Abstract
Phenazines are redox-active small molecules that play significant roles in the interactions between pseudomonads and diverse eukaryotes, including fungi. When Pseudomonas aeruginosa and Candida albicans were cocultured on solid medium, a red pigmentation developed that was dependent on P. aeruginosa phenazine biosynthetic genes. Through a genetic screen in combination with biochemical experiments, it was found that a P. aeruginosa-produced precursor to pyocyanin, proposed to be 5-methyl-phenazinium-1-carboxylate (5MPCA), was necessary for the formation of the red pigmentation. The 5MPCA-derived pigment was found to accumulate exclusively within fungal cells, where it retained the ability to be reversibly oxidized and reduced, and its detection correlated with decreased fungal viability. Pyocyanin was not required for pigment formation or fungal killing. Spectral analyses showed that the partially purified pigment from within the fungus differed from aeruginosins A and B, two red phenazine derivatives formed late in P. aeruginosa cultures. The red pigment isolated from C. albicans that had been cocultured with P. aeruginosa was heterogeneous and difficult to release from fungal cells, suggesting its modification within the fungus. These findings suggest that intracellular targeting of some phenazines may contribute to their toxicity and that this strategy could be useful in developing new antifungals.
Many diverse bacterial species secrete diffusible, redox-active phenazine compounds. Over 100 phenazine derivatives are produced by different bacterial species, with pseudomonads, streptomycetes, and Burkholderia spp. included among the best-known phenazine producers. Phenazines have antibiotic properties toward bacterial and eukaryotic species, and the side chain substituents on the phenazine backbone contribute to the biological activities of specific compounds. The production of phenazines has been shown to be important for antagonistic interactions among microbes. For example, phenazine-1-carboxylate (PCA) secreted by Pseudomonas fluorescens contributes to biocontrol activity against fungal phytopathogens such as Gaeumannomyces graminis (46, 47), and phenazine-1-carboxamide produced by Pseudomonas chlororaphis PCL1391 is essential for inhibition of the fungus Fusarium oxysporum, which causes tomato root rot (6). Many toxic effects have been reported for different phenazines, and much of their toxicity depends on their redox activity and their ability to generate reactive oxygen species (21, 22, 30, 42).
Pseudomonas aeruginosa, a common gram-negative soil bacterium and an opportunistic human pathogen, is well known for its ability to produce a blue phenazine, called pyocyanin, which is toxic to numerous bacteria and fungi and damages mammalian cells (21, 24, 35, 42, 52). P. aeruginosa culture supernatants also contain PCA, 1-hydroxyphenazine, and phenazine-1-carboxamide. In addition, P. aeruginosa can produce two red pigments, aeruginosins A and B (5-methyl-7-amino-1-carboxymethylphenazinium betaine and 5-methyl-7-amino-1-carboxy-3-sulfo-methylphenazinium betaine, respectively), after prolonged incubation. Unlike the other phenazines produced by P. aeruginosa, aeruginosins A and B are highly water soluble, and their biological activities are much less well characterized (15, 20).
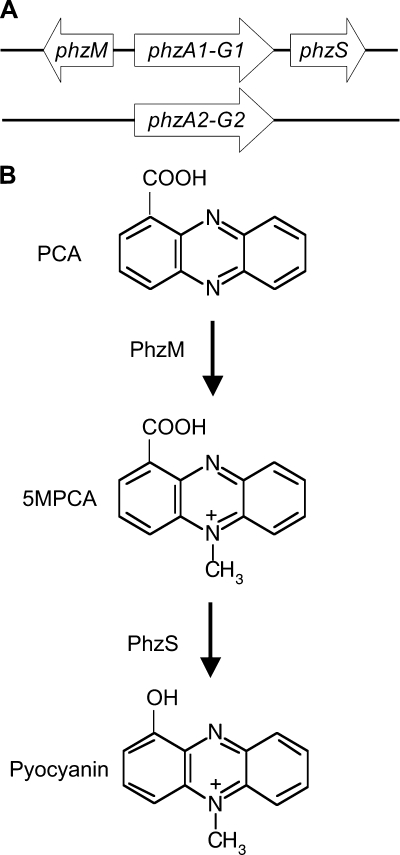
Pseudomonads synthesize PCA from chorismate by the products of the genes within the phzABCDEFG operon (32, 33) (Fig. 1A). In the P. aeruginosa genome, there are two highly similar phzABCDEFG operons, phzA1 to phzG1 and phzA2 to phzG2 (45). The production of pyocyanin from PCA requires two additional enzymes, namely, PhzM, which catalyzes methylation at N-5, yielding the proposed intermediate 5-methyl-phenazine-1-carboxylate (5MPCA) (32), and PhzS, which catalyzes the conversion of the 1-carboxylate moiety to a hydroxyl group (32) (Fig. 1B). The phzM and phzS genes are adjacent to the phzA1-to-phzG1 operon (Fig. 1A) (45). While both its precursor, PCA, and its derivative, pyocyanin, are detected at near millimolar concentrations in culture supernatants, the PhzM intermediate, proposed to be 5MPCA, has not been detected in supernatants and has been proposed to be unstable (4, 13, 39). In P. aeruginosa, phenazine production is controlled by a number of regulators, including those involved in cell density-dependent signaling, referred to as quorum sensing. Mutants that are unable to participate in signaling via C4-acylhomoserine lactone, synthesized by RhlI (3, 25), or the Pseudomonas quinolone signal (PQS) are defective in pyocyanin production (12).
FIG. 1.
P. aeruginosa phenazine biosynthetic genes and structures of pyocyanin and its immediate precursors. (A) P. aeruginosa has two redundant operons encoding the enzymes necessary for PCA production (phzABCDEFG). phzM and phzS are present as single copies. (B) Proposed biosynthetic pathway modified from reference 32. The 5MPCA intermediate has not been detected in P. aeruginosa cultures, while PCA and pyocyanin are readily detected in culture supernatants (4). Aeruginosin A has an amino substitution at position 7, and aeruginosin B has amino and sulfonate substitutions at positions 7 and 3, respectively.
Numerous reports indicate that P. aeruginosa and Candida albicans can coexist in a variety of different opportunistic infections (1, 10, 16, 36), and a number of different molecular interactions between these two organisms have been described (18, 19, 23, 24). Here we report the formation of a red pigment in P. aeruginosa-Candida albicans cocultures grown on solid medium. Through a combination of genetic, biochemical, and microscopic experiments, it was determined that a pyocyanin precursor, 5MPCA, was necessary and likely sufficient for the formation of the red pigmentation. Further characterization showed that the red pigment accumulated within fungal cells, where it remained redox active, and that its formation correlated with decreased fungal viability. We propose that the intracellular accumulation of a 5MPCA-derived product within target cells may represent an important aspect of phenazine-mediated antagonism between P. aeruginosa and other species, including fungi.
MATERIALS AND METHODS
Strains and growth conditions.
All strains used in these studies are included in Table 1. Fungal strains were grown at 30°C on YPD (2% peptone, 1% yeast extract, 2% glucose) solidified, when required, with 2% agar. Strains of Pseudomonas spp. and Escherichia coli were grown on LB, also at 30°C. All clinical isolates were obtained in compliance with federal guidelines and institutional policies. Liquid cultures were aerated in a roller drum. For assessment of swimming motility, P. aeruginosa strains were inoculated into LB containing 0.3% agar from a freshly streaked LB-grown culture, followed by incubation at room temperature for 6 to 24 h. Pyocyanin production by P. aeruginosa transposon mutants was determined by growth in LB medium for 16 h at 37°C with vigorous aeration.
TABLE 1.
