an active yjbE gene, confirming the nonpolar deletion of yjbE. The cellular aspect of the complemented phenotype is also shown in Fig. 4B.
FIG. 4.
(A) Fluorescence microscopy of rpoS::Tn10 and ΔyjbEFGH rpoS::Tn10 mutants harboring pES2 (recA′::gfp). (B) Light microscopy of ΔyjbE rpoS::Tn10 harboring pDEW201 (vector only) or pDEW/yjbF (containing yjbE upstream of yjbF′).
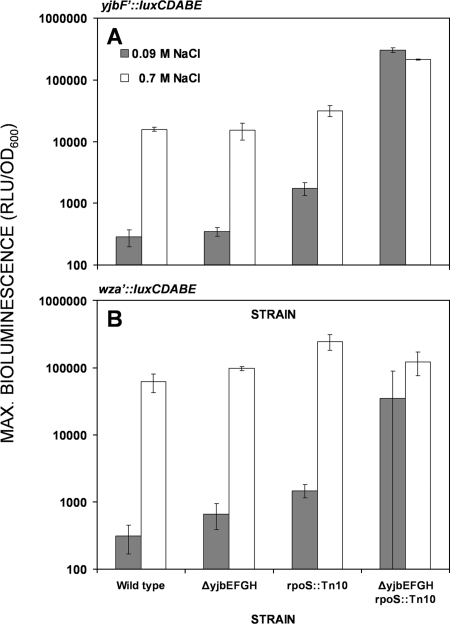
Activity of both yjb and wca promoters is enhanced in the ΔyjbEFGH rpoS::Tn10 mutant.
To explain the NaCl-independent mucoid colony appearance exhibited by the ΔyjbEFGH rpoS::Tn10 mutant, the activities of both promoters (yjb and wca) were assayed in the wild type and its three mutants, ΔyjbEFGH, rpoS::Tn10, and ΔyjbEFGH rpoS::Tn10. The background activity of both promoters in the rpoS mutant was five- to sixfold higher than in the wild-type strain, suggesting that the molecular mechanism that promotes these overinductions is osmotic-stress independent. As might be expected, the background activity (in 0.09 M NaCl) of both promoters was much higher in the ΔyjbEFGH rpoS::Tn10 double mutant than in their rpoS::Tn10 ancestor (Fig. 5). When induced by 0.7 M NaCl, the maximal activity was similar to (yjb) or twice as high as (wza) the background activity. The latter observation appears to be significant in spite of a large standard deviation in the wza′::lux background measurements. In either case, the response of the double mutant does not fully explain the 10-fold-higher carbohydrate content (CA-EPS) of this strain grown on 0.7 M NaCl (Table 4).
FIG. 5.
Maximal activities of the yjbF′::luxCDABE (A) and wza′::luxCDABE (B) transcriptional fusions in the wild type and ΔyjbEFGH, rpoS::Tn10, and ΔyjbEFGH rpoS::Tn10 mutants. Error bars show standard deviations. Max, maximum; RLU, relative light units.
Inactivation of either yjb or wca impaired growth in an rpoS-deficient strain.
The growth capabilities of all of the strains were tested in LB broth supplemented with 0.09 M (Fig. 6A) or 0.7 M (Fig. 6B) NaCl. While the wild-type strain and the ΔPwza and the ΔyjbEFGH mutants exhibited no differences in their growth characteristics, the rpoS mutant grew more slowly in the saline medium. Its two derived double mutants, ΔPwza rpoS::Tn10 and ΔyjbEFGH rpoS::Tn10, exhibited even slower growth: ΔPwza affected growth only in the presence of 0.7 M NaCl, while the effect of ΔyjbEFGH was observed also in the low NaCl medium. The effect of osmotic stress on ΔPwza rpoS::Tn10 growth is also shown in Fig. 3B, where this mutant appears to be reduced in comparison to all other strains on the plate.
FIG. 6.
