Abstract
Enzymes of the AlkB and CYP153 families catalyze the first step in the catabolism of medium-chain-length alkanes, selective oxidation of the alkane to the 1-alkanol, and enable their host organisms to utilize alkanes as carbon sources. Small, gaseous alkanes, however, are converted to alkanols by evolutionarily unrelated methane monooxygenases. Propane and butane can be oxidized by CYP enzymes engineered in the laboratory, but these produce predominantly the 2-alkanols. Here we report the in vivo-directed evolution of two medium-chain-length terminal alkane hydroxylases, the integral membrane di-iron enzyme AlkB from Pseudomonas putida GPo1 and the class II-type soluble CYP153A6 from Mycobacterium sp. strain HXN-1500, to enhance their activity on small alkanes. We established a P. putida evolution system that enables selection for terminal alkane hydroxylase activity and used it to select propane- and butane-oxidizing enzymes based on enhanced growth complementation of an adapted P. putida GPo12(pGEc47ΔB) strain. The resulting enzymes exhibited higher rates of 1-butanol production from butane and maintained their preference for terminal hydroxylation. This in vivo evolution system could be useful for directed evolution of enzymes that function efficiently to hydroxylate small alkanes in engineered hosts.
Microbial utilization and degradation of alkanes was discovered almost a century ago (27). Since then, several enzyme families capable of hydroxylating alkanes to alkanols, the first step in alkane degradation, have been identified and categorized based on their preferred substrates (30). The soluble and particulate methane monooxygenases (sMMO and pMMO) and the related propane monooxygenase and butane monooxygenase (BMO) are specialized on gaseous small-chain alkanes (C1 to C4), while medium-chain (C5 to C16) alkane hydroxylation seems to be the domain of the CYP153 and AlkB enzyme families.
Conversion of C1 to C4 alkanes to alkanols is of particular interest for producing liquid fuels or chemical precursors from natural gas. The MMO-like enzymes that catalyze this reaction in nature, however, exhibit limited stability or poor heterologous expression (30) and have not been suitable for use in a recombinant host that can be engineered to optimize substrate or cofactor delivery. Alkane monooxygenases often cometabolize a wider range of alkanes than those which support growth (12). We wished to determine whether it is possible to engineer a medium-chain alkane monooxygenase to hydroxylate small alkanes, thereby circumventing difficulties associated with engineering MMO-like enzymes as well as investigating the fundamental question of whether enzymes unrelated to MMO can support growth on small alkanes.
The most intensively studied medium-chain alkane hydroxylases are the AlkB enzymes (2, 20, 29), especially AlkB from Pseudomonas putida GPo1 (13, 28, 32, 35). While most members of the AlkB family act on C10 or longer alkanes, some accept alkanes as small as C5 (30). A recent study (12) indicated that AlkB from P. putida GPo1 may also be involved in propane and butane assimilation. AlkB selectively oxidizes at the terminal carbon to produce the 1-alkanols. No systematic protein engineering studies have been conducted on this di-iron integral membrane enzyme, although selection and site-directed mutagenesis efforts identified one amino acid residue that sterically determines long-chain alkane degradation (35).
The most recent addition to the known biological alkane-hydroxylating repertoire is the CYP153 family of heme-containing cytochrome P450 monooxygenases. Although their activity was detected as early as 1981 (1), the first CYP153 was characterized only in 2001 (16). Additional CYP153 enzymes were identified and studied more recently (9, 10, 31). These soluble class II-type three-component P450 enzymes and the AlkB enzymes are the main actors in medium-chain-length alkane hydroxylation by the cultivated bacteria analyzed to date (31). CYP153 monooxygenases have been the subject of biochemical studies (9, 16, 19), and their substrate range has been explored (10, 14). Known substrates include C5 to C11 alkanes. The best-characterized member, CYP153A6, hydroxylates its preferred substrate octane predominantly (>95%) at the terminal position (9).
Recent studies have shown that high activities on small alkanes can be obtained by engineering bacterial P450 enzymes such as P450cam (CYP101; camphor hydroxylase) and P450 BM3 (CYP102A; a fatty acid hydroxylase) (8, 36). The resulting enzymes, however, hydroxylate propane and higher alkanes primarily at the more energetically favorable subterminal positions; highly selective terminal hydroxylation is difficult to achieve by engineering a subterminal hydroxylase (22). We wished to determine whether a small-alkane terminal hydroxylase could be obtained instead by directed evolution of a longer-chain alkane hydroxylase that exhibits this desirable regioselectivity. For this study, we chose to engineer AlkB from P. putida GPo1 and CYP153A6 from Mycobacterium sp. strain HXN-1500 (9, 33) to enhance activity on butane. Because terminal alkane hydroxylation is the first step of alkane catabolism in P. putida GPo1, we reasoned that it should be possible to establish an in vivo evolution system that uses growth on small alkanes to select for enzyme variants exhibiting the desired activities.
The recombinant host Pseudomonas putida GPo12(pGEc47ΔB) was engineered specifically for complementation studies with terminal alkane hydroxylases and was used previously to characterize members of the AlkB and CYP153 families (26, 31). This strain is a derivative of the natural isolate P. putida GPo1 lacking its endogenous OCT plasmid (octane assimilation) (5) but containing cosmid pGEc47ΔB, which carries all genes comprising the alk machinery necessary for alkane utilization, with the exception of a deleted alkB gene (34). We show that this host can be complemented by a plasmid-encoded library of alkane hydroxylases and that growth of the mixed culture on butane leads to enrichment of novel butane-oxidizing terminal hydroxylases.
MATERIALS AND METHODS
Strains and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Luria-Bertani broth (23) and modified M9 medium with 1.5% yeast extract (8) supplemented with appropriate antibiotics, or E2 (15) and M9 (17) minimal media supplemented with carbon sources, were used for growth. All cultures were grown aerobically at 30°C (P. putida) or 37°C (Escherichia coli). Antibiotic concentrations were 100 μg/ml ampicillin, 15 μg/ml tetracycline, 20 μg/ml chloramphenicol, and 10 μg/ml gentamicin for E. coli cultures and 50 μg/ml for pseudomonads. Bacterial strains were grown on solid E2 minimal medium with liquid alkanes (Sigma-Aldrich) provided through the gas phase as described previously (26). Solid minimal medium growth tests on gaseous alkanes (Sigma-Aldrich) were conducted in gas-tight plastic containers (GasPak 150 large anaerobic vented system; VWR), pressurized at 20 lb/in2 for 20 s (ethane and propane) or 10 lb/in2 for 6 s (butane). Liquid minimal medium cultures growing on alkanes were shaken in custom-made gas-tight flasks with 1% liquid alkane (pentane and octane) in a reservoir or in gas-tight serum bottles (Alltech), pressurized with gaseous alkanes as described above. For growth tests of P. putida GPo12(pGEc47ΔB) on alkanols (Sigma-Aldrich), cells from an LB preculture were washed three times with M9 medium and used to inoculate the 5-ml M9 main cultures in 14-ml tubes (Greiner) to an optical density (OD) of 0.1, then grown at 30°C with continuous shaking. Libraries of P. putida GPo12(pGEc47ΔB) strains expressing AlkB or CYP153A6 variants were precultured on E2 minimal medium plates with antibiotics and 0.2% (wt/vol) citrate as carbon source and then enriched for improved strains through continuous growth in liquid E2 minimal medium with small-chain-length alkanes as the sole carbon source, as described above.