Bacterial and fungal strains used in this study
| Strain | Description | DH no.a | Source or reference |
|---|---|---|---|
| Pseudomonas aeruginosa strains | |||
| PA14 WT | WT | 123 | 41 |
| PA14 phzM::TnM | TnM mutant, pyocyanin negative | 693 | 28 |
| PA14 phzS::TnM | TnM mutant, pyocyanin negative | 698 | 28 |
| PA14 WT/pUCP26 | WT with empty plasmid from reference 51 | 942 | 51 |
| PA14 phzM::TnM/pUCP26 | Mutant with empty plasmid from reference 51 | 944 | This study |
| PA14 phzM::TnM/pUCP-M | Mutant complemented with the phzM gene on a plasmid (32) | 945 | This study |
| PA14 phzS::TnM/pUCP26 | Mutant with empty plasmid from reference 51 | 946 | This study |
| PA14 phzS::TnM/pUCP-S | Mutant complemented with the phzS gene on a plasmid (32) | 947 | This study |
| PA14 Δphz | In-frame deletion mutant of phzA1 to phzG1 and phzA2 to phzG2 | 933 | 9 |
| PA14 flgK::Tn5 | Tn5 mutant, nonmotile | 37 | |
| PA14 pqsA::Tn5 | Tn5 mutant, lacks PQS | This study | |
| PAO1 WT | WT | 20 | 45 |
| PAO1 ΔphzM | Mutant lacking the phzM gene, pyocyanin negative | 296 | 32 |
| PAO1 ΔphzS | Mutant lacking the phzS gene, pyocyanin negative | 295 | 32 |
| Clinical isolates | Isolates from respiratory sputum | 211 to 228 and 74 | This study |
| Other Pseudomonas strains | |||
| Pseudomonas fluorescens SWB25 | 245 | G. O'Toole lab | |
| Pseudomonas putida KT2440 | 468 | G. O'Toole lab | |
| Pseudomonas chlororaphis PCL1391 | 469 | G. O'Toole lab | |
| Fungal strains | |||
| C. albicans SC5314 | WT | 65 | 11 |
| C. albicans tup1/tup1 mutant | BCa2-10; tup1/tup1 URA3/ura3 | 36 | 2 |
| Saccharomyces cerevisiae Σ1278b | Σ1278b | 347 | F. Winston lab |
| S. cerevisiae BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | 195 | F. Winston lab |
| S. cerevisiae BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 196 | F. Winston lab |
| Environmental yeast isolate | 121 | This study | |
| Escherichia coli strains | |||
| DH5α/pUCP-M | 32 | ||
| DH5α/pUCP26 | 32 |
From our lab collection.
P. aeruginosa-C. albicans cocultures.
P. aeruginosa was inoculated onto preformed lawns of C. albicans SC5314 or a tup1/tup1 mutant, either by using a sharp toothpick or spotting 10-μl drops of a 7 × 107-CFU/ml suspension onto the surface of the plate-grown fungal culture. The C. albicans lawns were prepared by spreading 350 μl of a YPD-grown overnight C. albicans culture onto the plate by use of glass beads, followed by incubation at 30°C for 48 h. After inoculation with P. aeruginosa, the cocultures were incubated at 30°C for an additional 24 to 96 h.
Complementation analysis was performed on filter-grown C. albicans cultures so that the C. albicans lawns could be transferred to fresh antibiotic-containing medium (150 μg/ml tetracycline) to maintain selection for the complementing plasmids over the course of the experiment. In these assays, a sterile Nylon 66 Plus Transphor transfer membrane was applied to the surface of a YPD plate, followed by inoculation with the C. albicans tup1/tup1 mutant as described above. Prior to inoculation with P. aeruginosa, the nylon filter was transferred to a fresh YPD plate containing tetracycline. The C. albicans lawns were inoculated with 10-μl drops of P. aeruginosa cultures diluted to an absorbance (optical density at 600 nm [OD600]) of 0.01. Plates were incubated at 30°C for 48 h prior to being photographed.
To achieve larger amounts of red-pigmented cocultures, full-plate cocultures were prepared by flooding established lawns of the C. albicans tup1/tup1 mutant with a dilute bacterial suspension (OD600, 0.05), followed by incubation at 30°C for 24 to 48 h. To visualize the redox activity of the red pigment, growth from two full-plate cocultures was scraped from the surface of the agar and resuspended in 50 mM potassium phosphate buffer, pH 7. Fungal cells were sedimented by centrifugation for 3 min at 3,000 × g and then washed free of bacteria by repeating the procedure until the supernatant was no longer turbid. Bacteria were sedimented from the first wash by centrifugation at 4,500 × g for 10 min. To completely reduce or oxidize the suspension of fungal cells from the coculture, a few crystals of dithionite or 10 μl of a 3% solution of H2O2 was added to a 1-ml cell suspension.
Genetic screen for mutants defective in red pigment formation.
A P. aeruginosa strain PA14 Tn5 mutant library containing ∼9,000 random insertion mutants (29) was screened on 2-day lawns of the filamentous C. albicans tup1/tup1 mutant (2) grown on YPD. Inocula from frozen stocks stored in 96-well plates were first grown on LB agar for 24 h at 30°C or 48 h at room temperature prior to transfer of the P. aeruginosa strains to the fungal lawns by use of a 48-prong inoculation device (Dan-Kar Corp.). After inoculation with P. aeruginosa, cocultures were incubated at 30°C. Mutants with altered zones of pigmentation were retrieved from the master plate and retested at least three times in triplicate. The identities of the mutants were determined by arbitrary PCR as described previously (38).
Pyocyanin production in single-species and mixed-species cultures.
Pyocyanin levels in C. albicans-P. aeruginosa cocultures were assessed by extracting the blue pigment from plates. In these assays, lawns of the C. albicans tup1/tup1 mutant were grown on YPD for 48 h at 30°C as described above, except that one-half of the plate was covered with sterile cellophane before spreading of the fungal inoculum. The cellophane, together with cells growing on it, was peeled off after growth of the fungus to provide an area of “conditioned” medium for measuring pyocyanin production in the absence of fungus. Three or four well-separated drops of a suspension of P. aeruginosa PA14 were added to both the lawn and the lawn-free areas as described above. After different periods of growth, sections of the agar plate that encompassed the entire region of bacterial growth surrounding the inoculation point (1.5 cm2), including the underlying agar, were removed, transferred to tubes, and vortexed vigorously in 2 ml of sterile water. Numbers of viable P. aeruginosa cells were determined by plating serial dilutions of this suspension on LB agar. To recover the pyocyanin, the agar suspension was then mixed with 1 ml chloroform and incubated at 4°C in the dark until the agar fragments were no longer blue. The chloroform layer was recovered, and the aqueous layer was reextracted with 1 ml chloroform. Pyocyanin levels were determined spectrophotometrically at 690 nm (ΕmM at 690 nm = 4.2) (43) after extracting the combined chloroform layers two times with 0.4 ml 50 mM HCl, combining the extracts, and neutralizing the suspension with 200 μl of 0.1 M ammonium acetate, pH 7.
Fungal viability assays.
To assess the viability of C. albicans during coculture with P. aeruginosa, established lawns of yeast-form C. albicans SC5314 grown for 48 h were spotted with suspensions of the P. aeruginosa strains as described above and further incubated for various intervals. Agar cores (4-mm diameter) from the inoculated areas or control regions were taken in triplicate with inverted Pasteur pipettes after defined times of incubation. For a given strain at a given time point, the triplicate samples were taken from three distinct inoculation points. Each of the cores was vortexed vigorously in 1 ml sterile water, and the total number of fungal cells was determined microscopically using a hemocytometer. Numbers of viable C. albicans cells were determined by serial dilution followed by plate counts, using YPD plates containing 150 μg/ml tetracycline to inhibit bacterial growth. Viable bacterial cell counts were obtained by similar plating of cells on LB agar medium lacking antibiotics. For methylene blue staining, similar suspensions of cocultures (2.5 μl) were mixed with 0.05% methylene blue (2.5 μl) and incubated for 10 min prior to examination by bright-field or differential interference contrast microscopy.
Epifluorescence microscopy of red-pigmented fungal cells.
Cocultures of P. aeruginosa and C. albicans yeast cells were grown as described above for the viability assays. A small amount of the coculture growth was removed with a micropipette tip and resuspended in 500 μl of 50 mM phosphate buffer. Suspensions were incubated with 0.03% H2O2 for at least 20 min to ensure that pigments were oxidized before being viewed with a Zeiss filter set 20 (excitation, 550 nm; emission, 580 to 640 nm). Fixation of the yeast cells with 0.4% formaldehyde did not interfere with fluorescence and stabilized the fluorescence for more than 3 days. All images were captured using a fixed capture time and light intensity.
Preparation and spectral analysis of purified phenazines and fungus-associated red pigment.