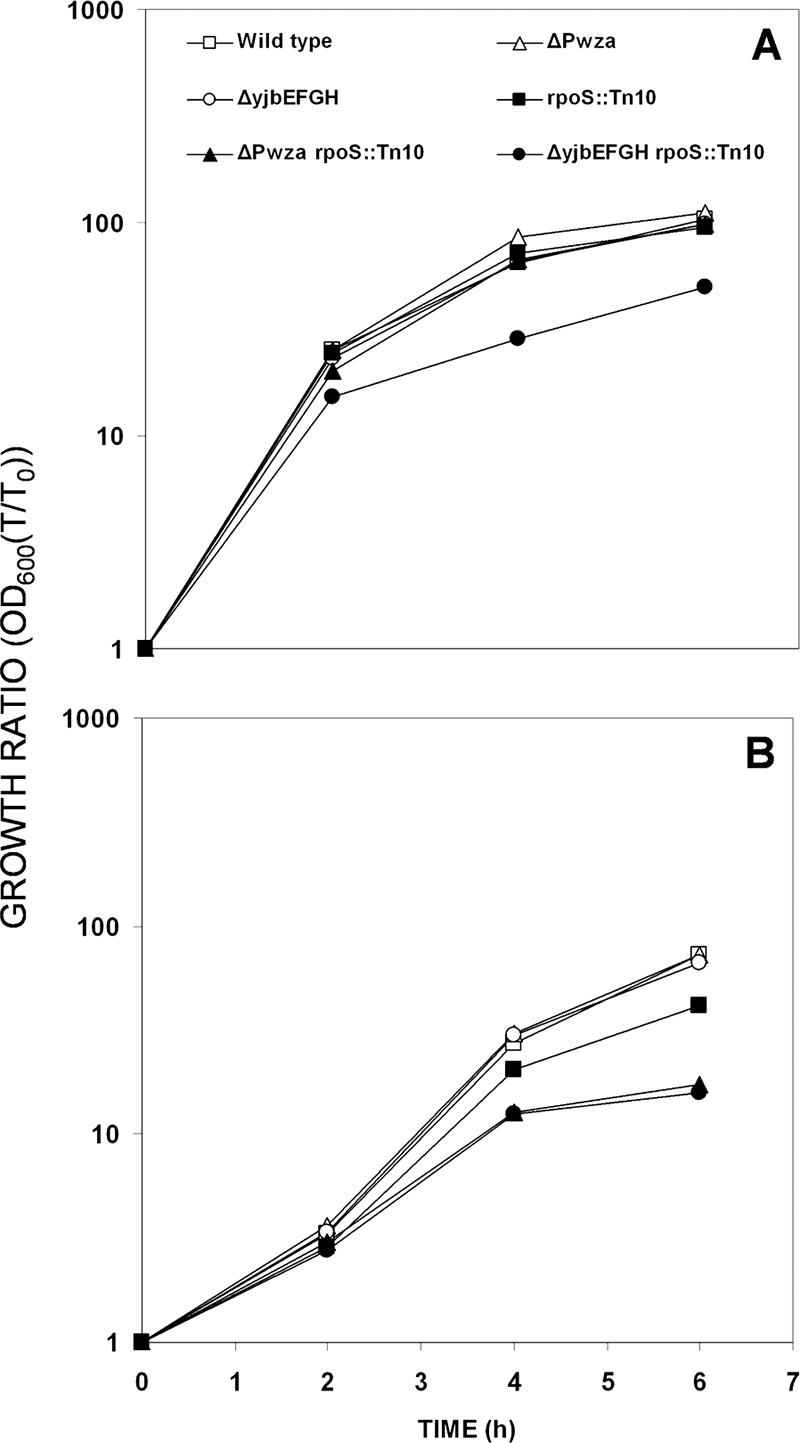
Growth in LB medium supplemented with 0.09 M (A) or 0.7 M (B) NaCl. The strains are listed in panel A. Results shown are averaged curves of the results of at least three repeat experiments.
Direct microscopic counts indicated that the measured OD600 values displayed in Fig. 6 are valid for comparisons between the tested strains: an OD600 of 1 represents 2.8 × 108 (rpoS::Tn10), 2.6 × 108 (ΔPwza rpoS::Tn10), and 2.2 × 108 (ΔyjbEFGH rpoS::Tn10) cells ml−1. The filamentous phenotype described above (Fig. 4) had started to emerge in the ΔyjbEFGH rpoS::Tn10 double-mutant culture only upon entry to stationary phase and was almost unnoticeable during the course of the experiment.
DISCUSSION
Natural selection for loss of RpoS activity has been shown to occur in the course of nutrient-limited growth and has been also detected among laboratory stocks of enteric bacteria and natural isolates. Since fully or partially inactive rpoS alleles are thus common, it may be hypothesized that some aspects of low-RpoS or RpoS-free physiology have evolved to compensate for the loss of the physiological functions that are normally RpoS dependent.