TABLE 1.
Strains and plasmids used or constructed in this work
| Strain or plasmid | Relevant genotype or characteristics | Reference and/or source |
|---|---|---|
| E. coli strains | ||
| CC118(RK600) | Helper strain for triparental mating | 6 |
| DH5α | Cloning strain | Invitrogen |
| JS200(pEP Pol I) | Mutator strain | Addgene; 3 |
| XL1-Red | Mutator strain | Invitrogen |
| P. putida strains | ||
| GPo12(pGEc47ΔB) | GPo1 cured of the OCT plasmid, transformed with pGEc47ΔB; contains all genes necessary for growth on alkanes, except for the alkane hydroxylase gene | 26 |
| Palk1 | GPo12(pGEc47ΔB) adapted to growth on gaseous alkanes with pCom10_alkB | This study |
| Pcyp1 | GPo12(pGEc47ΔB) adapted to growth on gaseous alkanes with pCom8_cyp153A6 | This study |
| Pcyp2 | Pcyp1 further adapted to growth on gaseous alkanes with pCom8*_cyp153A6-BMO1 | This study |
| Plasmids | ||
| pCom8 | Broad-host-range expression vector with PalkB; GmroriT alkS | 25 |
| pCom8* | pCom8 vector adapted for improved complementation for small-chain-length alkane-dependent host growth | This study |
| pCom10 | Broad-host-range expression vector with PalkB; KanroriT alkS | 25 |
Transformation, cloning, and mutagenesis.
E. coli DH5α strains used for plasmid amplification were transformed by electroporation (23), E. coli BL21(DE3) (Novagen) was used for protein expression by heat shock according to the manufacturer's manual, and P. putida GPo12(pGEc47ΔB) strains were transformed by triparental mating with the appropriate E. coli DH5α donor and E. coli CC118(pRK600) mediator strain (6). Plasmids pCom8_cyp153A6 and pCom10_alkB were obtained from the B. Witholt (now retired) laboratory, ETH Zurich. Sequencing of pCom8_cyp153A6 revealed it to be missing the first 9 nucleotides of the coding sequence of the monooxygenase gene compared to the published sequence (9). The CYP153A6 parent enzyme and its variants are therefore three N-terminal amino acids shorter than the published sequence but are otherwise identical and obviously functional.
Mutagenesis of pCom plasmids was performed in E. coli XL1-Red strains according to the manufacturer's manual (Stratagene) and in E. coli JS200(pEP Pol I) as described previously (3). Mutated alkB genes or the CYP153A6 gene fdrA6 and fdxA6 operons were cloned into the original pCom10 or into pCom8* plasmids as EcoRI-HindIII- or KpnI-digested fragments, respectively. Restriction enzymes were obtained from Roche Molecular or New England Biolabs. Plasmid pCom8_alkBFG was constructed by amplifying the alkBFG operon from the plasmid pblaP4_alkJBFG-luxAB (18) using the primers alkBFG_1 (GATCTACATATGCTTGAGAAACACAGAGTTCTGGATTC) and alkBFG_2 (GATCTACCCGGGTCACTTTTCCTCGTAGAGCACATAGTC) in a standard PCR method. The resulting fragment was cloned into the pCom8 vector using the NdeI and XmaI restriction sites introduced by the primers. The resulting plasmid was digested with SpeI, and the resulting alkB-containing 3.7-kb fragment was replaced with the appropriate fragment from SpeI-digested pCom10_alkB, pCom10_alkB-BMO1, and pCom10_alkB-BMO2 by cloning, resulting in pCom8_alkBFG, pCom8_alkB-BMO1_alkFG, and pCom8_alkB-BMO2_alkFG. Successful cloning was verified by sequencing.
Protein expression and bioconversions.
E. coli BL21(DE3) cells expressing AlkB variants were precultured in LB medium at 37°C with shaking at 250 rpm for 24 h. LB cultures (120 ml) in 1,000-ml flasks were inoculated to an OD at 600 nm (OD600) of 1.0 and grown at 37°C, 250 rpm, for 2.5 h. The cultures were then set to 25°C at 200 rpm and induced with 0.4 mM dicyclopropylketone (Sigma-Aldrich) after 30 min. The cultures were centrifuged (10 min, 3,300 × g, room temperature) 20 h later and the cell pellets resuspended in an equal volume of 100 mM KPi buffer (pH 7.0). For cell dry weight determinations, 10-ml cell suspensions were washed once with distilled water and the cell pellets were dried for 3 days at 80°C. For bioconversion experiments with liquid alkanes, 250 μl of the alkane and 1% (vol/vol) glycerol were added to a 1-ml cell suspension in a glass vial (Kimax), capped, and incubated at a 60° angle at 25°C and 200 rpm. After 60 min, reactions were stopped by addition of 200 μl of 1 N HCl. Pentane bioconversion samples were then left open at 60°C for 1 h to allow the substrate to evaporate and subsequently pelleted, filtered, and subjected to gas chromatography analysis. Octane bioconversion products were extracted by adding 250 μl hexane, vortexing for 30 s, and centrifuging at 14,000 rpm for 10 min. Alkanols were then detected in the top organic layer.
For bioconversions of gaseous alkanes, 80 ml of cell suspension and 15 μl of antifoam (Sigma-Aldrich) were stirred in a 100-ml bioreactor (Ochs-labor) at 25°C. Propane or butane was mixed in a 1:3 ratio with air, and the mixture was fed to the cells at an inlet gas flow rate of approximately 10 liter h−1. The reaction was started by addition of glycerol to a final concentration of 1% (vol/vol). After 30 min, 1-ml samples were taken from the cell suspension as well as from the wash fraction of a bubbler installed downstream of the bioreactor, combined, centrifuged, filtered, and subjected to gas chromatographic analysis.