PCA was prepared from spent culture supernatants of P. aeruginosa strains by published methods (32). The product of PhzM activity on PCA was prepared as described previously (32), by incubating PCA with E. coli DH5α/pUCP-M, which contains a plasmid carrying the phzM gene. Cells from 50 ml of an overnight culture of this E. coli strain in LB with tetracycline (15 μg/ml) at 37°C were harvested by centrifugation, washed once in M63 medium (34) containing 0.2% glucose, and then resuspended in 5 ml of the same medium amended with 1 mM methionine and 0.4 mM PCA. The suspension was shaken vigorously at 37°C for 6 to 7 h. As controls, E. coli cells containing the pUCP26 vector and incubated with PCA and E. coli/pUCP-M cells incubated without PCA were treated identically. E. coli cells were removed by centrifugation, and the supernatant was acidified to pH 5 with 2 M HCl before being extracted three times with 3 ml chloroform to remove unreacted PCA. Recovery and quantitation of PCA in the chloroform fraction indicated that approximately 5 to 15% of PCA had been consumed in E. coli/pUCP-M compared to that in control cultures. The aqueous fraction of the extracted medium was neutralized with 2 M NH4OH and concentrated under reduced pressure to a final volume of 200 μl. The solution containing the PhzM product was deep red, while the control solution was pale yellow.
Pyocyanin was extracted from 24-h LB-grown cultures of P. aeruginosa PA14 by cycling three times between chloroform at neutral pH and 20 mM HCl (7). Aeruginosins A and B were recovered as water-soluble red pigments from 6-day supernatants from P. aeruginosa PA14 cultures grown in Holliman's medium (20). Approximately 200 ml of the culture supernatant was applied to chromatographic columns. Aeruginosin A bound weakly to a 2.5- by 15-cm column of the C18 hydrophobic interaction medium LRP-2 (Whatman) equilibrated with 20 mM NH4HCO3, from which it was recovered by elution with 10% acetonitrile in 20 mM NH4HCO3 in a volume of about 50 ml. Aeruginosin B, which passed through the hydrophobic interaction column, bound to a column (1.5 by 12 cm) of DEAE Sephadex in 20 mM NH4HCO3, from which it was eluted with a gradient of NH4HCO3 (20 to 200 mM) in a volume of about 25 ml. The aeruginosin-containing eluates were concentrated under reduced pressure. The UV-visible spectra of both compounds corresponded to those previously reported (15, 20).
The fungus-associated red pigment was partially purified from the fungal fraction from full-plate cocultures (described above). The purification of the pigment is described in more detail in Results. The approximate concentration of the red pigment was determined using the extinction coefficient for aeruginosins A and B (15, 20). The absorbance and fluorescence spectra were obtained using a 1-cm-path-length quartz cuvette in a SpectraMax M5 (Molecular Devices) spectrophotometer.
Treatment of yeast with the PhzM product.
In order to determine if the E. coli-synthesized product of PhzM activity on PCA was sufficient to lead to red pigmentation and intracellular fluorescence of C. albicans SC5314 yeast cells, 50 μl of aqueous extract (described above) was added to 0.5 ml of overnight C. albicans culture grown in liquid YPD (OD600 of 15). As controls, C. albicans cultures received equivalent volumes of aqueous extracts from E. coli/pUCP26 incubated with PCA and E. coli/pUCP-M incubated without the PCA precursor. After 24 h at 30°C, the C. albicans suspensions were collected by centrifugation, washed two times with 50 mM phosphate buffer, pH 7, and resuspended in 100 μl of 50 mM phosphate buffer for analysis by visual inspection, epifluorescence microscopy, and enumeration of the number of CFU.
RESULTS
Red pigment formation in P. aeruginosa-fungal cocultures.
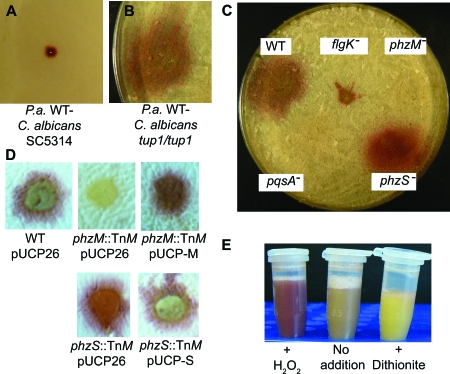
When P. aeruginosa strain PA14 was point inoculated onto established lawns of C. albicans strain SC5314 yeast and further incubated at 30°C, a red-pigmented zone developed around the inoculation point within 12 h and continued to develop over 48 h (Fig. 2A). Red pigmentation also developed when P. aeruginosa was inoculated onto established lawns of filamentous C. albicans cells, as demonstrated using the constitutively filamentous C. albicans tup1/tup1 mutant (Fig. 2B) (2). Inoculation of P. aeruginosa onto a lawn of the filamentous C. albicans strain led to a larger zone of pigmentation than that formed on C. albicans yeast (Fig. 2A and B). Neither P. aeruginosa nor C. albicans produced any red pigmentation when grown alone under similar conditions. While the red pigmentation was observed regardless of whether the cocultures were grown on rich or minimal agar plates, i.e., YPD or M63 medium with 0.2% glucose, respectively (34), no obvious red pigmentation was observed when P. aeruginosa and C. albicans were grown together in liquid cultures of the same medium composition.
FIG. 2.
Plate cocultures of C. albicans with P. aeruginosa strains. (A to C) C. albicans lawns were grown for 48 h at 30°C on YPD plates before point inoculation from LB agar-grown P. aeruginosa strains. Plates were photographed after coculture for 48 h. (A) C. albicans SC5314 with P. aeruginosa (P.a.) PA14 WT. (B) C. albicans tup1/tup1 mutant with P. aeruginosa PA14 WT. (C) C. albicans tup1/tup1 mutant with PA14 WT, flgK::Tn5, pqsA::TnM, phzS::TnM, and phzM::TnM strains. (D) Complementation of mutations in phzM::TnM and phzS::TnM strains in the coculture assay. P. aeruginosa strains were inoculated onto the C. albicans tup1/tup1 lawns as 10-μl drops and incubated at 30°C. Vector controls for WT, phzM::TnM, and phzS::TnM/(pUCP26) strains were also included. (E) Redox activity of fungal-associated pigment. C. albicans tup1/tup1 cells from two plate-grown P. aeruginosa PA14 WT cocultures were separated from the bacteria by centrifugation, resuspended in 5 ml 50 mM phosphate buffer, pH 7, and divided into three tubes. The suspensions were photographed several minutes after aeration (center), after the addition of 20 μl 3% hydrogen peroxide (left), and after the addition of dithionite (right).
To determine if the development of red pigmentation was specific to P. aeruginosa and C. albicans, other strains of P. aeruginosa, other Pseudomonas species, and other fungal strains were analyzed in coculture assays. A similar, but paler, red pigmentation was observed when P. aeruginosa strain PA14 was cultured on lawns of Saccharomyces cerevisiae BY4742, BY4741, and Σ1278b and three unidentified environmental yeast-like fungi isolated from soil samples and plant material (data not shown). Upon coculture with the C. albicans tup1/tup1 mutant, P. aeruginosa strain PAO1 and 17 of 19 clinical P. aeruginosa isolates gave rise to red pigmentation to various degrees. No red pigmentation was observed upon incubation of Pseudomonas putida, Pseudomonas fluorescens, or Pseudomonas chlororaphis on lawns of C. albicans (data not shown).
Identification of genes involved in coculture pigmentation.
To identify P. aeruginosa genes involved in the formation of red pigmentation, strains from a collection of P. aeruginosa strain PA14 Tn5 transposon mutants (29) were screened, with 48 mutants per plate, in the coculture plate assay described above. An initial screen of approximately 9,000 random Tn5 insertions found mutants with an altered pattern of pigmentation (more than 75 mutants), 5 mutants that lacked pigmentation, and 2 mutants with increased red pigmentation.
Role of swimming motility in coculture interactions.
All of the P. aeruginosa mutant strains categorized as having an altered pattern of red pigmentation gave rise to a compact red ring around the point of inoculation which was much smaller than that formed by the wild-type (WT) strain on lawns of the C. albicans tup1/tup1 mutant. Mutants in this class included flgK::Tn5 (Fig. 2C), fleN::Tn5, flgF::TnM, flgM::TnM, fliF::TnM, and fliD::TnM mutants (data not shown). All of these transposon mutants are predicted to lack a functional flagellum, and they did not swim in a 0.3% agar swim assay (data not shown). When nonmotile P. aeruginosa mutants were tested in cocultures with C. albicans SC5314 yeast, there were no obvious differences between the mutants and the WT; all remained at the point of inoculation (Fig. 2A and data not shown). These data suggest that flagellar motility is necessary for P. aeruginosa movement across C. albicans filaments and that P. aeruginosa is incapable of this flagellum-dependent motility across lawns of C. albicans in the yeast form. When P. aeruginosa mutants defective in type IV pilus-mediated twitching motility (pilB::Tn5 or pilC::Tn5 mutant) or in the production of rhamnolipids (rhlA::Gm mutant), which are necessary for swarming motility on an agar surface, were assayed on lawns of the filamentous C. albicans tup1/tup1 mutant, the zones of red pigmentation were indistinguishable from those formed by WT P. aeruginosa strain PA14 (Fig. 2B and data not shown) (37, 40).