RpoS has been shown to be an important player in E. coli's osmotic-stress response (22, 61). It activates the transcription of a large number of genes that provide osmoprotection, as well as cross-protection against several other stress factors. The synthesis or uptake of compatible solutes, one of the most important responses of cells against osmotic stress, is partially mediated by RpoS-dependent genes. These include otsA, otsB, and treA (21), involved in trehalose synthesis and metabolism. Nevertheless, the growth of rpoS mutants in a hyperosmotic medium, even if slower, was demonstrated and can be attributed to the function of the organic compatible solutes transport systems ProP and ProU, as well as to the high-affinity K+ transport system KdpFABC and to the BetTIBA system implicated in choline metabolism (9, 63). All of these systems were expressed in an RpoS-independent manner in response to osmotic stress. From our data it is clear that rpoS-deficient strains overproduce EPS in response to osmotic stress and that the deletion of yjb or wca operons attenuates the already impaired osmotolerance of the mutant.
The CA-EPS has been reported to endow the cells with some stress protection, including against extreme osmotic stress in E. coli O157:H7 (6, 34, 35). It has been demonstrated (6) that a CA-EPS-deficient strain lost viability faster than the wild-type strain in the course of a 2-day exposure to 1.5 M or more NaCl. Here we demonstrated that CA-EPS was overproduced in an rpoS mutant in response to osmotic stress and that its deficiency limited growth in a saline medium only in an rpoS-deficient strain. Many clinical isolates of enteric bacteria were found to be mucoid due to CA-EPS production (20). Furthermore, some Escherichia coli O157:H7 isolates were reported to be mucoid only upon growth on medium containing high salt concentrations (26). In view of the increasing reports of the prevalence of mutated rpoS genotypes among similar isolates, it may be speculated that these two phenomena, mucosity and loss of rpoS functionality, may be coupled, and that the mucoid appearance is a consequence of the rpoS mutation.
Several studies have reported varied osmotolerance of clinical E. coli isolates (29). It has been shown (9) that an E. coli CFT073 lacking rpoS, proP, and proU simultaneously is not limited in growth or virulence in high-osmolality human urine, suggesting that it possesses additional osmoregulatory systems. It will be interesting to determine whether a deletion of yjb or wca will affect CFT073's growth ability and/or its virulence.
Overproduction of CA-EPS that results in a mucoid colony appearance has been reported to emerge in response to the inactivation of lon (36), a treatment that also resulted in UV sensitivity and cell filamentation after UV treatment. Lon is an ATP-dependent protease (4, 7) that, in addition to its role in the degradation of misfolded proteins following heat shock, has a regulatory function (55). Both cell elongation and mucoid colony phenotype were linked to lon-specific target proteins RcsA (54) and SulA (37). RcsA is the unstable auxiliary DNA binding protein of the Rcs system (33, 54). SulA is a cell division inhibitor induced as a part of the SOS DNA repair response (47). It binds to FtsZ, a key protein in cell division which is responsible for septum formation (2, 23), and represses cell division (38). Hence, it is likely that in the yjb rpoS mutants, both phenotypes are mediated by the Lon protease. We have sequenced the lon allele in this double mutant, as well as assayed lon′::lux activity (not shown), but did not find alteration in sequence or reduced induction.
Our results indicate that there may be a group of genes that are of much greater importance for stress resistance and growth in an rpoS mutant than in the wild type. In the present communication, this is demonstrated by the two EPS production operons, wca and yjb. These two operons are similarly regulated, and their expression is significantly enhanced in the rpoS mutant. While the stress resistance capabilities endowed by EPS overproduction are unclear (6), our results clearly show that growth of the mutants impaired in EPS production is inhibited in saline (both wca and yjb) or even salt-free (yjb) medium.
It is tempting to hypothesize that these operons, along with other stress-responsive RpoS-independent systems, have evolved in order to cope with the prevalence of low levels of RpoS or rpoS mutations that may have been necessitated by the need for improved nutrient scavenging (14). EPS overproduction may be an example of a mechanism that has evolved to allow cells that have become rpoS deficient to grow and cope with osmotic upshifts while enjoying a better nutrient-scavenging capability due to the overexpression of RpoD-dependent uptake and nutrient utilization systems.
