Abstract
Spliced-leader-associated RNA (SLA1) guides the pseudouridylation at position −12 (relative to the 5′ splice site) of the spliced-leader (SL) RNA in all trypanosomatid species. Nevertheless, the exact role of this RNA is currently unknown. Here, we demonstrate that the absence of pseudouridine on Leptomonas collosoma SL RNA has only a minor effect on the ability of this RNA to function in trans splicing in vivo. To investigate the possible role of SLA1 during SL RNA biogenesis, the structure of the SL RNA was examined in permeable Trypanosoma brucei cells depleted for CBF5, the H/ACA pseudouridine synthase, lacking SLA1. Our results suggest that in the absence of SLA1, the SL RNA secondary structure is changed, as was detected by differential sensitivity to oligonucleotide-directed RNase H cleavage, suggesting that the association of SLA1 maintains the SL RNA in a structural form which is distinct from the structure of the SL RNA in the steady state. In T. brucei cells depleted for the SL RNA core protein SmD1, SL RNA first accumulates in large amounts in the nucleus and then is expelled to the cytoplasm. Here, we demonstrate by in vivo aminomethyltrimethyl UV cross-linking studies that under SmD1 depletion, SLA1 remains bound to SL RNA and escorts the SL RNA to the cytoplasm. In situ hybridization with SLA1 and SL RNA demonstrates colocalization between SLA1 and the SL RNA transcription factor tSNAP42, as well as with Sm proteins, suggesting that SLA1 associates with SL RNA early in its biogenesis. These results demonstrate that SLA1 is a unique chaperonic RNA that functions during the early biogenesis of SL RNA to maintain a structure that is most probably suitable for cap 4 modification.
H/ACA RNA is a group of small nucleolar RNAs that direct pseudouridylation on rRNA and snRNAs. In almost all eukaryotes, these RNAs consist of two stem-loop structures connected by a single-stranded hinge and a tail region carrying the conserved H (AnAnnA) and ACA boxes. Two short motifs of the snoRNA base pair with the target site flanking the uridine to be isomerized (9, 25). In trypanosomes, these guide RNAs have a unique structure, composed of a single stem-loop, and carry an AGA instead of an ACA box (15, 16). All of the H/ACA molecules identified so far in trypanosomes can guide rRNA pseudouridylation; however, molecules that guide analogous modifications on U snRNAs (U1 to U5) have not been identified to date. In higher eukaryotes, the guide RNAs that direct such modifications are localized in special subnuclear domains, the Cajal bodies (6). In trypanosomes, all mRNAs are processed by trans splicing. During the trans-splicing reaction, a common spliced-leader (SL) sequence of 39 nucleotides (nt) is added to the pre-mRNA from a small RNA, the SL RNA. A novel snoRNA, termed SLA1 (SL-associated RNA), in Trypanosoma brucei was initially discovered by virtue of its efficient cross-linking with the bifunctional reagent aminomethyltrimethyl (AMT) to the SL RNA (39). It was originally proposed that SLA1 is the trypanosome U5 snRNA; however, phylogenetic analysis did not support this hypothesis (27). After identification of the true U5 snRNA (4, 8, 42), the actual function of SLA1 remained unresolved.
The function of SLA1 as an H/ACA RNA that guides pseudouridylation at position −12 of the SL RNA was proposed based on a series of findings (17). All trypanosomatid SL RNAs carry a pseudouridine (ψ) at position −12 (relative to the 5′ splice site). In addition, a potential base pairing interaction between SLA1 and SL RNA is in agreement with the canonical rules for guiding pseudouridylation by H/ACA RNAs. Third, mutation in the proposed SL RNA interaction domain abolishes ψ formation (17). The final proof that SLA1 is indeed a bona fide H/ACA was obtained by silencing the pseudouridine synthase CBF5 by RNA interference and demonstrating that, in these silenced cells, the SLA1 is destabilized similarly to all other H/ACA RNAs that guide pseudouridylation on rRNA (3). CBF5 silencing induces trans-splicing defects at the first step of splicing. Interestingly, the modification of cap 4 was perturbed in the CBF5-silenced cells (3), suggesting that SLA1 may function as a chaperone to maintain the proper secondary structure of SL RNA, essential for recognition by the methyltransferases (1, 22, 28, 44) that mediate RNA cap 4 synthesis.
SL RNA is transcribed by RNA polymerase II in a very special nuclear compartment near the nucleolus described for Trypanosoma cruzi (7). Recently, the Leishmania tarentolae SL RNA genes and SL RNA were also shown to reside in a similar specialized domain (13). In T. brucei, the existence of two RNA polymerase II macrofoci colocalizing with SL RNA transcription domains was described. It was suggested that the two foci may represent the two alleles carrying the tandem arrays of the SL RNA gene clusters (37). We recently demonstrated that assembly of SL RNA with Sm proteins takes place in the nuclear compartment, where SL RNA transcription take place, and proposed that SL RNA transcription, modifications, and assembly take place within this subnuclear domain, the SL RNP factory (31). Indeed, when Sm assembly is blocked by RNA interference of Sm proteins, the SL RNA first accumulates in this compartment, but it is then exported to the cytoplasm (45) and accumulates in huge cytoplasmic “speckles” (5, 19). The SL RNA that accumulates in the cell under Sm depletion is found in a small RNP, which we termed SL RNP-C. The SL RNA in these particles lacks the cap 4 +4 modification and has a longer 3′ tail (19, 45). This SL RNA is a dead-end product that does not return to the nucleus when Sm protein synthesis resumes (5, 19). SL RNP-C thus represents a novel disposal mechanism to remove defective SL RNA from the nucleus without degrading it.
In this study, we investigated the role of SLA1 and the ψ it guides in the process of trans splicing. We provide evidence that SL RNA lacking ψ has only a minor effect on trans splicing in vivo. SLA1 does not interact with SL RNA during the splicing reaction but early in SL RNA biogenesis. Oligonucleotide-directed RNase H cleavage mapping of nascently transcribed SL RNA synthesized in a permeable-cell system suggests that the interaction of SLA1 with SL RNA maintains SL RNA in a distinct conformation. In vivo AMT UV cross-linking and in situ hybridization in T. brucei cells silenced for SmD1 show that SLA1 remains associated with the SL RNA under Sm depletion and migrates with it to the cytoplasm, suggesting that under normal conditions, SLA1 interacts with the SL RNA in its complex biogenesis pathway until it assembles with Sm proteins. SLA1 normally resides in the nucleus in two places: (i) the SL RNP factory, where SL RNA is transcribed and assembled with Sm proteins, as was visualized by colocalization of SLA1 with SL RNA, Sm proteins, and the SL RNA transcription factor tSNAP42; and (ii) in a special site situated near the nucleolus and distinct from the site where snoRNAs involved in rRNA processing are localized. This study provides the first example of an H/ACA-like RNA that functions as a chaperone in addition to the modification that it guides.