To determine the relationship between the area of the P. aeruginosa colony on the C. albicans filaments and the size of the zone of the red pigment, samples from plate-grown fungal cocultures with either a WT or nonmotile (flgK) strain were taken at 2-mm intervals from the point of inoculation and plated onto medium selective for P. aeruginosa. These assays showed that bacteria were present throughout the red-pigmented zone and that no bacteria were detected outside the red-pigmented area, indicating that the red pigmentation developed only in regions where the bacterial and fungal cells were in close proximity and that the red pigment did not diffuse away from the coculture region.
Involvement of phenazine-related genes in red pigment formation.
Those mutants that lacked pigment production were altered in quorum sensing (three independent hits in pqsA, one in its regulator, mvfR, and one in rhlR). The two mutants with increased pigment production had insertions in phzF1 and phzS, which both encode enzymes involved in pyocyanin biosynthesis. To further characterize the role of quorum-sensing regulation in phenazine production, mutants involved in quorum sensing and phenazine biosynthesis from the PA14 nonredundant mutant collection (28) were analyzed. Again, several mutants defective in signaling by PQS (pqsA::TnM and mvfR::TnM mutants) or C4-homoserine lactone (rhlR::TnM mutant), two quorum-sensing molecules, completely lacked red pigmentation upon coculture with C. albicans (Fig. 2C and data not shown). A mutant with a disrupted rhlI gene had a more variable phenotype, and lasI and lasR mutants produced pigment on fungal lawns. None of these mutants produced pyocyanin in single-species P. aeruginosa cultures.
A P. aeruginosa phzM::TnM mutant, which lacks the phenazine biosynthetic enzyme necessary for methylation of PCA (Fig. 1B), formed no color upon coculture with C. albicans (Fig. 2C). In contrast, the phzS::TnM mutant, which is defective in reduction of the 1-carboxylate group of PCA to an alcohol (Fig. 1B), produced more red pigmentation than the WT strain (Fig. 2C and D). Complementation analysis indicated that red pigmentation was largely restored to the phzM mutant by providing the phzM gene in trans and that the hyperpigmentation phenotype of the P. aeruginosa phzS::TnM mutant was corrected by providing the phzS gene on a plasmid (Fig. 2D). Complementation of the phzS mutant also restored the production of pyocyanin in cocultures (Fig. 2D). The phzH mutant, which converts PCA to phenazine-1-carboxamide, produced slightly more red pigment than the WT strains did (data not shown) (32). While mutants defective in the phzABCDEFG biosynthetic genes involved in PCA biosynthesis were not identified in our assays as being defective in pigment production, likely due to the fact that there are redundant phzABCDEFG operons (Fig. 1A), the P. aeruginosa PA14 ΔphzA1-G1 ΔphzA2-G2 mutant (Δphz mutant) (9) did not give rise to red pigmentation upon coculture with C. albicans (data not shown). Experiments with P. aeruginosa strain PAO1 similarly showed that the ΔphzM strain completely lacked production of the red pigment and that the PAO1 ΔphzS strain produced more red pigment than the WT strain did (data not shown). Because the fungus-associated red pigment required functional phzABCDEFG genes and phzM, but not phzS or phzH, we hypothesized that this pigment was derived from the PhzM product, proposed to be 5MPCA.
Our genetic experiments indicated that red pigmentation was dependent on a precursor to pyocyanin but not on pyocyanin itself. To determine if pyocyanin formation was negatively impacted by growth with C. albicans, its levels were determined in single-species and mixed-species cultures. Quantitative analysis of pyocyanin levels in agar core samples from P. aeruginosa-C. albicans cocultures grown on plates for 24 h detected 9.6 ± 1 nmol pyocyanin per core. The amount of pyocyanin was similar (9.7 ± 1.2 nmol pyocyanin per core) for single-species P. aeruginosa cultures grown on medium conditioned by C. albicans but in the absence of fungal cells. The numbers of P. aeruginosa CFU per unit area based on the core diameter were similar for both cultures (1.56 × 106 ± 0.45 × 106/mm2 and 1.2 × 106 ± 0.36 × 106/mm2, respectively) at the 24-h time point. After 48 h, the levels of pyocyanin were higher in cocultures (106.8 ± 8.5 nmol per core) than the amounts in P. aeruginosa single-species cultures (39.3 ± 6.3 nmol per core), even though the numbers of P. aeruginosa CFU per unit area were similar (1.65 × 106 ± 1.6 × 106/mm2 and 1.64 × 106 ± 0.66 × 106/mm2 in cocultures and single-species cultures, respectively). No red coloration was observed when P. aeruginosa was grown on C. albicans-conditioned medium without fungal cells. Because pyocyanin formation was similar or greater in P. aeruginosa-C. albicans cocultures than in single-species P. aeruginosa cultures, we propose that production of a 5MPCA product other than pyocyanin in cocultures is not likely due solely to inhibition of the final step involved in pyocyanin production.
Redox activity of the red pigment.
When P. aeruginosa-C. albicans cocultures were harvested from the surface of the agar medium and resuspended in phosphate buffer, the cell suspension was similarly red pigmented (Fig. 2E). The color of the cell suspension changed from red to buff upon standing at room temperature for several minutes, and the red pigment was restored upon vigorous aeration. Similarly, the redness was intensified upon the addition of dilute H2O2, and the cell suspension converted from red to buff upon addition of a few crystals of dithionite (Fig. 2E). Color changes upon addition of reducing and oxidizing agents could be observed repeatedly using the same coculture suspension, suggesting that the pigment could be reversibly oxidized and reduced.
Correlation between pigment production and fungal viability in coculture.
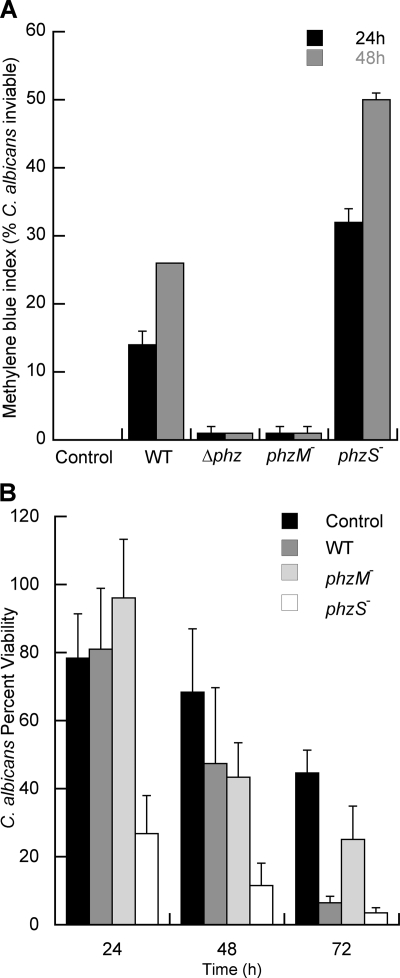
To determine if the formation of red pigmentation correlated with an altered viability of C. albicans SC5314, the survival of C. albicans was assessed by both methylene blue staining, which is indicative of a cell being metabolically inactive or membrane compromised, and comparing the total number of yeast cells determined by microscopic direct counts to the numbers of CFU. In C. albicans control cultures and C. albicans SC5314 cocultures with the Δphz or phzM::TnM strain, <1% of the cells were stained by methylene blue even after 48 h of coincubation (Fig. 3A). Yeast cells incubated with WT P. aeruginosa exhibited 14 and 26% methylene blue staining at 24 and 48 h, respectively. Cocultures with the phzS mutant, which led to increased red pigmentation, contained approximately twice as many methylene blue-stained cells (32 and 50% at 24 and 48 h, respectively).
FIG. 3.