Supplementary Material
Footnotes
Published ahead of print on 7 November 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bhagwat, A. A., J. Tan, M. Sharma, M. Kothary, S. Low, B. D. Tall, and M. Bhagwat. 2006. Functional heterogeneity of RpoS stress tolerance of enterohemorrhagic Escherichia coli strains. Appl. Environ. Microbiol. 72:4978-4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi, E. F., and J. Lutkenhaus. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161-164. [DOI] [PubMed] [Google Scholar]
- 3.Carballes, F., C. Bertrand, J. P. Bouche, and K. Cam. 1999. Regulation of Escherichia coli cell division genes ftsA and ftsZ by the two-component system rcsC-rcsB. Mol. Microbiol. 34:442-450. [DOI] [PubMed] [Google Scholar]
- 4.Charette, M., G. W. Henderson, and A. Markovitz. 1981. ATP hydrolysis-dependent activity of the lon(capR) protein of E. coli K12. Proc. Natl. Acad. Sci. USA 78:4728-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, G., C. L. Patten, and H. E. Schellhorn. 2004. Positive selection for loss of RpoS function in Escherichia coli. Mutat. Res. 455:193-203. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J., S. M. Lee, and Y. Mao. 2004. Protective effect of exopolysaccharide colanic acid of Escherichia coli to osmotic and oxidative stress. Int. J. Food Microbiol. 93:281-286. [DOI] [PubMed] [Google Scholar]
- 7.Chung, C. H., and A. L. Goldberg. 1981. The product of the lon (capR) gene in Escherichia coli is the ATP-dependent protease, protease La. Proc. Natl. Acad. Sci. USA 78:4931-4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conter, A., R. Sturny, C. Gutierrez, and K. Cam. 2002. The RcsCB His-Asp phosphorelay system is essential to overcome chlorpromazine-induced stress in Escherichia coli. J. Bacteriol. 184:2850-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Culham, D. E., A. Lu, M. Jishage, K. A. Krogfelt, A. Ishihama, and J. M. Wood. 2001. The osmotic stress response and virulence in pyelonephritis isolates of Escherichia coli: contributions of RpoS, ProP, ProU and other systems. Microbiology 147:1657-1670. [DOI] [PubMed] [Google Scholar]
- 10.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dhar, A. K., M. M. Roux, and K. R. Klimpel. 2001. Detection and quantification of infectious hypodermal and hematopoietic necrosis virus and white spot virus in shrimp using real-time quantitative PCR and SYBR green chemistry. J. Clin. Microbiol. 39:2835-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farewell, A., K. Kvint, and T. Nystrom. 1998. Negative regulation by RpoS: a case of sigma factor competition. Mol. Microbiol. 29:1039-1051. [DOI] [PubMed] [Google Scholar]
- 14.Ferenci, T. 2008. The spread of a beneficial mutation in experimental bacterial population: the influence of the environment and genotype on the fixation of rpoS mutations. Heredity 100:446-452. [DOI] [PubMed] [Google Scholar]
- 15.Ferrières, L., and D. J. Clarke. 2003. The RcsC sensor kinase is required for normal biofilm formation in Escherichia coli K-12 and controls the expression of a regulon in response to growth on solid surface. Mol. Microbiol. 50:1665-1682. [DOI] [PubMed] [Google Scholar]
- 16.Ferrières, L., S. N. Aslam, R. M. Cooper, and D. J. Clarke. 2007. The yjbEFGH locus in Escherichia coli K-12 is an operon encoding proteins involved in exopolysaccharide production. Microbiology 153:1070-1080. [DOI] [PubMed] [Google Scholar]
- 17.Francez-Charlot, A., B. Laugel, A. Van Gemert, N. Dubarry, F. Wiorowski, M. P. Castanié-Cornet, C. Gutierrez, and K. Cam. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49:823-832. [DOI] [PubMed] [Google Scholar]
- 18.Goller, C., X. Wang, Y. Itoh, and T. Romeo. 2006. The cation-responsive protein NhaR of Escherichia coli activates pgaABCD transcription, required for production of the biofilm adhesion poly-β-1,6-N-acetyl-d-glucosamine. J. Bacteriol. 188:8022-8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gottesman, S., P. Trisler, and A. Torres-Cabassa. 1985. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. J. Bacteriol. 162:1111-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant, W. D., I. W. Sutherland, and J. F. Wilkinson. 1969. Exopolysaccharide colanic acid and its occurrence in the Enterobacteriaceae. J. Bacteriol. 100:1187-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hengge-Aronis, R., W. Klein, R. Lange, M. Rimmele, and W. Boos. 1991. Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J. Bacteriol. 173:7918-7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hengge-Aronis, R., R. Lange, N. Henneberg, and D. Fischer. 1993. Osmotic regulation of rpoS-dependent genes in Escherichia coli. J. Bacteriol. 175:259-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higashitani, A., N. Higashitani, and K. Horiuchi. 1995. A cell division inhibitor SulA of Escherichia coli directly interacts with FtsZ through GTP hydrolysis. Biochem. Biophys. Res. Commun. 209:198-204. [DOI] [PubMed] [Google Scholar]