Bioconversions with CYP153A6 variants were conducted as described above, with the following modifications: cells induced for expression of CYP153 were grown in modified M9 medium with 1.5% yeast extract for 14 h, and the buffer used for bioconversions was modified M9 medium prepared without nitrogen. Gaseous alkane and air were introduced at 1:1 (vol/vol) through a gas dispenser. Bioconversions of liquid and gaseous alkanes were started by addition of 20 mM glucose and stopped after 60 min. The concentration of properly folded P450 protein was determined from the CO difference spectrum of 5×-concentrated cell extract after sonication, removal of cell debris, and bubbling CO into the sample. Addition of reducing agent to a concentrated cell extract was not necessary and in fact decreased CO binding. To assess P450 stability, CO-binding studies were performed on aliquots of cell extracts that had been stored at 25°C for up to 24 h.
RESULTS
P. putida GPo12(pGEc47ΔB) grows on short-chain 1-alkanols but not the 2-alkanols.
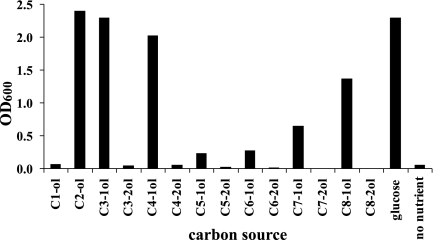
P. putida GPo12(pGEc47ΔB) was shown in previous studies to grow on medium-chain-length alkanols like 1-octanol and on the corresponding alkane only when complemented by a terminal alkane hydroxylase (26, 31). To determine whether this strain could be used to select or screen for improved terminal alkane hydroxylation activity, we tested its ability to grow with the primary and secondary C1 to C8 alkanols as sole carbon sources (Fig. 1). No growth was observed on any of the secondary alcohols or on methanol during an 18-day period. Ethanol, 1-propanol, and 1-butanol supported relatively strong growth, comparable to that of the positive control grown on glucose. Slow growth was observed on 1-pentanol, 1-hexanol, and 1-octanol. The results indicated that the terminal hydroxylation products of all the short-chain n-alkanes except for methane are readily utilized as carbon sources, while subterminal oxidation products (the sec-alkanols) are not. Thus, this strain should be suitable for growth-based screening and selection for terminal hydroxylation of alkanes of long, medium, and short chain lengths.
FIG. 1.
Growth of P. putida GPo12(pGEc47ΔB) with primary and secondary linear short and medium chain-length alkanols. The OD600 of the cultures was measured after 18 days of growth in liquid M9 minimal medium with 0.5% (vol/vol) primary (CX-1-ol; X = number of carbon atoms) or secondary (CX-2ol) alcohols as carbon source dissolved in a 5% (vol/vol) organic layer of heptamethylnonane. Alcohols smaller than five carbon atoms were added without organic solvent. Cultures with 0.5% (wt/vol) glucose or no added nutrient served as controls.
Creation of gene libraries through random plasmid mutation.
It was not efficient to use error-prone PCR to randomly mutate the target genes, as cloning of PCR products into the pCom vector yielded fewer than 2,000 transformants, much less than the hundreds of millions of mutants that can be evaluated in principle using a growth selection. Mutant libraries were therefore constructed by complementing P. putida GPo12(pGEc47ΔB) strains with randomly mutated plasmids encoding AlkB or CYP153A6. The drawbacks of including mutations that affect the vector (antibiotic resistance and origin of replication) rather than just the inserted genes are compensated for by the large size and ease of library construction. Mutator strains were used to generate the plasmid libraries and to increase diversity both available mutator strains were used, Escherichia coli XL1-Red (Stratagene) and E. coli JS200 pEP polymerase I (Pol I) (4). E. coli XL1-Red has deficiencies in the DNA repair mechanism that lead to a 5,000-fold increase in the general mutation rate (11) (Stratagene manual). E. coli JS200 pEP Pol I expresses an engineered mutator DNA polymerase I which mainly amplifies plasmid DNA, with lower reliability, thus introducing mutations in the plasmid DNA (4). The nucleotide mutation level in XL1-Red after 2 weeks of continuous culturing was approximately 0.1/kb, while four rounds of mutation in JS200 pEP Pol I yielded up to 0.4/kb. Cultures of both mutator strains were combined and the mutated plasmids were transformed into P. putida GPo12(pGEc47ΔB) through triparental mating with the helper strain E. coli CC118(pRK600) (31). The growth selection was performed by culturing the resulting strain library in minimal medium with an alkane as sole carbon source for up to 3 weeks, as described in Materials and Methods.
P. putida GPo12(pGEc47ΔB) strains grow faster on butane when complemented with evolved AlkB and CYP153A6 variants.
A mixed culture containing alkane hydroxylase variants will become enriched in strains best adapted to use alkanes as their sole carbon source. However, not only can beneficial mutations in the hydroxylase gene lead to improved growth, but also adaptations of the host and vector will do so as well. For P. putida GPo12(pGEc47ΔB) complemented by CYP153 genes, it had been observed that the host had to be adapted through prolonged cultivation on alkanes to obtain significant growth on these substrates (31), without any mutations occurring in the CYP153 genes themselves. To test whether host adaptation was also occurring in our experiments, 21 single colonies obtained from the first round of enrichment cultures were compared to the parent strain in plate growth tests (data not shown). Solid medium growth tests were chosen over liquid medium tests due to the growth instability of liquid minimal medium cultures of P. putida GPo12(pGEc47ΔB), which often show different growth rates between replicates or, occasionally, failure to grow at all. The host, vector, and operons of the best mutants were analyzed individually by comparing them in growth tests to their wild-type counterparts (see Fig. S1 in the supplemental material). To identify and analyze potentially adapted hosts, the adapted recombinant strain was cured of the plasmid and transformed with the appropriate wild-type plasmid. Adapted vectors were isolated from the strains, and the alkB gene or the CYP153A6 gene fdrA6 and fdxA6 operons were replaced with the wild-type sequence by cloning before being mated into the wild-type host. Furthermore, potentially improved hydroxylase genes were recloned into a wild-type vector and transferred into a wild-type host.