Survival of C. albicans in coculture with strains of P. aeruginosa PA14. Established lawns of C. albicans SC5314 yeast were inoculated with P. aeruginosa suspensions. Core samples from plates were taken for counts of total cells by microscopy and of viable cells (CFU) after incubation for various times. (A) C. albicans methylene blue staining after incubation alone or with P. aeruginosa PA14 WT, Δphz (which lacks the genes necessary to synthesize PCA), phzM::TnM, and phzS::TnM strains. (B) Survival of C. albicans after 72 h alone (control) or in coculture with the P. aeruginosa PA14 WT, phzM::TnM, or phzS::TnM strain. Percent viability represents the fraction of the total cells that gave rise to visually detectable colonies within 24 h. These experiments were repeated as three completely separate experiments, with similar results each time.
Viability determined by combining direct counts with determination of CFU strongly supported the results from the methylene blue staining experiments. In the absence of bacteria, C. albicans yeast viability was 86% at the 72-h time point, as measured by these means (Fig. 3B). When C. albicans was cocultured with the P. aeruginosa PA14 strains, the viability in the presence of P. aeruginosa was reduced to 12% for WT P. aeruginosa and 3% for the phzS mutant. In contrast, C. albicans cocultured with the phzM strain had 76% viability. Analysis of variance found the differences between the WT and mutant strains to be statistically significant (P < 0.05). The differences in fungal killing were not due to differences in P. aeruginosa growth, as the numbers of P. aeruginosa CFU were equivalent (9.6 × 107 to 10.4 × 107 CFU per core) in all of the cocultures included in this experiment. These data indicate that the presence of the phzM gene, which leads to the formation of 5MPCA, is required for much of the killing of C. albicans by P. aeruginosa in this assay.
Accumulation of red pigment within fungal cells.
Several pieces of evidence suggested that the red pigment that formed in P. aeruginosa-C. albicans cocultures was present only in association with the fungal cells. First, while pyocyanin diffused into the agar below the P. aeruginosa-C. albicans coculture, the red pigmentation was observed only in the layer of cells atop the agar medium in cocultures of up to 7 days old and did not diffuse into the surrounding agar (data not shown). Second, centrifugal separation of bacteria from the fungal cells showed that the red pigmentation was associated with the fungal pellet and was not present in the bacterium-containing fractions. No red coloration was observed in water or phosphate buffer washes of the fungal pellet, suggesting that the pigment was not capable of readily diffusing away from fungal cells (data not shown). The red pigmentation of the fungal cell fraction was redox sensitive (Fig. 2E). Third, when an established lawn of the C. albicans tup1/tup1 mutant was covered with cellophane prior to inoculation with P. aeruginosa, removal of the bacterial colony after a 24-h growth period revealed red pigmentation only of the fungal cells (data not shown). Culturing experiments confirmed that P. aeruginosa and C. albicans remained separate over the course of the cellophane separation experiment. Experiments describing the release and purification of the red pigment from fungal cells are described below.
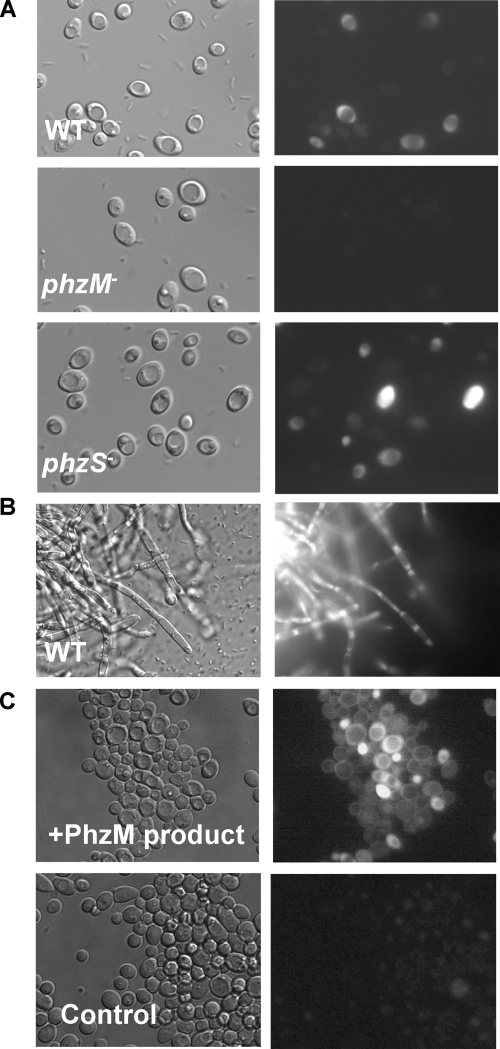
Visualization of cells grown in coculture with P. aeruginosa by epifluorescence microscopy provided additional information regarding the location of the red pigmentation that formed in P. aeruginosa-C. albicans cocultures. After incubation of WT P. aeruginosa on lawns of C. albicans yeast, a small amount of the coculture was resuspended in buffer and then observed by epifluorescence microscopy. C. albicans that had been incubated with WT P. aeruginosa exhibited bright intracellular fluorescence (Fig. 4A). The intracellular fluorescence was evident at 24 h, and the percentage of fluorescing cells increased with time. C. albicans cells from cocultures with the P. aeruginosa phzS::TnM strain, which led to increased production of the red pigment, were also brightly fluorescent (Fig. 4A). In contrast, only a faint background fluorescence was observed with C. albicans grown either in the absence of P. aeruginosa or in the presence of the P. aeruginosa phzM::TnM strain, which does not support red pigmentation (Fig. 4A). In many C. albicans cells from cocultures with WT P. aeruginosa, the red fluorescence seemed brightest in the cytoplasm (Fig. 4A), but in very brightly fluorescing cells, such as those detected in cocultures with the phzS::TnM strain, the fluorescence appeared throughout the cell (Fig. 4A). The level of fluorescence per cell was not even across the population. Incubation of the constitutively filamentous tup1/tup1 mutant with WT P. aeruginosa, but not the Δphz or phzM mutant, also yielded brightly fluorescent cells (Fig. 4B). The addition of dithionite, the reducing agent that changed the visual pigmentation of the culture from red to colorless (Fig. 2E), also suppressed fluorescence (data not shown).
FIG. 4.
Epifluorescence microscopy of C. albicans grown in coculture with P. aeruginosa or in medium containing the E. coli-synthesized PhzM product. For each pair of images, identical fields were photographed using differential interference contrast and Zeiss set 20 fluorescence optics for rhodamine. (A) C. albicans SC5314 from 72-h cocultures with P. aeruginosa PA14 WT, P. aeruginosa phzM::TnM, or P. aeruginosa phzS::TnM strain. (B) C. albicans tup1/tup1 mutant from 72-h cocultures with P. aeruginosa PA14 WT. No fluorescence was observed in similar cocultures with a P. aeruginosa ΔphzM strain (not shown). (C) C. albicans SC5314 after 24 h of incubation with either ∼200 μM 5MPCA prepared by incubating E. coli/pUCP-M (expressing the phzM gene) with PCA (+PhzM product) or extracts from a control preparation from E. coli/pUCP26 incubated with PCA (control).
Effects of PhzM product on red pigmentation, intracellular fluorescence, and viability of fungal cells.
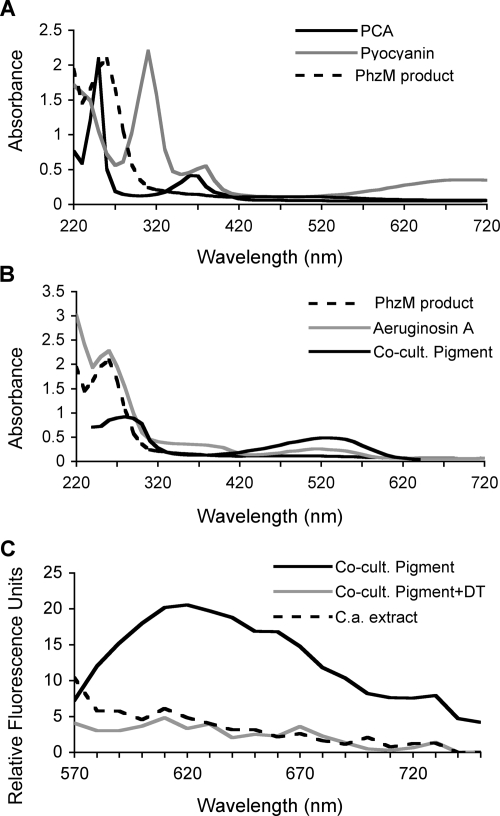
Because cocultures with the P. aeruginosa phzM::TnM mutant formed no red pigment, while those with the phzS mutant overproduced the red pigment, we hypothesized that the PhzM product, proposed to be 5MPCA, is responsible for the red pigmentation. To test this hypothesis directly, the PhzM product was synthesized by feeding PCA to resting cells of an E. coli strain carrying the phzM gene on a high-copy-number plasmid, using a previously published protocol (32). The supernatants from E. coli/pUCP-M incubated with PCA were yellow-orange after 4 h and developed a reddish color after 8 h of incubation. The colored product(s) remained in the aqueous phases during chloroform extraction to remove the unreacted PCA. Quantification of the PCA extracted from the suspensions at the end of the incubation period suggested that ∼10% of the PCA was acted upon by PhzM. The spectrum for the aqueous extracts from PCA incubation with PhzM was distinct from the spectrum for PCA (Fig. 5A).