MATERIALS AND METHODS
Oligonucleotides for generation of mutated SL RNA.
The oligonucleotides used for generation of mutated SL RNA are as follows: 34378, 5′-CCCCGTATCATCGATAGACCCCCGC-3′, sense, complementary to positions −63 to −38 upstream of the Leptomonas collosoma SL RNA +1 gene, used to clone the PCR product carrying SL RNA mutations; 19487, 5′-CGGGATCCGGACCCCCCAAAGGCCCCCCA-3′, antisense, complementary to positions +127 to +147 downstream of the L. collosoma SL RNA +1 gene, carrying a BamHI site (underlined), used to clone the PCR product carrying SL RNA mutations; 16354, 5′-AATGAAGTATTGAAACTGTTCT-3′, antisense, complementary to L. collosoma SL RNA from positions +19 to +40 downstream of the SL RNA +1 gene, bearing G31A and U30A mutations (underlined), used to clone the PCR product carrying SL RNA mutations; 16356, 5′-AATGAAGTATGGAAACTGTTCT-3′, antisense, specific to L. collosoma SL RNA from positions +19 to +40 downstream of the SL RNA +1 gene, bearing G31A and U30C mutations (underlined), used to clone the PCR product carrying SL RNA mutations; and 16355, 5′-AATGAAGTATCGAAACTGTTCT-3′, antisense, complementary to L. collosoma SL RNA from positions +19 to +40 downstream of the SL RNA +1 gene, bearing G31A and U30G mutations (underlined), used to clone the PCR product carrying SL RNA mutations.
Oligonucleotides for splint labeling.
The oligonucleotides used for splint labeling are as follows: 6211, GTTTTTTTTTTGAGTCTCGCTCTCCAGTTTC, antisense, complementary to the T. brucei SLA1 3′ end (positions 52 to 71), containing a poly(T) tail (underlined), used for splint labeling of SLA1 snoRNA; 84752, GTTTTTTTTTGGAGGTCATCCGACCCC, antisense, complementary to the T. brucei SL RNA 3′ end (positions 120 to 137), bearing a poly(T) tail (underlined), used for splint labeling of SL RNA; 13290, GTTTTTTTTTTAAAGAGTGGAGGTCATCCGAC, antisense, complementary to the T. brucei SL RNA 3′ end plus 6 nt (positions 123 to 143), containing a poly(T) tail (underlined), used for splint labeling of the extended SL RNA in SmD1-silenced cells; and 55448, GTTTTTGACACCCCAATGTTTAAACGCTAA, antisense, complementary to the T. brucei U5 3′ end (positions 39 to 62), containing a poly(T) tail (underlined), used for splint labeling of U5.
Oligonucleotides for in vitro RNase H cleavage of splint-labeled, cross-linked species.
The oligonucleotides used for in vitro RNase H cleavage of splint-labeled, cross-linked species are as follows: 3426, 5′-GTACCTGCTACTTT-3′, antisense, complementary to T. brucei SLA1, from positions 30 to 43; 9091, 5′-TGCGTCTGTTGGCCC-3′, antisense, complementary to the SL RNA, from positions 65 to 79; and 3427, 5′-GACACCCCAAAGTTT-3′, antisense, complementary to T. brucei U5 snRNA, from positions 48 to 62.
Oligonucleotides for primer extension.
The oligonucleotides used for primer extension are as follows: 9825, 5′-GCCCGAAAGCTCGGTC-3′, antisense, complementary to positions 80 to 95 of the L. collosoma SL RNA; 36815, 5′-GAGGGAGGAATGAGGTGAGC-3′, antisense, complementary to neo/hygro mRNA, positions 4996 to 5016 on the pX vector; 1097, 5′-GAACCCCCGCTTGTC-3′, antisense, complementary to T. brucei 7SL RNA at positions 61 to 75; 36591, 5′-CAGGGCATAAAATAGTAC-3′, antisense, complementary to TB11Cs2C2 at positions 73 to 90; and 12407, 5′-AGCTATATCTCTCGAA-3′, antisense, complementary to T. brucei U6 snRNA from positions 80 to 95.
Oligonucleotides for synthesis of T7 PCR products for in vitro transcription.
The oligonucleotides used for synthesis of T7 PCR products for in vitro transcription are as follows: 21635, 5′-AAAGCTCTTTTATGTAGTGTGCG-3′, sense, specific to T. brucei SLA1 snoRNA 5′ from positions 1 to 23; 8657, 5′-TTAATACGACTCACTATAGGGAGAGAGTCTCGCTCTCCAGTTTC-3′, antisense, specific to T. brucei SLA1 (positions 52 to 71), carrying the T7 promoter sequence (underlined); 8977, sense, 5′-AACTAACGCTATTATTAGAACAGTTTCTGTACTAT-3′, specific to T. brucei SL RNA 5′ from positions 1 to 35; 0244C02, 5′-TTAATACGACTCACTATAGGGAGAAAAAAAATAAAAAAAATA-3′, antisense, complementary to positions 143 to 161 with respect to the +1 position of the SL RNA transcript bearing the T7 promoter (underlined); 81971, 5′-AAGACCGTACTCTGAACAGAATCG-3′, sense, specific to T. brucei U3 snoRNA from positions 1 to 24, used for synthesis of the U3 PCR product for in vitro transcription; and 81970, 5′-TTAATACGACTCACTATAGGGAGAATCCTTCTGGAACCGGCTC-3′, antisense, specific to T. brucei U3 snoRNA from positions 120 to 138, carrying the T7 promoter sequence (underlined), used for synthesis of U3 PCR product for in vitro transcription.
Oligonucleotides for in vivo RNase H cleavage in permeable cells.