Comparison of all the resulting strains in growth assays led to the identification of several improved hosts and vectors. Strain Pcyp1, an adapted P. putida GPo12(pGEc47ΔB) strain, showed faster growth on pentane than its parent when complemented by the wild-type pCom8_cyp153A6 plasmid. Further adaptation of Pcyp1 led to Pcyp2, which again grew faster on pentane. Similarly, strain Palk1 showed improved growth on propane and butane when transformed with wild-type pCom10_alkB. For the CYP153A6 system, adapted plasmid pCom8* enabled faster growth of P. putida GPo12(pGEc47ΔB) on pentane, even when it contained the wild-type operon. Sequencing showed no mutation of the CYP operon in pCom8*_cyp153A6 or in the sequence 500 nucleotides up- and downstream from the operon. For the AlkB system, no improved vectors were obtained. The adapted host and vector components were specific for the particular system used, i.e., strain Palk1 did not show improved growth on short-chain-length alkanes compared to the wild-type host when complemented with CYP153A6. Likewise, Pcyp1, Pcyp2, and pCom8* were only adapted for their specific systems. The nature of the mutations and how they benefit growth on short-chain alkanes is unknown.
In addition to creating these adapted hosts and plasmid, the first rounds of in vivo-directed evolution generated enzyme variants AlkB-BMO1 (butane monooxygenase) and CYP153A6-BMO1, both of which conferred improved growth on butane. Sequencing revealed a single nucleotide mutation in each: mutation of the codon CTA to GTA led to the single amino acid substitution L132V in AlkB-BMO1 and, through a GCA-to-GTA change, the substitution A94V in CYP153A6-BMO1 (A97V in the published sequence reported in reference 9) (see Materials and Methods).
All the adapted components were combined and evaluated in plate growth tests (Table 2; Fig. 2). Palk1 expressing AlkB-BMO1 showed a significant increase in the rate of growth on butane compared to Palk1 expressing wild-type AlkB. In contrast, growth rates on pentane and octane were reduced. No significant growth improvement on propane was observed, and neither enzyme supported growth on ethane. These results suggest that the L132V mutation in AlkB-BMO1 specifically improves activity toward butane. A similar result was found for Pcyp2 (pCom8*_cyp153A6-BMO1) recombinants, which grew more slowly on octane than Pcyp2 (pCom8*_cyp153A6) but faster on pentane. The CYP153A6-BMO1 variant also supported growth on butane, which wild-type CYP153A6 does not. Thus, the A94V mutation appears to specifically enhance activity on the smaller alkanes.
TABLE 2.
Relative growth of adapted P. putida GPo12(pGEc47ΔB) strains, expressing CYP153A6 and AlkB variants, on minimal medium plates with alkanes as the sole carbon source
| Complementing alkane monooxygenase | Days required for growth to full lawn with selected carbon source
|
||||
|---|---|---|---|---|---|
| Ethane | Propane | Butane | Pentane | Octane | |
| AlkB wild type | NGa | 5 | 5 | 3 | 2 |
| AlkB-BMO1 | NG | 5 | 3 | 6 | 7 |
| AlkB-BMO2 | NG | 5 | 2 | 4 | 8 |
| CYP153A6 wild type | NG | NG | NG | 2 | 2 |
| CYP153A6-BMO1 | NG | NG | 5 | 1.5 | 5 |
NG, no growth detected during 3-week observation.
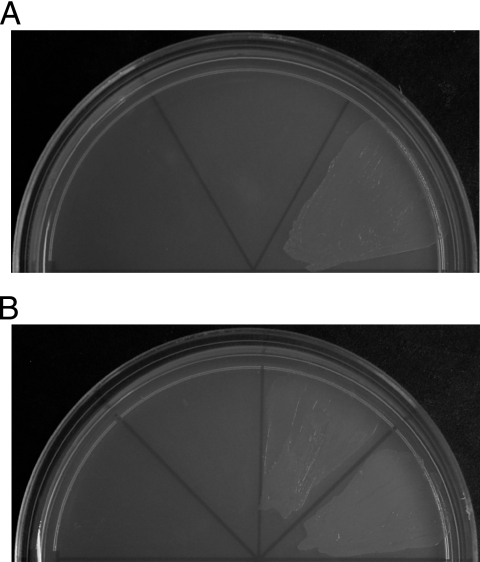
FIG. 2.
Growth of P. putida GPo12(pGEc47ΔB) strains on alkanes. (A) Strains of adapted P. putida GPo12(pGEc47ΔB) complemented by an empty plasmid or expressing CYP153A6 wild-type or CYP153A6-BMO1 (left, middle, and right sections) were grown for 5 days with butane as sole carbon source. (B) Growth of adapted P. putida GPo12(pGEc47ΔB) strains complemented by an empty plasmid, expressing the AlkB wild type, or its variants, BMO1 and BMO2, on butane after 2 days.
Plasmids pCom8*_cyp153A6-BMO1 and pCom10_alkB-BMO1 were subjected to a second round of mutagenesis and mated into Pcyp2 and Palk1, respectively. A further-improved AlkB monooxygenase, AlkB-BMO2, was obtained after enrichment and screening. Sequencing revealed a total of three nucleotide mutations, all resulting in amino acid substitutions in AlkB-BMO2: V129M (GTG to ATG), L132V (CTA to GTA), and I233V (ATC to GTC). To ensure comparison in identical genetic backgrounds, the mutated gene was recloned into a wild-type pCom10 vector, mated into fresh Palk1, and compared in growth tests to Palk1 expressing wild-type AlkB and AlkB-BMO1 (Table 2; Fig. 2). Compared to its parent AlkB-BMO1, AlkB-BMO2 performed even better in growth complementation studies with butane. Growth on pentane and octane was also improved but was still inferior to that obtained with the wild-type enzyme. Thus, the mutations V129M and I233V improved the overall activity of AlkB-BMO2 compared to its parent, AlkB-BMO1. Enrichment and screening yielded no additional improvement in the CYP153A6 system.
Whole-cell bioconversions in E. coli confirmed improved performance of AlkB-BMO1, -BMO2, and CYP153A6-BMO1 for conversion of butane to 1-butanol.
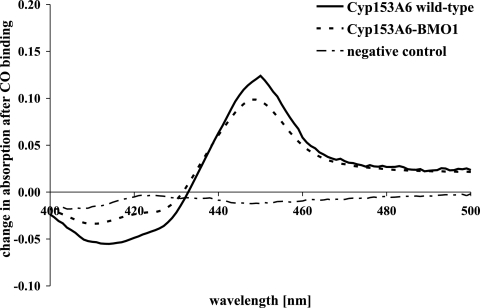
In order to quantify the effects of the mutations on enzyme performance, whole-cell bioconversions were performed using growth-arrested E. coli BL21(DE3) cells containing the CYP153A6 variants expressed from pCom8*. Although pCom plasmids are not efficient expression platforms, we nonetheless observed CYP153A6 expression in E. coli, as indicated by the CO difference spectral peak at 450 nm (Fig. 3). In contrast, no 450-nm signal was observed for cells harboring the empty pCom8* plasmid. Cytochrome P450s are notoriously difficult to express, and it has been reported that the CO-binding activity of CYP153A6 is lost shortly after cell disruption at room temperature, even when the enzyme is isolated from its native host (33). However, we found CYP153A6 was expressed well in E. coli DH5α and showed a stable CO difference spectrum for hours at room temperature if protease inhibitor was added before cell disruption (data not shown). Using E. coli BL21(DE3) cells, which are deficient in the Lon and OmpT proteases, eliminated the need to add protease inhibitor for stable CYP153A6 expression. Cell extract from these cultures, expressing CYP153A6 or CYP153A6-BMO1, retained full CO-binding capacity for 24 h when stored at 25°C. At 45°C, CO-binding capacity decreased with time, showing a half-life of 638 min (±68 min [standard error]) for cell extract containing CYP153A6-BMO1 and 367 min (±58 min) with CYP153A6 (see Fig. S2 in the supplemental material).