FIG. 5.
Spectra of late intermediates in pyocyanin biosynthesis and the coculture pigment. All solutions were in 0.1 M NH4HCO3 and were normalized at the UV maximum. (A) Absorption spectra of PCA, the E. coli-synthesized PhzM product, and pyocyanin. (B) Absorption spectra of 5MPCA, aeruginosin A, and the partially purified coculture pigment. (C) Fluorescence emission spectra of partially purified red pigment from C. albicans cells grown in coculture with P. aeruginosa (solid lines). Excitation was done at 550 nm. The pigment was analyzed before (black line) and after (gray line) the addition of a few crystals of sodium dithionite (DT). A comparable fraction from C. albicans (C.a.) cells grown in the absence of P. aeruginosa was also analyzed (dashed line).
When C. albicans SC5314 yeast cells were incubated in YPD medium with the PCA-free aqueous extracts containing the PhzM product for 24 h at 30°C, the C. albicans cells developed a reddish color that was visible by eye in the cell pellet after centrifugation, even after repeated washing with buffer. No pigmentation was observed in the C. albicans cell pellets after incubation with aqueous extracts from E. coli/pUCP-M cultures that did not receive PCA or from the vector control strain, E. coli/pUCP26, incubated with PCA. Epifluorescence microscopy of yeast cells incubated with extracts containing the PhzM product found that 8.5% ± 2.3% of cells fluoresced brightly and the remaining cells had increased fluorescence (Fig. 4C). The lack of fluorescence in yeast cells from the above-described control cultures confirmed that the 5MPCA product was necessary for development of the intracellular fluorescence (Fig. 4C). After 48 h, C. albicans yeast cells incubated with the PhzM product had reduced viability (36.7% ± 3.2%) in comparison to C. albicans yeast cells incubated with the control extract from E. coli/pUCP26 with PCA (52.7% ± 11.6% viable).
Comparison of 5MPCA-derived pigment within C. albicans cells to known P. aeruginosa phenazines.
To determine the nature of the red-pigmented product observed within fungal cells, suspensions of C. albicans from P. aeruginosa-C. albicans cocultures were lysed and the red-pigmented products were analyzed. While the red product 5MPCA and derivatives synthesized from PCA by E. coli/pUCP-M are highly soluble in water, the red pigment within fungal cells was poorly soluble in either aqueous solutions (water or acidic or basic buffers) or organic solvents (chloroform, ethyl acetate, or ethanol). Mechanical or enzymatic and chemical disruption of cells, followed by centrifugation of the cell lysate at 10,000 × g, led to sedimentation of the majority of the pigment, yielding only a pale pink supernatant. Conditions that promote yeast autolysis, including incubation of dense cell suspensions at 37°C for 24 to 48 h, led to further, but far from complete, release of pigment in a time-dependent manner. Heat inactivation of the cell suspension prior to incubation under autolysis conditions prevented release of red-pigmented products into the aqueous supernatant. The observations suggest that pigment is aggregated or polymerized inside the fungus but can be released slowly by the action of endogenous fungal enzymes.
Partial purification of red pigment from autolyzed suspensions of the C. albicans tup1/tup1 mutant from P. aeruginosa cocultures could be achieved by size-exclusion chromatography on Bio Gel P-2 (molecular weight, 200 to 2,000; Bio-Rad). The red fractions emerged close to the void volume, whereas pyocyanin and vitamin B12 eluted in later fractions that corresponded to their predicted molecular weights of 210 and 1,355, respectively (49). These data suggest that the red pigment molecule(s) is of a substantially larger size than that of other phenazines known to be produced by P. aeruginosa. For spectrophotometric analysis, the red fractions were further purified using a C18 hydrophobic interaction column (LRP-2; Whatman) in 1 M NH4CO3, followed by elution with 10% acetonitrile. The red fraction had a single broad absorbance maximum at ∼530 nm in the visible region (Fig. 5B) but lacked the maximum at ∼390 nm, which is characteristic of red P. aeruginosa phenazines aeruginosins A (Fig. 5B) and B (not shown), as reported previously by Holliman et al. (15, 20). The spectrum of the red pigment isolated from fungal cells also differed from those of other known P. aeruginosa phenazines, including PCA, pyocyanin, and the products of PhzM activity on PCA produced in E. coli (Fig. 5).
To determine if the fluorescence observed within C. albicans yeast cells from cocultures by epifluorescence microscopy could be attributed to the red pigment that was partially purified from C. albicans, the fluorescence spectrum of the partially purified material was measured with an excitation wavelength of 550 nm (Fig. 5C). Red fluorescence was detected with a maximum at 620 nm when the sample was aerated. Consistent with what was observed in the C. albicans whole-cell suspensions, the fluorescence was not observed in the same sample after reduction with dithionite (Fig. 5C). A comparable preparation from C. albicans cultures grown in the absence of P. aeruginosa yielded fractions with no detectable fluorescence (Fig. 5C).
While we were not able to determine the identity of the fungally associated pigment that formed upon incubation of C. albicans with the products of PhzM activity on PCA- or 5MPCA-producing strains, largely due to its insolubility, it is clear that it has properties that are distinct from those of the phenazines present in single-species P. aeruginosa cultures. We could detect this red pigment within C. albicans cells only after coculture with P. aeruginosa or upon incubation with the PhzM product, leading us to hypothesize that the pigment results from an activity on 5MPCA that occurs within the fungal cell. Incubation of fungal cells with the partially purified red pigment, aeruginosins A or B, PCA, or pyocyanin at concentrations of up to 1 mM did not give rise to red-pigmented fungal cells with intracellular fluorescence. It is important, however, that while treatment of the partially purified red pigment with 1 mg/ml proteinase K, DNase, RNase, boiling, acid (1 M HCl), base (1 M NaOH), or sodium dodecyl sulfate (1%) did not liberate a homogenous low-molecular-weight species, we cannot rule out the alternative hypothesis which states that 5MPCA or a derivative thereof is unaltered but aggregated in some way.
DISCUSSION
The studies reported here show that the P. aeruginosa product of PhzM activity on PCA, proposed to be 5MPCA, leads to the accumulation of a red-pigmented, redox-active compound within fungal cells. Like the C. albicans cells from cocultures, the partially purified pigment exhibited a red coloration that was colorless upon reduction and could readily be oxidized by aeration or the addition of hydrogen peroxide (Fig. 2E). Both the pigmentation in association with fungal cells and the partially purified pigment exhibited a red fluorescence only in an oxidized state. P. aeruginosa PA14 and PAO1 mutants that were defective in phzM, the gene responsible for the methylation of PCA to form 5MPCA, were incapable of red pigment formation. Furthermore, pseudomonads such as Pseudomonas putida and Pseudomonas fluorescens, whose genomes do not contain phzM (53), did not induce the accumulation of pigment on C. albicans lawns. In contrast, deletion of the phzS gene, which encodes the enzyme that acts upon 5MPCA to form pyocyanin, led to hyperpigmentation (Fig. 2), suggesting that the formation of pyocyanin competed for the 5MPCA pool. Disruption of the phzH gene, which encodes an enzyme that acts on PCA (32), also led to increased red pigment formation, suggesting that PhzH competes with PhzM for the PCA pool within cocultures.