The oligonucleotides used for in vivo RNase H cleavage in permeable cells are as follows: 192787-oligo A, 5′-AGCGTTAGTT-3′, antisense, complementary to T. brucei SL RNA from positions 1 to 10; 191055-oligo Z, 5′-CTAATAATAGCG-3′, antisense, complementary to T. brucei SL RNA from positions 7 to 18; 192053-oligo H, 5′-ACAGAAACTG-3′, antisense, complementary to T. brucei SL RNA from positions 21 to 30; and 192788-oligo C, 5′-GCTTCTCATAC-3′, antisense, complementary to T. brucei SL RNA from positions 41 to 50.
Generation and preparation of YFP-tSNAP42 parasites.
To generate the YFP-tSNAP42 protein (where YFP is yellow fluorescent protein), a PCR fragment was amplified using the oligonucleotides specified above covering position 1 (first ATG) and position 830. The vector p2828 YFP-tSNAP42 was digested with HindIII and NotI, and the fragment coding for tSNAP42 was cloned using the same sites (14). This manipulation created an N-terminal fusion, and the HindIII site was used for fusing the gene and YFP. For integration, the plasmid was linearized with BamHI at position 626 from the ATG codon. Clonal transgenic cell lines were selected on blasticidin.
Growth and RNA preparation.
L. collosoma and T. brucei cells were grown as described previously (19, 20). Cells were harvested at log phase, and total RNA was prepared with TRIzol reagent (Sigma) according to the manufacturer's protocol.
Plasmid construction and transformation.
Mutations in SL RNA were generated by site-directed mutagenesis, as previously described (41). The mutated SL RNA gene was further subcloned into the pX-neo expression vector. Cell lines were selected on 25 μg/ml G418 and further selected at higher G418 concentrations to obtain cells that strongly express the tagged SL RNA gene (41). For cloning into the expression vector, the PCR products carrying the mutated SL RNA genes were digested with XhoI (5′ end) and BamHI (3′ end) and cloned into a pX vector. All mutations were confirmed by DNA sequencing.
Northern and primer extension analyses.
RNA samples (5 μg) were fractionated on a 10% polyacrylamide, 7 M urea denaturing gel and electroblotted onto a nylon membrane (Hybond; Amersham Biosciences). Hybridization with labeled oligonucleotides was performed at 37°C with 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS), 5× Denhardt's solution, and 100 μg/ml salmon sperm DNA. Primer extension was performed using end-labeled oligonucleotides (1 × 105 cpm/pmol). After an annealing step at 60°C for 15 min, the sample was kept on ice for 1 min. Next, 1 U of reverse transcriptase (Expand RT; Roche Molecular Biochemicals) and 1 U of RNase inhibitor (Promega) were added, and extension was performed at 42°C for 90 min. The reaction product was analyzed on a 6% polyacrylamide denaturing gel next to products from DNA sequencing reactions performed with the same primer.
Poisoning primer extension.
The poisoning primer extension used for examining the utilization of mutated SL RNA in trans splicing was performed as described for the primer extension assay, but dideoxy CTP was used instead of dCTP. The reaction product was analyzed on a 6% polyacrylamide denaturing gel, as previously described (41).
Splint labeling of small RNAs and identification of cross-linking bands by RNase H cleavage.
For labeling, 10 to 20 μg of total RNA was mixed with 30 to 100 pmol of oligonucleotides and heated for 2 min at 85°C in 50 mM Tris-HCl (pH 7.8), 10 mM MgCl2, and 1 mM dithiothreitol. The annealing reaction mixture was quenched on ice for 30 min, 50 μCi of [α-32P]dATP (3,000 Ci/mmol) and 5 U of T7 DNA polymerase (Sequenase version 2.0; Amersham Biosciences) were added, and the reaction mixture was then incubated for 1 h at 37°C (12, 18). RNA was separated on a 6% polyacrylamide, 7 M urea gel. For identification of cross-linked species, the splint-labeled bands were excised from the gel and eluted for 2 h in 10 mM Tris-HCl (pH 8); after phenol extraction and ethanol precipitation, they were subjected to RNase H cleavage with antisense SLA1-, SL RNA-, and U5 snRNA-specific oligonucleotides. The reaction products were separated on 6% polyacrylamide, 7 M urea gels.
UV cross-linking of living cells treated with AMT.
T. brucei cells were harvested at 1 × 107 cells/ml and washed with phosphate-buffered saline (PBS). Cells (5 × 109) were concentrated and incubated on ice. AMT (Sigma) was added to the cells at a concentration of 0.2 mg/ml (18, 39). Cells treated with AMT were kept on ice and irradiated using a UV lamp at 365 nm with an intensity of 10 mW/cm2 for 30 min. The cells were deproteinized with proteinase K (2 mg/ml) in 1% SDS. RNA was prepared from the cells with TRIzol reagent and subjected to the splint labeling technique.
Cellular fractionation of UV-cross-linked cells.
UV-cross-linked cells were subjected to cellular fractionation as previously described (19). The nuclear and cytoplasmic fractions were deproteinized with proteinase K (2 mg/ml) in 1% SDS and then extracted with phenol and precipitated with ethanol. This RNA was labeled by splint labeling.
Oligonucleotide-directed RNase H cleavage in permeable T. brucei cells.
Permeabilization of procyclic trypanosome cells was performed by a procedure similar to that described by Tschudi and Ullu (34, 35). The only deviation from the published protocol is that 1× transcription buffer was used (150 mM sucrose, 20 mM potassium glutamate, 10 mM HEPES-KOH [pH 7.9], 2.5 mM MgCl2, 1 mM dithiothreitol, 10 μg/ml leupeptin). DNA oligonucleotides specific to various regions of the T. brucei SL RNA (see the oligonucleotide list) were added to permeable cells at a final concentration of 50 pg/ml in the presence of 10 U/ml of RNase H (New England BioLabs). RNA was prepared from the cells with TRIzol reagent and was separated on a 6% polyacrylamide, 7 M urea denaturing gel.
In vitro transcription of biotinylated and DIG-labeled antisense probes for in situ hybridization.
The PCR products from the coding regions of U3, SLA1 (TB11Cs2H1), and SL RNA were synthesized using antisense oligonucleotides containing T7 promoters at the 3′ end. These PCR products were used as a template for an in vitro transcription reaction carried out using a Megascript T7 kit (Ambion). The reaction was performed according to the manufacturer's instructions. For labeling the RNA probe with biotin, biotin-16-UTP (Roche) was added to the nucleotide mixture at a final concentration of 200 μM (at a ratio of 1:1.5 of modified to unmodified nucleotides). For labeling the RNA probe with digoxigenin (DIG), DIG RNA labeling mix (Roche), which contains DIG-11-UTP (at a final concentration of 167 μM, a ratio of 1:2 of modified to unmodified nucleotides), was used according to the manufacturer's instructions. The labeled RNA probes were extracted with phenol and were ethanol precipitated.