FIG. 3.
CO difference spectra of lysed E. coli BL21(DE3) cell suspensions. E. coli BL21(DE3) cultures expressing CYP153A6 or CYP153A6-BMO1 were concentrated 5:1 in buffer, and UV-VIS spectra were obtained from the cell extracts after CO saturation. The peaks correspond to 0.21 μM and 0.17 μM folded P450 for the CYP153A6 and CYP153A6-BMO1 samples, respectively. Cells carrying the empty vector treated in the same way served as the negative control.
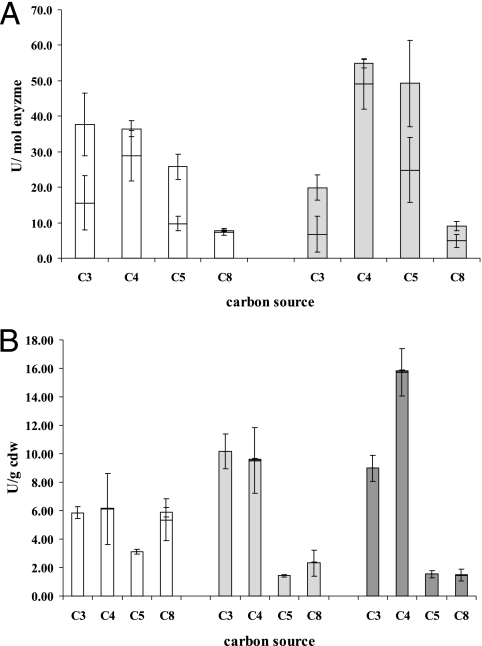
The typical concentration of folded CYP153A6 in the cell suspensions used for bioconversions was 0.1 to 0.2 μM, with CYP153A6-BMO1 usually expressing ∼20% less than its parent, CYP153A6 (Fig. 3). For both enzymes, we observed no significant decrease in apparent P450 concentration after the 60-min bioconversion reactions (data not shown). Bioconversion studies showed significantly altered activity and selectivity for CYP153A6-BMO1 that closely followed its growth complementation performance (Fig. 4A). Control bioconversions performed under the same conditions but using transformants of empty pCom8* vector did not produce alkanols (data not shown). Butane bioconversions yielded an averaged total concentration of 393 μM 1-butanol in the aqueous phase after 60 min with CYP153A6-BMO1, versus 277 μM with wild-type CYP153A6 (Table 3). From the concentration of product formed divided by the concentration of folded P450, the average turnover rate of CYP153A6-BMO1 in 1-butanol production was 49 min−1, a 75% increase compared to 28 min−1 for CYP153A6. Interestingly, the selectivity for terminal hydroxylation also increased with the variant, from 78% to 89% of total alkanol product. The A94V mutation also improved activity and selectivity for conversion of pentane to 1-pentanol but had the opposite effect with propane and octane.
FIG. 4.
Bioconversions of alkanes to 1- and 2-alkanols. Whole-cell bioconversions were carried out with at least two replicates using resting E. coli BL21(DE3) cells after expression of CYP153A6 (A) and AlkB (B) variants. Relative enzymatic activities in the aqueous cell suspension were calculated from the alcohol product formed per minute and the enzyme concentration (A) or total cell dry weight (B). The lower graphs represents relative activities for 1-alkanol, and the upper part shows 2-alkanol production (if detected). Sole added carbon sources were propane (C3), butane (C4), pentane (C5), and octane (C8). White graphs depict wild-type activity, light gray shows the BMO1 activity, and dark gray shows the BMO2 variant activities.
TABLE 3.
Absolute product concentrations after bioconversion in resting E. coli BL21(DE3) cells
| Substratea | Total hydroxylated product (μM)b
|
||||
|---|---|---|---|---|---|
| CYP153A6 | CYP153A6- BMO1 | AlkB | AlkB-BMO1 | AlkB-BMO2 | |
| Propane | 421 (59) | 244 (68) | 625 (0) | 1,048 (0) | 973 (0) |
| Butane | 356 (22) | 439 (11) | 639 (1) | 1,045 (1) | 1,586 (1) |
| Pentane | 260 (62) | 374 (49) | 606 (0) | 294 (0) | 317 (0) |
| Octane | 78 (3) | 69 (46) | 1,122 (9) | 487 (2) | 308 (1) |
Bioconversions with propane and butane were performed in a bioreactor for 30 min with AlkB variants and for 60 min with CYP153A6 variants. Bioconversion mixtures with pentane and octane were shaken in a glass vial for 60 min for all enzymes.
Values in parentheses are the percentage of 2-alkanol in the total hydroxylated product. Only 1- and 2-alkanols were formed in detectable amounts.
Since E. coli cells expressing only AlkB showed no product formation, bioconversions were performed using cells transformed with pCom8_alkBFG. Minak-Bernero et al. (18) demonstrated that the AlkBFG system, including the monooxygenase AlkB and the nonessential and essential rubredoxins AlkF and AlkG, is functional in E. coli, without the need for the rubredoxin reductase AlkT. Results of bioconversions using AlkB, AlkB-BMO1, and AlkB-BMO2 (Fig. 4B) showed that the activity of the variants was greater than that of the wild type on butane, the substrate used for in vivo evolution, while performance on pentane and octane was decreased. The evolved variants also showed increased activity on propane, the only change in activity that was not reflected in a similar observable change in growth complementation performance.
In 30 min, butane bioconversions utilizing AlkB, AlkB-BMO1, or AlkB-BMO2 produced on average 630 μM, 1030 μM, and 1580 μM 1-butanol in the aqueous phase, respectively. The dry cell weights of the cell suspensions used for the bioconversions ranged from 3.0 to 4.0 g/liter. The activities (in μmol of 1-butanol min−1 g [cell dry wt]−1) thus increased from 6.1 to 9.5 and 15.7 units, respectively. Bioconversions with AlkB variants were highly regioselective, producing no detectable 2-alkanol from propane and pentane and very little 2-alkanol from butane and octane (Table 3).