The putative intermediate 5MPCA, which is responsible for the coculture pigment, has not been studied extensively. While its immediate precursor, PCA, and its immediate derivative, pyocyanin, accumulate in P. aeruginosa cultures, 5MPCA has not been detected in supernatants (4, 14). Because pyocyanin levels were higher in P. aeruginosa-C. albicans cocultures than in P. aeruginosa cultures grown in the absence of C. albicans but on C. albicans-conditioned medium, it does not appear that the release of the PhzM product was simply due to inhibition of the last PhzS-dependent step in pyocyanin production by C. albicans. In vitro assays with purified PhzM and PhzS indicate that PhzM activity is greatly enhanced or dependent on PhzS (39), suggesting that PhzM reaction kinetics are favorable only when the product, 5MPCA, is consumed. Furthermore, 5MPCA synthesized as a phenazinium chloride was found to be stable in acid but not tractable at neutral pH (14). The fact that the fungally associated red pigment formed only when P. aeruginosa and C. albicans were grown in close contact with one another leads us to speculate that C. albicans uptake and subsequent modification of a small extracellular pool of 5MPCA may increase PCA conversion to 5MPCA and the formation of 5MPCA-derived products. In a medium that supports growth of P. aeruginosa on the surfaces of C. albicans hyphae (8), the fungal pellet develops a distinct pink coloration, and red fluorescent C. albicans cells were observed upon coculture with WT P. aeruginosa and phzS mutant strains (data not shown). The role that fungal cells play in promoting the formation and release of the 5MPCA intermediate is a subject of our future research.
The exact chemical nature of this coculture red pigment extracted from fungi remains elusive. Its UV-visible spectrum and solubility characteristics are distinct from those of phenazines known to be produced by monocultures of P. aeruginosa, including PCA, pyocyanin, and aeruginosins A and B, and from the product in extracts from PCA-fed cultures of E. coli carrying the phzM gene (15, 20) (Fig. 5A and B). 5MPCA is converted to a red phenazine, aeruginosin A, upon incubation with late P. aeruginosa culture supernatants or through a chemical reaction with concentrated ammonia (14). A similar, soluble red phenazine accumulates in supernatants of phzS mutant cultures, as shown previously (32) and confirmed by our laboratory (data not shown). While aeruginosin A and the red pigment in phzS culture medium are very soluble in liquid and agar media, the red phenazine derivative formed in yeast was located exclusively in yeast cells and did not diffuse into the surrounding medium. Incubation of C. albicans with purified aeruginosins did not give rise to red-pigmented yeast (data not shown). Nevertheless, the absorption spectrum of the coculture pigment does resemble that of aeruginosin in that it has an absorption maximum in the 520-nm range.
The concentration of the coculture pigment in the fungal cell fraction may indicate that fungal enzymes or other factors in yeast participate in the formation of the red pigment. At this time, however, we know little about the factor or factors that lead to the formation of the red product with poor solubility and an apparent molecular weight that is significantly larger than that of the 5MPCA precursor. Because formation of the red pigment is observed upon coculture with numerous fungi, including S. cerevisiae, any modifications are not unique to C. albicans. The red pigmentation that develops in ade2 mutants of both S. cerevisiae and C. albicans grown in media with limiting concentrations of adenine, due to the conjugation of the toxic adenine biosynthetic precursors with glutathione (5, 44), is distinct from the pigment we observe in P. aeruginosa-C. albicans cocultures. The red pigment in ade2 mutants is not redox sensitive (50). The addition of adenine to the agar medium does not alter pigmentation of C. albicans in our experiments (data not shown).
Previous reports have demonstrated the toxicities of pyocyanin and 1-hydroxyphenazine on C. albicans and S. cerevisiae, with MICs in the 100- to 500-μM range (24, 42). Our studies indicate that the product of PhzM activity on PCA also leads to decreased fungal viability. Because the precise nature of the 5MPCA product has not been described and purified preparations are reported to be unstable at neutral pH (14), direct quantitative comparison of its lethal effect on yeast to that of other phenazines has not been performed. The phzS mutant, which leads to increased C. albicans red pigmentation and fluorescence compared to those of C. albicans cocultured with WT P. aeruginosa, is also more lethal for yeast than the WT strain (Fig. 4). These data indicate that the putative product of the PhzM enzyme, 5MPCA, is more important for yeast killing than pyocyanin when the two species are grown in close proximity to one another. While pyocyanin has previously been demonstrated to play an important role in P. aeruginosa virulence (24, 42), previous studies by Lau et al. found that both the phzM and phzS mutants were attenuated in lung infections in vivo. It is interesting, however, that the phzM mutant exhibited a greater defect, as determined by competitive index, than the phzS mutant in these in vivo assays (26). Experiments with both partially purified pigment and whole C. albicans cells showed that the fungally associated red pigment retained redox activity within the fungus despite any modifications that may occur within the fungal cell (Fig. 2E and 5C). Reaction with oxygen is important for the toxicity of many different phenazines toward a variety of species (31), and we hypothesize that the redox activity of the fungally associated pigment contributes to the decreased C. albicans viability that we observe in its presence.
While many phenazines have been shown to have toxic effects toward a variety of species, including fungi, it is not yet known if previously characterized phenazine antibiotics are modified or sequestered by other organisms (22, 27, 31, 48). One report describes the production of an extracellular red pigment, with different properties from the pigment described here, that forms in cocultures of Aspergillus sclerotiorum and Pseudomonas chlororaphis, suggesting that other fungi may be able to modify certain bacterially produced phenazines (17); the biological activity of the modified P. chlororaphis phenazine is not known. One can envision many ways in which modification or aggregation of phenazines after secretion by the producing microbe could enhance or reduce the toxicity of the antibiotic, and these processes may be important factors to consider in the design of phenazine-producing biocontrol strains.
Acknowledgments
This paper is dedicated to Jane Gibson, an inspirational scientist, for whom this paper is published posthumously.
We thank Nicholas Jacobs for his important intellectual contributions to this project. We thank Linda Thomashow and Dimitri Mavrodi for providing P. aeruginosa PAO1 ΔphzS and ΔphzM strains and the complementation plasmids. We thank Dianne Newman for providing the PA14 ΔphzA1-G1ΔphzA2-G2 strain, Joseph Schwartzman and the DHMC clinical lab for providing P. aeruginosa clinical isolates, and George O'Toole for use of the P. aeruginosa PA14 Tn5 mutant library and for providing strains of other Pseudomonas species. We acknowledge Nida Intarapanich for preparing and analyzing the complemented strains and Brittany Ciesluk for aiding in the viability studies.
This work was funded by the Pew Biomedical Scholars Program (D.A.H.) and the Cystic Fibrosis Foundation (D.A.H.).
Footnotes
Published ahead of print on 14 November 2008.