In situ hybridization.
Cells were washed with PBS, mounted on poly-l-lysine-coated slides, and fixed with 8% formaldehyde in PBS at room temperature for 20 min. Then, cells were treated with 1% Triton X-100 for exactly 5 min. After being washed with PBS, cells were prehybridized with hybridization buffer (60% deionized formamide, 50 mM sodium phosphate, pH 7.2, 0.5 mg/ml salmon sperm DNA, 1 μg/ml tRNA, and 5× Denhardt's solution in 2× SSC) for 4 h at 72°C (humid chamber). The selected RNA probes were diluted to 40 ng/μl in 50% formamide, denatured for 5 min at 85°C, and immediately chilled on ice. Next, probes were added to the slides to a final concentration of 4 ng/μl in hybridization buffer (as specified above). The slides were covered, sealed, heated for 5 min at 80°C, and transferred immediately to 55°C for overnight incubation. Slides were opened, washed with 2× SSC for 5 min at room temperature, washed with 2× SSC for 30 min at 60°C, and finally washed with 2× SSC for 5 min at room temperature. Slides were blocked with 0.25% blocking solution (Roche) for 30 min at room temperature. Cells were then incubated for 45 min with 1:500-diluted Alexa Fluor 647 streptavidin conjugate (Molecular Probes) for the detection of biotinylated probes or with 1:200-diluted fluorescein isothiocyanate-conjugated goat anti-DIG (Roche) for the detection of DIG-labeled probes. When both probes were used in the same experiment, cells were simultaneously incubated with both detection reagents. Slides were washed with 2× SSC, and the cells were incubated with 1:2,000-diluted 4′,6′-diamidino-2-phenylindole (DAPI) for 5 min (to stain the nucleus and kinetoplast). The cells were visualized under a Zeiss LSM 510 META inverted microscope.
Immunofluorescence assay combined with in situ hybridization.
In situ hybridization was performed as described above. After a final wash with 2× SSC, cells were washed with PBS and were then incubated with PBS containing 10% fetal calf serum at room temperature for 30 min. Then, cells were incubated with 1:400-diluted primary anti-Nhp2p antibodies (30) for 1 h, washed with PBS, and then reacted with anti-immunoglobulin G conjugated to fluorescein isothiocyanate (Jackson ImmunoResearch). To stain the nucleus and kinetoplast, the cells were incubated with DAPI for 5 min. The cells were visualized under a Zeiss LSM 510 META inverted microscope.
RESULTS
Mutations that abolish the pseudouridylation of L. collosoma SL RNA have only a minor effect on utilization of this RNA in trans splicing.
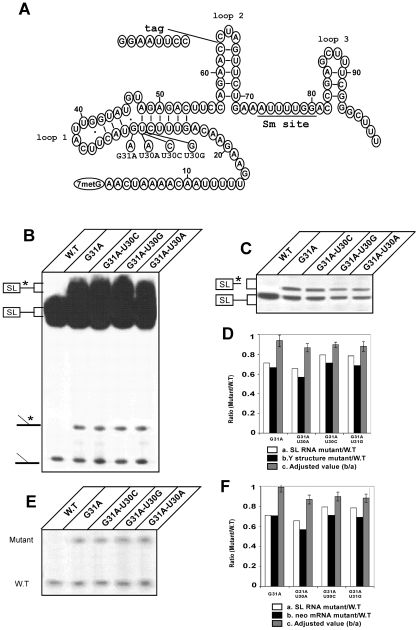
To directly address the role of ψ in trans splicing, we generated mutations to change the U30 that undergoes pseudouridylation (at position −12 relative to the 5′ splice site) to an A, C, or G. To this end, we utilized the tagged SL RNA molecule expressed in the monogenetic trypanosomatid L. collosoma (10, 20, 41). The mutations in the SL RNA were generated in the context of our model SL RNA gene, tagged at both the intron (an insertion of 8 nt in loop II) and the exon (G31A, a G-to-A substitution at position 31), and are indicated schematically in Fig. 1A. The mutated SL RNA genes were expressed in the episomal pX-neo expression vector, and cell lines were selected under high concentrations of G418. We first compared the levels of expression of the different mutations at position U30 with those of the tagged wild-type (pseudouridylated) molecules. Northern analysis using an oligonucleotide complementary to the SL RNA intron (Fig. 1C) demonstrated that all of the mutated SL RNAs were properly expressed, albeit with minor differences. These minor differences were taken into account when calculating the utilization of these mutated SL RNAs in trans splicing (Fig. 1B and E). Primer extension was used to monitor the levels of SL RNA and cap 4 formation. The multiple bands represent stops at the modified cap 4 structure (Fig. 1B), and no difference in the pattern of the stops appeared under shorter exposure (data not shown). To calculate the trans-splicing efficiency of the mutated SL RNAs, the level of the Y structure was compared to the level in the parental G31A mutation by densitometry. In all of the mutations, the tagged Y structure was formed, albeit with slight differences in efficiency. To normalize the utilization of the above-mentioned mutations in trans splicing, we first determined for each cell line the ratio between the levels of expression of the mutated SL RNA and the wild-type SL RNA by using the data from the Northern analysis shown in Fig. 1C. We next calculated the ratio between the Y-structure intermediates derived from the mutated and wild-type SL RNAs (Fig. 1D) and corrected this value by the expression coefficient of each of the SL RNA mutations. The corresponding values are indicated as adjusted values (Fig. 1Dc). The results suggest that the utilization of the SL RNA mutations in trans splicing is affected only slightly relative to that of the control, suggesting only a minor effect of these mutations on their utilization in the first step of the reaction.
FIG. 1.