DISCUSSION
Advantages and drawbacks of in vivo evolution of terminal alkane hydroxylases.
An in vivo-directed evolution system with selection for terminal alkane hydroxylase activity has been developed and applied to engineering enzymes from the AlkB and CYP153 families. The goal of this work was to increase activity on short-chain alkanes while maintaining these enzymes' remarkable preferences for the thermodynamically disfavored terminal position, thereby taking a first step toward engineering small-alkane hydroxylases that can be expressed in a recombinant bacterial host amenable to further engineering.
Evolution in vivo using growth selection enables searches through as many as 108 mutants in a simple flask culture while exerting selection pressure for many useful enzyme characteristics simultaneously (e.g., substrate specificity, regioselectivity, specific activity, coupling of cofactor utilization to product formation, or expression level). In this particular case, the Pseudomonas host used for the directed evolution only utilizes primary alcohols, allowing selection directly for terminal hydroxylation. This is the only method we know of for efficient, high-throughput selection or screening of this important activity. Previous efforts to engineer P450 BM3 or P450cam for alkane hydroxylation resulted in enzyme variants mainly targeting subterminal positions (8, 36). The present system promises to be useful for the directed evolution of a range of alkane hydroxylases, since P. putida GPo12(pGEc47ΔB) can be complemented for growth on alkanes by many different AlkB and CYP153 genes (26, 31). Furthermore, growth tests showed that the host can utilize 1-alkanols down to ethanol, potentially enabling enrichment for activity on alkanes as small as ethane.
This growth selection system could handle a mutation rate perhaps 10 times greater than the 0.1 to 0.4 nucleotide changes per kb obtained using the mutator strains. The P. putida GPo12 organism and the components used in this study, however, are somewhat difficult to handle with regard to growth stability, expression, mutation rates, cloning, and transformation efficiency. Furthermore, in vivo evolution also has limits for identifying moderately improved variants, since enrichment to pure culture may require weeks or months of continuous culturing, depending on the actual doubling time on a given substrate and the level of improvement over the parent. This makes the stepwise accumulation of small improvements more challenging.
AlkB enables P. putida GPo12(pGEc47ΔB) to grow on propane and butane and can be improved in this function.
Our results prove that propane and butane are substrates of AlkB from P. putida GPo1, verifying earlier assumptions (12). No growth complementation was observed on ethane, and the E. coli system utilized in the bioconversion reactions could not be used to determine whether ethane is converted to ethanol by AlkB variants, because the glycerol added to allow NADH regeneration was fermented to ethanol by the E. coli BL21(DE3) host. Quantification of AlkB activity on ethane would require purification and reconstitution of AlkB and its electron transport system or genetic modification of the strain used for whole-cell bioconversions.
Second-generation variant AlkB-BMO2 supported significantly better growth and butane bioconversion, more than double that of wild-type AlkB. Although specific activity (moles of product/moles of enzyme/minute) could not be determined for the AlkB variants, absolute 1-butanol production rates in bioconversion reactions were significantly higher for the AlkB-BMO2 system (up to 1.6 mM in 30 min) than for CYP153A6-BMO1 (0.4 mM in 60 min). The higher product formation rate with the AlkB system was surprising, in view of the fact that it relied on a nonnative E. coli host reductase, while the CYP153A6 system contained the complete native electron transfer chain with its ferredoxin and ferredoxin reductase. In earlier studies, AlkB activity in E. coli cells increased from 3 U to 20 U when the specific reductase, encoded by alkT, was present (29). However, the significantly higher E. coli whole-cell bioconversion rates with AlkB variants did not translate into better growth complementation of adapted P. putida GPo12(pGEc47ΔB) strains. It is known that AlkB expresses 2- to 10-fold better in E. coli than in P. putida. On the other hand, AlkB has a five- to sixfold-higher specific activity in P. putida (28). Thus, a possible explanation for the growth complementation is that the CYP153 variants express better or have a higher specific activity in P. putida compared to the AlkB variants. Furthermore, the twice-adapted host Pcyp2 might contain chromosomal improvements that are able to compensate for the lower apparent butane oxidation activity of CYP153A6-BMO1 and thus achieve the same growth rate on butane as Palk1 expressing AlkB.
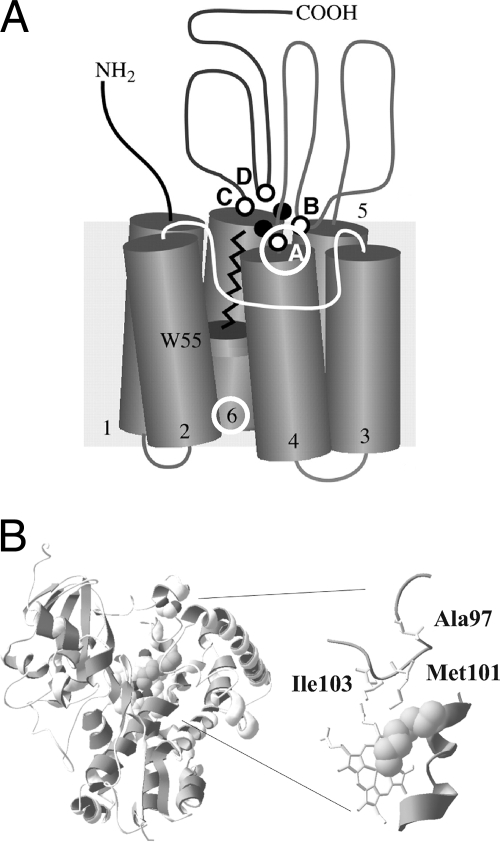
Although no crystal structures are available for any enzyme of the AlkB family, a topology model (34, 35) has been published. Mapping of the evolved AlkB variant mutations on that model (Fig. 5) showed that two of the three mutations generated in AlkB-BMO2, V129M and L132V, are close to the histidine-containing sequence motif A (H138EXXHK143), which is one of four highly conserved histidine-containing motifs in AlkB believed to coordinate the Fe ions and presumed to be part of or close to the active site (24, 35). Thus, a direct effect of V129M and L132V on the active site of AlkB seems possible. The third amino acid substitution, I233V, is located close to the periplasm in the sixth predicted transmembrane domain, distant from the active site.
FIG. 5.