REFERENCES
- 1.Bauernfeind, A., R. M. Bertele, K. Harms, G. Horl, R. Jungwirth, C. Petermuller, B. Przyklenk, and C. Weisslein-Pfister. 1987. Qualitative and quantitative microbiological analysis of sputa of 102 patients with cystic fibrosis. Infection 15:270-277. [DOI] [PubMed] [Google Scholar]
- 2.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105-109. [DOI] [PubMed] [Google Scholar]
- 3.Brint, J. M., and D. E. Ohman. 1995. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 177:7155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byng, G. S., D. C. Eustice, and R. A. Jensen. 1979. Biosynthesis of phenazine pigments in mutant and wild-type cultures of Pseudomonas aeruginosa. J. Bacteriol. 138:846-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudhuri, B., S. Ingavale, and A. K. Bachhawat. 1997. apd1+, a gene required for red pigment formation in ade6 mutants of Schizosaccharomyces pombe, encodes an enzyme required for glutathione biosynthesis: a role for glutathione and a glutathione-conjugate pump. Genetics 145:75-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chin, A. W. T. F., G. V. Bloemberg, I. H. Mulders, L. C. Dekkers, and B. J. Lugtenberg. 2000. Root colonization by phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol. Plant-Microbe Interact. 13:1340-1345. [DOI] [PubMed] [Google Scholar]
- 7.Cox, C. D. 1986. Role of pyocyanin in the acquisition of iron from transferrin. Infect. Immun. 52:263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cugini, C., M. W. Calfee, J. M. Farrow III, D. K. Morales, E. C. Pesci, and D. A. Hogan. 2007. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol. Microbiol. 65:896-906. [DOI] [PubMed] [Google Scholar]
- 9.Dietrich, L. E., A. Price-Whelan, A. Petersen, M. Whiteley, and D. K. Newman. 2006. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61:1308-1321. [DOI] [PubMed] [Google Scholar]
- 10.El-Azizi, M. A., S. E. Starks, and N. Khardori. 2004. Interactions of Candida albicans with other Candida spp. and bacteria in the biofilms. J. Appl. Microbiol. 96:1067-1073. [DOI] [PubMed] [Google Scholar]
- 11.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallagher, L. A., S. L. McKnight, M. S. Kuznetsova, E. C. Pesci, and C. Manoil. 2002. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 184:6472-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenhagen, B. T., K. Shi, H. Robinson, S. Gamage, A. K. Bera, J. E. Ladner, and J. F. Parsons. 2008. Crystal structure of the pyocyanin biosynthetic protein PhzS. Biochemistry 47:5281-5289. [DOI] [PubMed] [Google Scholar]
- 14.Hansford, G. S., F. G. Holliman, and R. B. Herbert. 1972. Pigments of Pseudomonas species. IV. In vitro and in vivo conversion of 5-methylphenazinium-1-carboxylate into aeruginosin A. J. Chem. Soc. (Perkin 1) 1:103-105. [DOI] [PubMed] [Google Scholar]
- 15.Herbert, R. B., and F. G. Holliman. 1969. Pigments of pseudomonas species. II. Structure of aeruginosin B. J. Chem. Soc. 18:2517-2520. [DOI] [PubMed] [Google Scholar]
- 16.Hermann, C., J. Hermann, U. Munzel, and R. Ruchel. 1999. Bacterial flora accompanying Candida yeasts in clinical specimens. Mycoses 42:619-627. [DOI] [PubMed] [Google Scholar]
- 17.Hill, J., and G. Johnson. 1969. Microbial transformation of phenazines by Aspergillus sclerotiorum. Mycologia 61:452-467. [PubMed] [Google Scholar]
- 18.Hogan, D. A., and R. Kolter. 2002. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science 296:2229-2232. [DOI] [PubMed] [Google Scholar]
- 19.Hogan, D. A., A. Vik, and R. Kolter. 2004. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 54:1212-1223. [DOI] [PubMed] [Google Scholar]
- 20.Holliman, F. G. 1969. Pigments of Pseudomonas species. I. Structure and synthesis of aeruginosin A. J. Chem. Soc. 18:2514-2516. [DOI] [PubMed] [Google Scholar]
- 21.Kanthakumar, K., G. Taylor, K. W. Tsang, D. R. Cundell, A. Rutman, S. Smith, P. K. Jeffery, P. J. Cole, and R. Wilson. 1993. Mechanisms of action of Pseudomonas aeruginosa pyocyanin on human ciliary beat in vitro. Infect. Immun. 61:2848-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerr, J. R. 2000. Phenazine pigments: antibiotics and virulence factors. Infect. Dis. Rev. 2:184-194. [Google Scholar]
- 23.Kerr, J. R. 1994. Suppression of fungal growth exhibited by Pseudomonas aeruginosa. J. Clin. Microbiol. 32:525-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerr, J. R., G. W. Taylor, A. Rutman, N. Hoiby, P. J. Cole, and R. Wilson. 1999. Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J. Clin. Pathol. 52:385-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Latifi, A., M. K. Winson, M. Foglino, B. W. Bycroft, G. S. Stewart, A. Lazdunski, and P. Williams. 1995. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 17:333-343. [DOI] [PubMed] [Google Scholar]
- 26.Lau, G. W., H. Ran, F. Kong, D. J. Hassett, and D. Mavrodi. 2004. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect. Immun. 72:4275-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laursen, J. B., and J. Nielsen. 2004. Phenazine natural products: biosynthesis, synthetic analogues, and biological activity. Chem. Rev. 104:1663-1686. [DOI] [PubMed] [Google Scholar]
- 28.Liberati, N. T., J. M. Urbach, S. Miyata, D. G. Lee, E. Drenkard, G. Wu, J. Villanueva, T. Wei, and F. M. Ausubel. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 103:2833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mah, T. F., B. Pitts, B. Pellock, G. C. Walker, P. S. Stewart, and G. A. O'Toole. 2003. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426:306-310. [DOI] [PubMed] [Google Scholar]
- 30.Mahajan-Miklos, S., M. W. Tan, L. G. Rahme, and F. M. Ausubel. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96:47-56. [DOI] [PubMed] [Google Scholar]
- 31.Mavrodi, D. V., W. Blankenfeldt, and L. S. Thomashow. 2006. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu. Rev. Phytopathol. 44:417-445. [DOI] [PubMed] [Google Scholar]
- 32.Mavrodi, D. V., R. F. Bonsall, S. M. Delaney, M. J. Soule, G. Phillips, and L. S. Thomashow. 2001. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 183:6454-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mavrodi, D. V., V. N. Ksenzenko, R. F. Bonsall, R. J. Cook, A. M. Boronin, and L. S. Thomashow. 1998. A seven-gene locus for synthesis of phenazine-1-carboxylic acid by Pseudomonas fluorescens 2-79. J. Bacteriol. 180:2541-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norman, R. S., P. Moeller, T. J. McDonald, and P. J. Morris. 2004. Effect of pyocyanin on a crude-oil-degrading microbial community. Appl. Environ. Microbiol. 70:4004-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nseir, S., E. Jozefowicz, B. Cavestri, B. Sendid, C. Di Pompeo, F. Dewavrin, R. Favory, M. Roussel-Delvallez, and A. Durocher. 2007. Impact of antifungal treatment on Candida-Pseudomonas interaction: a preliminary retrospective case-control study. Intensive Care Med. 33:137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 38.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to the study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 39.Parsons, J. F., B. T. Greenhagen, K. Shi, K. Calabrese, H. Robinson, and J. E. Ladner. 2007. Structural and functional analysis of the pyocyanin biosynthetic protein PhzM from Pseudomonas aeruginosa. Biochemistry 46:1821-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pukatzki, S., R. H. Kessin, and J. J. Mekalanos. 2002. The human pathogen Pseudomonas aeruginosa utilizes conserved virulence pathways to infect the social amoeba Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA 99:3159-3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 42.Ran, H., D. J. Hassett, and G. W. Lau. 2003. Human targets of Pseudomonas aeruginosa pyocyanin. Proc. Natl. Acad. Sci. USA 100:14315-14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reszka, K. J., Y. O'Malley, M. L. McCormick, G. M. Denning, and B. E. Britigan. 2004. Oxidation of pyocyanin, a cytotoxic product from Pseudomonas aeruginosa, by microperoxidase 11 and hydrogen peroxide. Free Radic. Biol. Med. 36:1448-1459. [DOI] [PubMed] [Google Scholar]
- 44.Sharma, K. G., R. Kaur, and A. K. Bachhawat. 2003. The glutathione-mediated detoxification pathway in yeast: an analysis using the red pigment that accumulates in certain adenine biosynthetic mutants of yeasts reveals the involvement of novel genes. Arch. Microbiol. 180:108-117. [DOI] [PubMed] [Google Scholar]
- 45.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 46.Thomashow, L. S., and D. M. Weller. 1988. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J. Bacteriol. 170:3499-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomashow, L. S., D. M. Weller, R. F. Bonsall, and L. S. Pierson. 1990. Production of the antibiotic phenazine-1-carboxylic acid by fluorescent Pseudomonas species in the rhizosphere of wheat. Appl. Environ. Microbiol. 56:908-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turner, J. M., and A. J. Messenger. 1986. Occurrence, biochemistry and physiology of phenazine pigment production. Adv. Microb. Physiol. 27:211-275. [DOI] [PubMed] [Google Scholar]
- 49.Watson, D., J. MacDermot, R. Wilson, P. J. Cole, and G. W. Taylor. 1986. Purification and structural analysis of pyocyanin and 1-hydroxyphenazine. Eur. J. Biochem. 159:309-313. [DOI] [PubMed] [Google Scholar]
- 50.Weisman, L. S., R. Bacallao, and W. Wickner. 1987. Multiple methods of visualizing the yeast vacuole permit evaluation of its morphology and inheritance during the cell cycle. J. Cell Biol. 105:1539-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.West, S. E., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81-86. [DOI] [PubMed] [Google Scholar]
- 52.Wilson, R., D. A. Sykes, D. Watson, A. Rutman, G. W. Taylor, and P. J. Cole. 1988. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect. Immun. 56:2515-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winsor, G., R. Lo, S. Sui, K. Ung, S. Huang, D. Cheng, W. Ching, R. Hancock, and F. Brinkman. 2005. Pseudomonas aeruginosa genome database and PseudoCAP: facilitating community-based, continually updated, genome annotation. Nucleic Acids Res. 33:D338-D343. [DOI] [PMC free article] [PubMed] [Google Scholar]