Mutated SL RNAs of L. collosoma that cannot undergo pseudouridylation are affected only slightly as trans-splicing substrates. (A) Sequences and secondary structures of L. collosoma SL RNA and its mutated derivatives. The loops, Sm site, intron tag, G31A substitution, and U30 substitutions are indicated. (B) Effects of U30 mutations on the first step of trans splicing. Total RNA from the indicated cell lines was primer extended with SL RNA oligonucleotide 9825. The cDNA products were separated on a denaturing 6% polyacrylamide gel. The 5′ end of the SL RNA is indicated. The positions of the cDNAs derived from full-length mutated SL RNA and its corresponding Y structure are marked with asterisks. (C) Northern analysis of SL RNAs bearing U30 mutations. Total RNA was separated on a 10% polyacrylamide denaturing gel and electroblotted onto a nylon membrane. The identities of the mutations are indicated at the top. The blot was hybridized with oligonucleotide 9825, which recognizes both the wild-type (W.T) and the mutated SL RNAs. (D) Quantitative analysis of utilization of U30 SL RNA mutations in the first step of splicing. SL RNA levels (mutated and wild type) were estimated by densitometry. (a) Ratio between the levels of mutated and wild-type SL RNAs, calculated from the data presented in Fig. 1B and C. (b) Ratio between the levels of the mutated and wild-type Y structures, calculated from the data presented in Fig. 1B. (c) Adjusted values accounting for differential expression, i.e., the Y-structure ratio divided by the SL RNA ratio. Error bars indicate standard deviations of the adjusted values (n = 5; P < 0.01) for all cases. (E) Effects of U30 mutations on the second step of trans splicing. Total RNA from the indicated cell lines was reverse transcribed with an end-labeled primer (36815), as described previously (41) and in Materials and Methods. The primer extension products were separated on a denaturing 6% polyacrylamide gel. The diagnostic cDNAs derived from the presence of mutated or wild-type SL RNA exons are indicated. (F) Quantitative analysis of utilization of U30 SL RNA mutations in the second step of trans splicing. The level of mature neo mRNA was estimated using densitometry. (a) Ratio between the levels of mutated and wild-type SL RNAs, calculated from the Northern analysis data presented in Fig. 1C. (b) Ratio between the levels of mutated and wild-type SL RNA stops, calculated from the data presented in Fig. 1E. (c) Adjusted values obtained by dividing the SL RNA stop ratio by the SL RNA ratio. Error bars indicate standard deviations of the adjusted values (n = 5; P < 0.01) for all cases.
We next examined whether SL RNA mutations affect SL RNA utilization in the second step of the reaction (Fig. 1E). As previously shown (10, 20, 41), in the presence of dideoxy CTP, two primer extension products are produced, (i) a 30-nt-long cDNA corresponding to termination at G31 and diagnostic of the wild-type SL RNA exon and (ii) a 36-nt-long cDNA corresponding to termination at G25 of the SL RNA exon and diagnostic of the DHFR-neo mRNA (encoded by the pX vector) derived from trans splicing of mutated SL RNA. The results, shown in Fig. 1E, suggest only minor changes in the efficiency of utilization of the U30 mutations in trans splicing. Quantitation of the data (Fig. 1F) shows that all of the mutations in U30 slightly affect trans splicing, as overall splicing levels were reduced by 10 to 20%. Such trans-splicing defects seem to be minor compared to the reduction of 50 to 70% observed when the same system was used to examine the role of the individual cap 4 nucleotides for trans splicing (20), suggesting that the pseudouridylation of the SL RNA is not critical for the function of SL RNA in trans splicing. Indeed, a similar mutation introduced in a domain covering the −12 position of L. tarentolae, which undergoes pseudouridylation, had only a minor effect on trans splicing (29).
SLA1 remains associated with SL RNA in cells depleted for SmD1.
After finding that ψ itself has a relatively minor effect on the function of SL RNA in trans splicing, we sought to explore the interaction of SLA1 with SL RNA. Our working hypothesis was that the main function of SLA1 might be to act as an SL RNA chaperone; SLA1 may interact with the SL RNA early in its biogenesis and fold the SL RNA in a conformation most suitable for interactions with the capping machinery and less favorable for recruitment into the spliceosome. To investigate this intriguing possibility, we utilized a T. brucei cell line that is blocked in SL RNA biogenesis due to depletion of an Sm core protein (5, 19, 31). We noticed that in Sm-depleted cells the SL RNA still undergoes pseudouridylation, suggesting that this modification takes place before Sm assembly (19). However, we further observed that only a fraction of the SL RNA that accumulates in these Sm-depleted cells is pseudouridylated (19). In fact, the level of the pseudouridylated SL RNA is lower than the level of SL RNA, suggesting that not all of the SL RNA under SmD1 depletion undergoes pseudouridylation. As mentioned above, during Sm depletion, the SL RNA first accumulates in the SL RNP factory and then migrates to the cytoplasm to form cytoplasmic “speckles” (5). It was interesting to determine whether SLA1 remains associated with SL RNA in SmD1-depleted cells. We therefore analyzed the association of SLA1 with SL RNA in cells depleted of SmD1 and compared it to the association in uninduced cells by using in vivo psoralen UV cross-linking. To detect SLA1, SL RNA, and their cross-linked products, we used the splint labeling approach with T. brucei (12, 18). In this technique, a primer complementary to the RNA carrying runs of T nucleotides at the 3′ end of the oligonucleotide is used as a template to incorporate [α-32P]ATP at the 3′ end of the RNA, using Sequenase (12). When RNA was labeled with an SLA1 splint labeling probe (Fig. 2A, lanes 1 to 4), two types of labeled RNAs were observed: the ∼83-nt RNA, which is the authentic SLA1 molecule, and higher-molecular-weight RNA bands, which appeared only upon UV cross-linking (Fig. 2A, lanes 2 and 4). In the SmD1-silenced cells, we observed labeling of the authentic SLA1 (whose level did not change in SmD1-depleted cells) and two high-molecular-weight, cross-linked species (Fig. 2A, lane 4). Both of these high-molecular-weight species appeared when the RNA was labeled with an SL RNA-specific probe (Fig. 2A, lanes 6 and 8), suggesting that these bands represent species resulting from cross-linking between SL RNA and SLA1. The uppermost band appeared mainly under SmD1 depletion and may represent multimeric cross-linked species that are favored only when the level of SL RNA is high. Note that the major SLA1-SL RNA cross-linked species changes in size and intensity during SmD1 silencing (Fig. 2A, lanes 4 and 8). The size of the cross-linked SLA1-SL RNA species present in the RNA from the SmD1-silenced cells was larger than that of the RNA extracted from the uninduced cells. Indeed, we have previously demonstrated by RNase protection assay that the SL RNA that accumulates in SmD1-silenced cells has a longer (by 5 nt) 3′ tail (19). To verify that these cross-linked species are indeed SLA1-SL RNA hybrids, the two cross-linked species were excised from the gel and subjected to oligonucleotide-directed RNase H cleavage with oligonucleotides specific to SL RNA (Fig. 2C) or SLA1 (Fig. 2D). The results suggest that only oligonucleotides specific to either SLA1 or SL RNA specifically cleaved the SLA1-SL RNA cross-linked species. Previously, SL RNA was also shown to interact with U5 snRNA (39). Indeed, such a cross-linked species is observed in Fig. 2A, lane 6, and Fig. 2C, lane 3, and comigrates with the cross-linked species that appears when the U5-specific oligonucleotide was used for splint labeling (Fig. 2A, lane 10). Note that no U5-SL RNA species was observed under SmD1 depletion (Fig. 2A, lane 8, and C, lane 7), because, in these cells, the level of U5 snRNA is markedly reduced and trans splicing is severely inhibited (19). These results demonstrate that SLA1 and U5 snRNA associate with SL RNA at different stages, either before (SLA1) or during (U5) splicing. SLA1 associates with SL RNA even when trans splicing is inhibited and before Sm assembly, suggesting that SLA1 functions early in SL RNA biogenesis, whereas U5 snRNA functions, as expected, during the trans-splicing reaction. The results also imply that SLA1 does not detach from the SL RNA if Sm assembly is blocked. In the Sm-depleted cells, SL RNA undergoes pseudouridylation until all of the SLA1 is bound by SL RNA, since SLA1 does not detach from SL RNA, and since the SLA1 level does not increase as SL RNA increases during Sm silencing, the majority of the SL RNA does not bind to SLA1 and does not undergo pseudouridylation. This explains our previous observation that the ratio between the stops at position −12 after CMC [N-cyclohexyl-N′-β-(4-methylmorpholinium)ethylcarbodimide p-tosylate] treatment and the extension at the 5′ end of the SL RNA (reflecting the amount of SL RNA) is reduced under Sm depletion (19).