Models for AlkB (A) and CYP153A6 (B). (A) Topology model of AlkB, taken from van Beilen et al. (35). Variants AlkB-BMO1 and AlkB-BMO2 with increased activity on butane carry L132V and V129M+L132V+I233V mutations (marked by white circles), respectively. Amino acids 129 and 132 are close to the first of four histidine-containing sequence motifs (A to D) that contribute to coordination of the Fe ions. Position 233 is located in the periplasmic end of the sixth transmembrane domain (numbers 1 to 6). (B) CYP153A6 structural model with bound substrate octane (spherical object), modified from Funhoff et al. (9). CYP153A6-BMO1 carries a mutation corresponding to A97V in the model.
CYP153A6-BMO1 constitutes the first functional butane monooxygenase of the CYP153 family.
Evolved CYP153A6-BMO1 is the first CYP153 shown to hydroxylate butane and enable its host to grow on this short-chain-length alkane. The gaseous alkanes are usually processed by distinct, specialized enzymes, like the di-iron methane and butane monooxygenases (30) that are unrelated to the cytochrome P450s. Complementation of P. putida GPo12(pGEc47ΔB) with various CYP153 wild-type enzymes resulted in growth on alkanes ranging from pentane to decane (31), but never on butane (unpublished results). And, until now, biotransformations using CYP153 enzymes have only demonstrated activity for hexane and longer alkanes (9, 10, 14). Here we demonstrated that CYP153A6 wild-type and CYP153A6-BMO1 act on butane and even propane, but only the BMO1 variant complemented growth of Pcyp2 on butane. The activity for 1-butanol formation of 49 min−1 (0.8 s−1) measured in CYP153A6-BMO1 assays is still less than that reported for the methane monooxygenase from Methylosinus trichosporium OB3b (8.8 s−1) (7), although these values were generated under very different reaction conditions. Not only does CYP153A6-BMO1 exhibit increased total activity on butane compared to its wild-type parent, but its preference for terminal hydroxylation also increased, resulting in only 11% 2-butanol formation compared to 22% for the wild-type enzyme. CYP153A6-BMO1 was converted into a genuine butane monooxygenase by in vivo-directed evolution.
CYP153A6-BMO1 contains a single amino acid substitution, which corresponds to A97V in the published structure model (9). This mutation, which has a slightly negative effect on expression level (the CYP153A6-BMO1 variant usually showed ∼80% of the CO-binding activity of the wild type) stabilized CYP153A6 and nearly doubled its half-life at 45°C. However, this effect is unlikely to explain the higher observed bioconversion rates, since both enzymes were stable at 25°C, and no loss of folded P450 was observed at the end of the bioconversions. These findings argue for a direct positive effect of the A97V substitution on the butane hydroxylation activity. Based on a proposed model of CYP153 (9), however, A97 is predicted to be located in a loop distant from the active site, pointing toward the protein surface and not the substrate channel (Fig. 5). Why the activity of CYP153A6 and CYP153A6-BMO1 on propane, as observed in the E. coli bioconversion experiments, is not accompanied by growth complementation of Pcyp2 on propane remains unknown. A minimum activity required for growth may not have yet been reached. It is also possible that the substrate is not available at a sufficient concentration or is toxic.
The appearance of improved variants of AlkB and CYP153A6 demonstrated that in vivo evolution in adapted P. putida GPo12(pGEc47ΔB) can be applied to very different monooxygenase enzymes. Despite the higher absolute activity and better regioselectivity of AlkB variants in the bioconversion experiments, the CYP153A enzymes nonetheless offer some important advantages for further engineering. CYP153 enzymes are soluble, and accurate determination of functional expression and concentration is easier due to observable CO binding. Furthermore, CYP153 proteins have been shown to function in whole-cell bioconversions as single-component, self-sufficient fusion proteins with the P450RhF reductase domain (14, 21) as well as with the CYP102A reductase domain (unpublished results). Further directed evolution of terminal alkane hydroxylases like AlkB or CYP153A6 should help us to better understand and utilize their remarkable catalytic activities.
Supplementary Material
Acknowledgments
We thank B. Witholt, A. Schmid, E. Funhoff, and M. Camps for providing materials or instructions.
D. J. Koch acknowledges grant KO 3503/1-1 from the Deutsche Forschungsgemeinschaft. This study was funded by Department of Energy award DE-FG02-06ER15762.
Footnotes
Published ahead of print on 14 November 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Asperger, O., A. Naumann, and H. P. Kleber. 1981. Occurrence of cytochrome P450 in Acinetobacter strains after growth on n-hexadecane. FEMS Microbiol. Lett. 11:309-312. [Google Scholar]
- 2.Baptist, J. N., R. K. Gholson, and M. J. Coon. 1963. Hydrocarbon oxidation by a bacterial enzyme system. I. Products of octane oxidation. Biochim. Biophys. Acta 73:1-6. [DOI] [PubMed] [Google Scholar]
- 3.Camps, M., and L. A. Loeb. 2003. Use of pol I-deficient E. coli for functional complementation of DNA polymerase, p. 11-18. In J. F. H. Arnold and G. Georgiou (ed.), Directed enzyme evolution: screening and selection methods. Humana, Totowa, NJ. [DOI] [PubMed]
- 4.Camps, M., J. Naukkarinen, B. P. Johnson, and L. A. Loeb. 2003. Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I. Proc. Natl. Acad. Sci. USA 100:9727-9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakrabarty, M., G. Chou, and I. C. Gunsalus. 1973. Genetic regulation of octane dissimilation plasmid in Pseudomonas. Proc. Natl. Acad. Sci. USA 70:1137-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duetz, W. A., J. B. van Beilen, and B. Witholt. 2001. Using proteins in their natural environment: potential and limitations of microbial whole-cell hydroxylations in applied biocatalysis. Curr. Opin. Biotechnol. 12:419-425. [DOI] [PubMed] [Google Scholar]
- 8.Fasan, R., M. M. Chen, N. C. Crook, and F. H. Arnold. 2007. Engineered alkane-hydroxylating cytochrome P450(BM3) exhibiting nativelike catalytic properties. Angew. Chem. Int. Ed. Engl. 46:8414-8418. [DOI] [PubMed] [Google Scholar]