- 24.Ionescu, M., A. Franchini, T. Egli, and S. Belkin. 2008. Induction of the yjbEFGH operon is regulated by growth rate and oxygen concentration. Arch. Microbiol. 189:219-226. [DOI] [PubMed] [Google Scholar]
- 25.Jishage, M., and A. Ishihama. 1997. Variation in RNA polymerase sigma subunit composition within different stocks of Escherichia coli W3110. J. Bacteriol. 179:959-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Junkins, A. D., and M. P. Doyle. 1992. Demonstration of exopolysaccharide production by enterohemorrhagic Escherichia coli. Curr. Microbiol. 25:9-17. [DOI] [PubMed] [Google Scholar]
- 27.Kelm, O., C. Kiecker, K. Geider, and F. Bernhard. 1997. Interaction of the regulator proteins RcsA and RcsB with the promoter of the operon for amylovoran biosynthesis in Erwinia amylovora. Mol. Gen. Genet. 256:72-83. [DOI] [PubMed]
- 28.King, T., S. Seeto, and T. Ferenci. 2006. Genotyping-by-environment interaction influencing the emergence of rpoS mutations in Escherichia coli population. Genetics 172:2071-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunin, C. M., T. H. Hua, L. Van Arsdale White, and M. Villarejo. 1992. Growth of Escherichia coli in human urine: role of salt tolerance and accumulation of glycine betaine. J. Infect. Dis. 166:1311-1315. [DOI] [PubMed] [Google Scholar]
- 30.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 31.Loewen, P. C., and B. L. Triggs. 1984. Genetic mapping of katF, a locus that with katE affects the synthesis of a second catalase species in Escherichia coli. J. Bacteriol. 160:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maeda, H., N. Fujita, and A. Ishihama. 2000. Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 28:3497-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Majdalani, N., and S. Gottesman. 2005. The Rcs phosphorelay: a complex signal transcription system. Annu. Rev. Microbiol. 59:379-405. [DOI] [PubMed] [Google Scholar]
- 34.Mao, Y., M. P. Doyle, and J. Chen. 2006. Role of colanic acid exopolysaccharide in the survival of enterohaemorrhagic Escherichia coli O157:H7 in simulated gastrointestinal fluids. Lett. Appl. Microbiol. 42:642-647. [DOI] [PubMed] [Google Scholar]
- 35.Mao, Y., M. P. Doyle, and J. Chen. 2001. Insertion mutagenesis of wca reduces acid and heat tolerance of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:3811-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Markovitz, A. 1964. Regulatory mechanisms for synthesis of capsular polysaccharide in mucoid mutants of Escherichia coli K12. Proc. Natl. Acad. Sci. USA 51:239-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizusawa, S., and S. Gottesman. 1983. Protein degradation in Escherichia coli: the lon gene controls the stability of the SulA protein. Proc. Natl. Acad. Sci. USA 80:358-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mukherjee, A., C. Cao, and J. Lutkenhaus. 1998. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc. Natl. Acad. Sci. USA 95:2885-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Notley-McRobb, L., T. King, and T. Ferenci. 2002. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J. Bacteriol. 184:806-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peleg, A., Y. Shifrin, O. Ilan, C. Nadler-Yona, S. Nov, S. Koby, K. Baruch, S. Altuvia, M. Elgrably-Weiss, C. M. Abe, S. Knutton, M. A. Saper, and I. Rosenshine. 2005. Identification of an Escherichia coli operon required for formation of O-antigen capsule. J. Bacteriol. 187:5259-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prigent-Combaret, C., G. Prensier, T. T. Le Thi, O. Vidal, P. Lejeune, and C. Dorel. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2:450-464. [DOI] [PubMed] [Google Scholar]
- 42.Rahn, A., and C. Whitfield. 2003. Transcriptional organization and regulation of the Escherichia coli K30 group 1 capsule biosynthesis (cps) gene cluster. Mol. Microbiol. 47:1045-1060. [DOI] [PubMed] [Google Scholar]