FIG. 2.
Identification and characterization of SLA1 cross-linked species in T. brucei. (A) Identification of SLA1, SL RNA, and U5 cross-linked species. SmD1 cells were grown with or without tetracycline (Tet + or −, respectively). AMT was added to cells 40 h after induction. Cells were either UV cross-linked for 30 min (365 nm) (UV +) or left untreated (UV −). RNA was extracted and subjected to splint labeling using oligonucleotide 6211, specific to SLA1 (lanes 1 to 4); oligonucleotide 84752, specific to SL RNA (lanes 5 and 6); oligonucleotide 13290, specific to SL RNA from SmD1 cells (lanes 7 and 8); or oligonucleotide 55448, specific to U5 snRNA (lanes 9 and 10). The reaction products were separated on a denaturing 6% polyacrylamide gel alongside a labeled pBR322/HpaII digest marker. The identities of the splint-labeled RNAs are indicated on the right. Nonspecific labeling is marked by an asterisk. (B) Cellular fractionation to determine the localization of SLA1/SL RNA cross-linked species. UV cross-linking was performed with SmD1 cells as described for panel A. Nuclear (Nuc, N) and cytoplasmic (Cyt, C) fractions were prepared from the cells as described in Materials and Methods, and RNA was subjected to splint labeling using oligonucleotide 6211, specific to the SLA1 3′ end (lanes 1 to 4); oligonucleotide 84752, specific to the SL RNA 3′ end (lanes 5 and 6); or oligonucleotide 13290, specific to the SL RNA extended 3′ end (lanes 7 and 8). The same RNA was subjected to primer extension using oligonucleotides complementary to 7SL RNA (1097), Tb11Cs2C2 (36591), and U6 snRNA (12407). The reaction products were separated on a denaturing 6% polyacrylamide gel alongside a labeled pBR322 marker. The values at left indicate the RNA sizes (in nucleotides). (C) Identification of SL RNA cross-linked species using oligonucleotide-directed RNase H cleavage. Splint-labeled SL RNA cross-linked species and splint-labeled SL RNA (as shown in panel A) from uninduced cells (lanes 1 to 6) and SmD1 induced cells (lanes 7 to 9) were excised from the gel and subjected to RNase H cleavage with an antisense oligonucleotide (Oligo) specific to SL RNA (lanes 2, 4, and 8), SLA1 (lanes 5 and 9), or U5 snRNAs (lane 6) or with no oligonucleotide as a control (lanes 1, 3, and 7). Reaction products were separated on a denaturing 6% polyacrylamide gel. The cross-linked species and their cleavage derivatives are marked with dashed lines, and their identities are indicated. (D) Identification of SLA1 cross-linked species using oligonucleotide-directed RNase H cleavage. The experiment was the same as that described for panel C, but the RNA was splint labeled with an SLA1-specific probe and the cross-linked species was cleaved with an antisense oligonucleotide specific to SLA1 (lanes 2 and 5) or SL RNA (lane 4) or with no oligonucleotide as a control (lanes 1 and 3).