- 9.Funhoff, E. G., U. Bauer, I. García-Rubio, B. Witholt, and J. B. van Beilen. 2006. CYP153A6, a soluble P450 oxygenase catalyzing terminal-alkane hydroxylation. J. Bacteriol. 188:5220-5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Funhoff, E. G., J. Salzmann, U. Bauer, B. Witholt, and J. B. van Beilen. 2007. Hydroxylation and epoxidation reactions catalyzed by CYP153 enzymes. Enzyme Microb. Technol. 40:806-812. [Google Scholar]
- 11.Glickman, B. W., and M. Radman. 1980. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc. Natl. Acad. Sci. USA 77:1063-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, E. L., and M. R. Hyman. 2006. Propane and n-butane oxidation by Pseudomonas putida GPo1. Appl. Environ. Microbiol. 72:950-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kok, M., R. Oldenhuis, M. P. G. van der Linden, P. Raatjes, J. Kingma, P. H. van Lelyveld, and B. Witholt. 1989. The Pseudomonas oleovorans alkane hydroxylase gene. Sequence and expression. J. Biol. Chem. 264:5435-5441. [PubMed] [Google Scholar]
- 14.Kubota, M., M. Nodate, M. Yasumoto-Hirose, T. Uchiyama, O. Kagami, Y. Shizuri, and N. Misawa. 2005. Isolation and functional analysis of cytochrome P450 CYP153A genes from various environments. Biosci. Biotechnol. Biochem. 69:2421-2430. [DOI] [PubMed] [Google Scholar]
- 15.Lageveen, R. G., G. W. Huisman, H. Preusting, P. E. F. Ketelaar, G. Eggink, and B. Witholt. 1988. Formation of polyester by Pseudomonas oleovorans: the effect of substrate on the formation and composition of poly-(R)-3-hydroxyalkanoates and poly-(R)-3-hydroxyalkenoates. Appl. Environ. Microbiol. 54:2924-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maier, T., H. H. Förster, O. Asperger, and U. Hahn. 2001. Molecular characterization of the 56-kDa CYP153 from Acinetobacter sp. EB104. Biochem. Biophys. Res. Commun. 286:652-658. [DOI] [PubMed] [Google Scholar]
- 17.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 18.Minak-Bernero, V., R. E. Bare, C. E. Haith, and M. J. Grossman. 2004. Detection of alkanes, alcohols, and aldehydes using bioluminescence. Biotechnol. Bioeng. 87:170-177. [DOI] [PubMed] [Google Scholar]
- 19.Müller, R., O. Asperger, and H. P. Kleber. 1989. Purification of cytochrome P-450 from n-hexadecane-grown Acinetobacter calcoaceticus. Biomed. Biochim. Acta 48:243-254. [PubMed] [Google Scholar]
- 20.Nieder, M., and J. Shapiro. 1975. Physiological function of the Pseudomonas putida PpG6 (Pseudomonas oleovorans) alkane hydoxylase: monoterminal oxidation of alkanes and fatty acids. J. Bacteriol. 122:93-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nodate, M., M. Kubota, and N. Misawa. 2006. Functional expression system for cytochrome P450 genes using the reductase domain of self-sufficient P450RhF from Rhodococcus sp. NCIMB 9784. Appl. Microbiol. Biotechnol. 71:455-462. [DOI] [PubMed] [Google Scholar]
- 22.Peters, M. W., P. Meinhold, A. Glieder, and F. H. Arnold. 2003. Regio- and enantioselective alkane hydroxylation with engineered cytochromes P450 BM-3. J. Am. Chem. Soc. 125:13442-13450. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Shanklin, J., and E. Whittle. 2003. Evidence linking the Pseudomonas oleovorans alkane omega-hydroxylase, an integral membrane diiron enzyme, and the fatty acid desaturase family. FEBS Lett. 545:188-192. [DOI] [PubMed] [Google Scholar]
- 25.Smits, T. H. M., M. A. Seeger, B. Witholt, and J. B. van Beilen. 2001. New alkane-responsive expression vectors for E. coli and Pseudomonas. Plasmid 46:16-24. [DOI] [PubMed] [Google Scholar]
- 26.Smits, T. H. M., S. B. Balada, B. Witholt, and J. B. van Beilen. 2002. Functional analysis of alkane hydroxylases from gram-negative and gram-positive bacteria. J. Bacteriol. 184:1733-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Söhngen, N. L. 1913. Benzin, Petroleum, Paraffinöl und Paraffin als Kohlenstoff- und Energiequelle für Mikroben. Zentralbl. Bacteriol. Parasitenk. 37:595-609. [Google Scholar]
- 28.Staijen, I. E., J. B. van Beilen, and B. Witholt. 2000. Expression, stability and performance of the three-component alkane mono-oxygenase of Pseudomonas oleovorans in Escherichia coli. Eur. J. Biochem. 267:1957-1965. [DOI] [PubMed] [Google Scholar]
- 29.van Beilen, J. B. 1994. Alkane oxidation by Pseudomonas oleovorans: genes and proteins. Ph.D. thesis. University of Groningen, Groningen, The Netherlands.
- 30.van Beilen, J. B., and E. G. Funhoff. 2007. Alkane hydroxylases involved in microbial alkane degradation. Appl. Microbiol. Biotechnol. 74:13-21. [DOI] [PubMed] [Google Scholar]
- 31.van Beilen, J. B., E. G. Funhoff, A. van Loon, A. Just, L. Kaysser, M. Bouza, R. Holtackers, M. Röthlisberger, Z. Li, and B. Witholt. 2006. Cytochrome P450 alkane hydroxylases of the CYP153 family are common in alkane-degrading eubacteria lacking integral membrane alkane hydroxylases. Appl. Environ. Microbiol. 72:59-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Beilen, J. B., J. Kingma, and B. Witholt. 1994. Substrate specificity of the alkane hydroxylase of Pseudomonas oleovorans GPo1. Enzyme Microb. Technol. 16:904-911. [Google Scholar]
- 33.van Beilen, J. B., D. Lüscher, R. Holtacker, U. Bauer, B. Witholt, and W. A. Duetz. 2005. Biocatalytic production of perillyl alcohol from limonene using a novel Mycobacterium cytochrome P450 alkane hydroxylase expressed in P. putida. Appl. Environ. Microbiol. 71:1737-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Beilen, J. B., D. Penninga, and B. Witholt. 1992. Topology of the membrane-bound alkane hydroxylase of Pseudomonas oleovorans. J. Biol. Chem. 267:9194-9201. [PubMed] [Google Scholar]
- 35.van Beilen, J. B., T. H. M. Smits, F. F. Roos, T. Brunner, S. B. Balada, M. Röthlisberger, and B. Witholt. 2005. Identification of an amino acid position that determines the substrate range of integral membrane alkane hydroxylases. J. Bacteriol. 187:85-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu, F., S. G. Bell, J. Lednik, A. Insley, Z. Rao, and L. Wong. 2005. The heme monooxygenases cytochrome P450cam can be engineered to oxidize ethane to ethanol. Angew. Chem. Int. Ed. 44:4029-4032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.