- 43.Robbe-Saule, V., G. Algorta, I. Rouihac, and F. Norel. 2003. Characterization of the RpoS status of clinical isolates of Salmonella enterica. Appl. Environ. Microbiol. 69:4352-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rozen, Y., T. K. Van Dyk, R. A. LaRossa, and S. Belkin. 2001. Seawater activation of Escherichia coli gene promoter elements: dominance of RpoS control. Microbiol. Ecol. 42:635-643. [DOI] [PubMed] [Google Scholar]
- 45.Sagi, E., N. Hever, R. Rosen, A. J. Bartolome, J. Rajan Premkumar, R. Ulber, O. Lev, T. Scheper, and S. Belkin. 2003. Fluorescence and bioluminescence reporter functions in genetically modified bacterial sensor strains. Sens. Actuators 90:2-8. [Google Scholar]
- 46.Samuel, S., and P. Reeves. 2003. Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydr. Res. 338:2503-2519. [DOI] [PubMed] [Google Scholar]
- 47.Schoemaker, J. M., R. C. Gayada, and A. Markovitz. 1984. Regulation of cell division in Escherichia coli: SOS induction and cellular location of the sulA protein, a key to lon-associated filamentation and death. J. Bacteriol. 158:551-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Sledjeski, D., and S. Gottesman. 1996. Osmotic shock induction of capsular synthesis in Escherichia coli K-12. J. Bacteriol. 178:1204-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stout, V. 1996. Identification of the promoter region for the colanic acid polysaccharide biosynthetic genes in Escherichia coli K-12. J. Bacteriol. 178:4273-4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stout, V., and S. Gottesman. 1990. RcsB and RcsC: a two-component regulator of capsule synthesis in Escherichia coli. J. Bacteriol. 172:659-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sutton, A., W. H. Hsing, K. E. Gibson, and A. Eisenstark. 2000. rpoS mutants in archival cultures of Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:4375-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takeda, S., Y. Fujisawa, M. Matsubara, H. Aiba, and T. Mizuno. 2001. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC → YojN → RcsB signaling pathway implicated in capsular synthesis and swarming behavior. Mol. Microbiol. 40:440-450. [DOI] [PubMed] [Google Scholar]
- 54.Torres-Cabassa, A. S., and S. Gottesman. 1987. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J. Bacteriol. 169:981-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsilibaris, V., G. Meanhaut-Michel, and L. Van Melderen. 2006. Biological role of the Lon ATP-dependent protease. Res. Microbiol. 157:701-713. [DOI] [PubMed] [Google Scholar]
- 56.Van Dyk, T. K., and R. A. Rosson. 1998. Photorhabdus luminescens luxCDABE probe vector, p. 85-95. In R. A. LaRossa (ed.), Bioluminescence methods and protocols. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 57.Van Dyk, T. K., Y. Wei, M. K. Hanafey, M. Dolan, M. I. J. Reeve, J. A. Rafalski, L. B. Rothman-Denes, and R. A. LaRossa. 2001. A genomic approach to gene fusion technology. Proc. Natl. Acad. Sci. USA 98:2555-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Virlogeux, I., H. Waxin, C. Ecobichon, J. O. Lee, and M. Y. Popoff. 1996. Characterization of the rcsA and rcsB genes from Salmonella typhi: rcsB through tviA is involved in regulation of Vi antigen synthesis. J. Bacteriol. 178:1691-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, X., J. F. Preston, and T. Romeo. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186:2724-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weber, A., and K. Jung. 2002. Profiling early osmostress-dependent gene expression in Escherichia coli using DNA macroarrays. J. Bacteriol. 184:5502-5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weber, H., T. Polen, J. Heuveling, V. F. Wendisch, and R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli σs-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wehland, M., and F. Bernhard. 2000. The RcsAB box. J. Biol. Chem. 275:7013-7020. [DOI] [PubMed] [Google Scholar]
- 63.Wood, J. M. 2006. Osmosensing by bacteria. Sci. STKE 357:pe43. [DOI] [PubMed] [Google Scholar]
- 64.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuval, B., R. Kaspi, S. Shloush, and M. S. Warburg. 1998. Nutritional reserves regulate male participation in Mediterranean fruit fly leks. Ecol. Entomol. 23:211-215. [Google Scholar]
- 66.Zambrano, M. M., D. A. Siegele, M. Almiron, A. Tormo, and R. Kolter. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757-1760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.