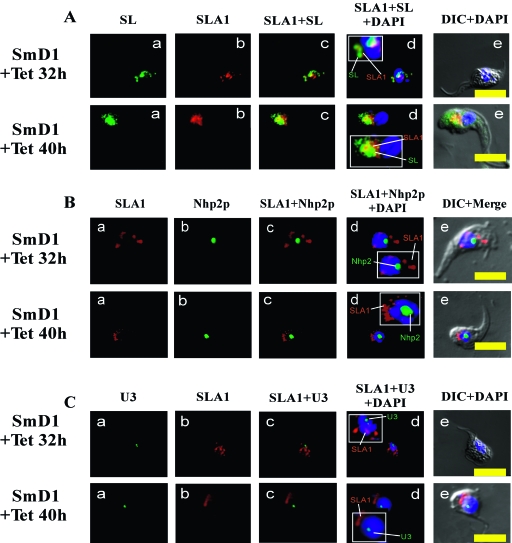
We have previously demonstrated that, in Sm-silenced cells, the SL RNA first accumulates in the nucleus and then migrates to the cytoplasm to form cytoplasmic “speckles.” It was therefore of great interest to examine whether SLA1 remains associated with SL RNA under these conditions and migrates with it. Such a finding would suggest that SLA1 escorts the SL RNA until it is assembled with Sm proteins. To investigate this possibility, we prepared nuclear and cytoplasmic fractions from cells that were treated with AMT and UV cross-linked. The RNA was labeled by splint labeling as described above. The results presented in Fig. 2B (lanes 7 and 8) show that, under SmD1 depletion (2 days after induction of silencing), SL RNA was found in the nucleus but was present to a greater degree in the cytoplasm. The high-molecular-weight, SLA1/SL RNA cross-linked species were detected mostly in the cytoplasm following SmD1 silencing (Fig. 2B, lane 8), suggesting that the SL RNA that migrates to the cytoplasm carries SLA1. This observation further indicates that SLA1 remains associated with SL RNA when Sm assembly is blocked. Indeed, this was also observed when SLA1 was splint labeled (Fig. 2B, lane 4). In comparing lanes 3 and 4 of Fig. 2B, it is clear that under Sm depletion, more of the SLA1 is localized in the cytoplasm than in the nucleus. Densitometric analysis shows that 72.3% ± 6.7% (n = 4; P < 0.01) of total SLA1 is located in the cytoplasm in Sm-depleted cells, whereas in uninduced cells 83.2% ± 3.6% (n = 5; P < 0.01) of SLA1 molecules are localized in the nucleus. The quality of the fractionation was verified by examining the levels of 7SL RNA (a cytoplasmic RNA), TB11Cs2C2 (C/D snoRNA, which is localized to the nucleolus), and U6 snRNA (which localizes to the nucleus). Note that SL RNA was shown to easily leak from the nuclear to the cytoplasmic compartment, as reported by us and others (19, 39). In SmD1-depleted cells, U6 snRNA was localized in the nucleus and did not leak to the cytoplasm (5, 46). The leakage of SL RNA might reflect our recent finding that the majority of the SL RNA is found in a special nuclear compartment, which might be more fragile than other parts of the nucleus. The data presented in Fig. 2B therefore provide clear evidence that SLA1 remains associated with SL RNA when Sm assembly is blocked. Note that the ratios between the nuclear and cytoplasmic SL RNAs vary over the course of the silencing process, as we noticed previously (19). After 32 h of silencing, SL RNA is still found in the nucleus, but at later stages, the majority of the SL RNA is expelled to the cytoplasm (see Fig. 4).
FIG. 4.
Localization of SLA1 in SmD1-silenced cells. After 32 or 40 h of tetracycline induction (+Tet), SmD1 cells were fixed with 8% (vol/vol) formaldehyde and hybridized with labeled RNA probes. (A) Localization of SLA1 relative to the SL RNA under SmD1 silencing. (a) In situ hybridization with DIG-labeled antisense SL RNA probe (green); (b) in situ hybridization with biotin-labeled antisense SLA1 probe (red); (c) merge of a and b; (d) nuclear staining with DAPI merged with a and b; (e) differential interference contrast (DIC) merged with nuclear staining with DAPI. (B) Localization of SLA1 relative to Nhp2p under SmD1 silencing. (a) In situ hybridization with biotin-labeled antisense SLA1 RNA probe (red); (b) immunofluorescence with anti-Nhp2 antibody (green); (c) merge of a and b; (d) nuclear staining with DAPI merged with a and b; (e) DIC merge of a, b, and d. (C) Localization of SLA1 relative to U3 under SmD1 silencing. (a) In situ hybridization of DIG-labeled U3 RNA probe (green); (b) in situ hybridization of biotin-labeled SLA1 antisense probe (red); (c) merge of a and b; (d) nuclear staining with DAPI merged with a and b; (e) DIC merged with nuclear staining using DAPI. All slides were visualized under a Zeiss LSM 510 META inverted microscope. Bar, 10 μm.
In situ hybridization of SLA1 demonstrates that SLA1 is localized at two nuclear domains and is transported to the cytoplasm during SmD1 silencing.
The data presented in Fig. 2 suggest that pseudouridylation takes place early in the biogenesis of the SL RNA and that, if Sm assembly is blocked, SLA1 remains associated with the SL RNA. We have recently demonstrated that SL RNA and Sm proteins colocalize in a special nuclear compartment where SL RNA transcription and Sm assembly take place (31). It was therefore of great interest to identify the site at which SLA1 is localized in the trypanosome nucleus. To address this question, we localized SLA1 by in situ hybridization using antisense biotinylated RNA that was detected with a streptavidin-Alexa Fluor 647 conjugate (see Materials and Methods). This probing system is more sensitive than the DIG system. This sensitive probe enabled us to detect SLA1, which is at least 10-fold less abundant than its target, SL RNA.
As an H/ACA-like RNA, SLA1 is also expected to reside inside or in the vicinity of the nucleolus. Indeed, our previous cellular fractionation studies suggested that SLA1 is found both in the nucleolus and in the nucleoplasm (17). We therefore examined the localization of SLA1 by in situ hybridization together with the nucleolar marker U3 snoRNA or antibodies specific for the H/ACA RNA binding protein Nhp2p, as well as with Sm-GFP protein (where GFP is green fluorescent protein) and the SL RNA transcription factor tSNAP42, to examine whether SLA1 resides in the nucleus with SL RNA, where transcription and Sm assembly of SL RNA may take place. SLA1 needs to be assembled with the H/ACA proteins, and this assembly may take place in a subnuclear domain analogous to the Cajal bodies.
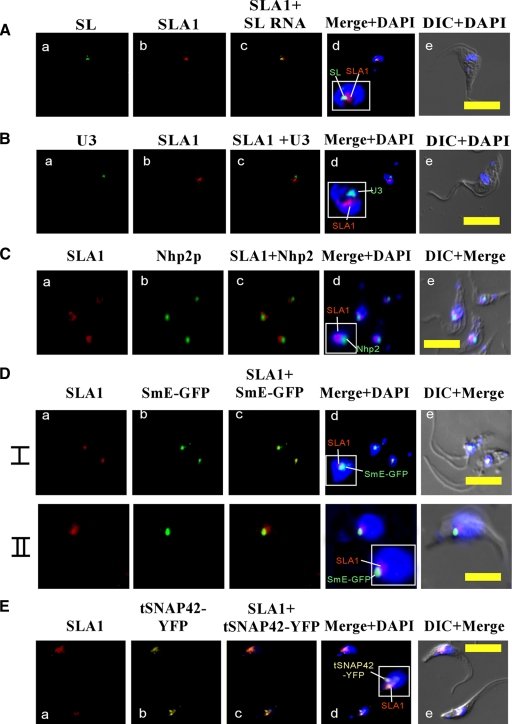
To examine the colocalization of SL RNA and SLA1, we performed in situ hybridization with labeled RNA probes specific to SL RNA and SLA1. Figure 3A (a to e) demonstrates that SLA1 is either colocalized with the SL RNA or surrounding it. Note that while it seems that these RNAs are found at equal levels, this is only due to the much more efficient detection by the biotin/avidin system; the true relative levels can be seen by specific labeling of these RNAs within total RNA (Fig. 2A). These data therefore suggest that SLA1 is either colocalized with SL RNA or found next to the SL RNA and near the nucleolus.
FIG. 3.
Localization of SLA1 in the nucleus. (A) Colocalization of SLA1 and SL RNA. (a) In situ hybridization with DIG-labeled antisense SL RNA probe (green); (b) in situ hybridization with biotin-labeled antisense SLA1 probe (red); (c) merge of a and b; (d) nuclear staining with DAPI merged with a and b; (e) differential interference contrast (DIC) merged with nuclear staining using DAPI. (B) Localization of SLA1 relative to the U3 snoRNA. (a) In situ hybridization of DIG-labeled U3 RNA probe (green); (b) in situ hybridization of biotin-labeled SLA1 antisense probe (red); (c) merge of a and b; (d) nuclear staining with DAPI merged with a and b; (e) DIC merged with nuclear staining with DAPI. (C) Localization of SLA1 relative to Nhp2p. (a) In situ hybridization with biotin-labeled antisense SLA1 RNA probe (red); (b) immunofluorescence with anti-Nhp2 antibody (green) (the antibodies [31] were diluted 1:200); (c) merge of a and b; (d) nuclear staining with DAPI merged with a and b; (e) DIC merge of a, b, and d. (D) Localization of SLA1 relative to SmE-GFP. Cells expressing the SmE-GFP construct (31) were fixed with 4% (vol/vol) formaldehyde and hybridized with labeled RNA probes (I and II represent two fields of the same experiment). (a) In situ hybridization with biotin-labeled antisense SLA1 RNA probe (red); (b) fluorescence of SmE-GFP (green); (c) merge of a and b; (d) nuclear staining with DAPI merged with a and b; (e) DIC merge of a, b, and d. (E) Localization of SLA1 relative to tSNAP42-YFP. Cells (see Materials and Methods) were fixed with 4% (vol/vol) formaldehyde and hybridized with labeled RNA probes. (a) In situ hybridization with biotin-labeled antisense SLA1 RNA probe (red); (b) fluorescence of tSNAP42-YFP (yellow); (c) merge of a and b; (d) nuclear staining with DAPI merged with a and b; (e) DIC merge of a, b, and d. All slides were observed under a Zeiss LSM 510 META inverted microscope. Bar, 10 μm.
To fine map the location of SLA1 in the nucleus/nucleolus, we determined SLA1 localization with respect to the nucleolar constituent U3, which functions in rRNA processing (Fig. 3B), and Nhp2p, which binds to all H/ACA RNAs (Fig. 3C). In situ hybridization with U3 snoRNA and SLA1 (Fig. 3B) suggests that SLA1 and U3 do not colocalize in the nucleolus. SLA1 seems to be localized at the border of the nucleolus, in a structure similar to Cajal bodies, whereas U3 snoRNA, which is involved in rRNA processing, is found inside the nucleolus. The same phenomenon was observed when in situ hybridization with SLA1 was performed in parallel to immunofluorescence with anti-Nhp2p antibodies. The results, shown in Fig. 3C, demonstrate (as expected) that Nhp2 is localized inside the nucleolus, where all of the H/ACA-like RNA species that are involved in rRNA processing and modification function. Note that we failed to detect Nhp2 colocalization with SLA1 by in situ techniques, although we previously demonstrated that Nhp2p binds to SLA1 (3). This might simply reflect the fact that the antibody is not sensitive enough to detect the protein bound to a single type of H/ACA RNA and can detect the protein only when bound to the entire repertoire (of ∼35 H/ACA species) that is concentrated at a single site in the nucleolus.
As suggested previously, transcription of SL RNA takes place in a distinct nuclear compartment (7, 13). We recently demonstrated that Sm assembly may take place in this compartment (31). To determine whether SLA1 is concentrated at this same nuclear compartment, we examined the colocalization of SLA1 with the Sm protein and the SL RNA-specific transcription factor tSNAP42. The results, presented in Fig. 3D (I and II), indicate that SLA1 colocalizes with Sm proteins. Further support for the existence of SLA1 in the SL RNP factory is presented in Fig. 3E, demonstrating the colocalization of SLA1 with the tSNAP42 transcription factor. Note that it is possible to observe either a single locus or two such loci in T. brucei (37). Interestingly, in most cases, we observed SLA1 in only a single locus, suggesting that only a single SL RNP factory may exist in the cell. In sum, the data presented in Fig. 3 demonstrate that SLA1 is present together with SL RNA, tSNAP42, and SmE in the nuclear domain, where SL RNA transcription, modification, and Sm assembly most probably take place. In addition, SLA1 is found near the nucleolus but not in association with U3 and Nhp2.
It was demonstrated previously that during SmD1 silencing the SL RNA first accumulates in the nucleus and then is expelled to the cytoplasm. This process can be followed by in situ hybridization of the SL RNA (5, 19). Based on the data shown in Fig. 2B, we expected to find the SLA1 in the cytoplasm together with SL RNA in SmD1-silenced cells. The migration of SLA1 to the cytoplasm is clearly visualized after 40 h of silencing (Fig. 4A). By this time point, all of the SLA1 and SL RNA has migrated from the nucleus to the cytoplasm. These data visually confirm our results shown in Fig. 2, i.e., that under Sm depletion SLA1 does not dissociate from the SL RNA but rather escorts it to the cytoplasm. Note that there is not a complete overlap between SL RNA and SLA1, most probably because there is less SLA1 than SL RNA in the silenced cells; hence, many of the SL RNP-C particles do not contain SLA1, but all SLA1 is found in SL RNP-C particles (Fig. 4A). In the silenced cells, SLA1 but not U3 snoRNA (Fig. 4C) or Nhp2 (Fig. 4B) migrates from the nucleus to the cytoplasm. These results further suggest that the migration of SLA1 together with SL RNA to the cytoplasm is a specific event induced by Sm silencing and does not represent nonspecific cellular leakage due to the major upheaval in mRNA processing in these silenced cells.
The SL RNA structure is altered in CBF5-silenced cells, which results in a marked reduction of SLA1.
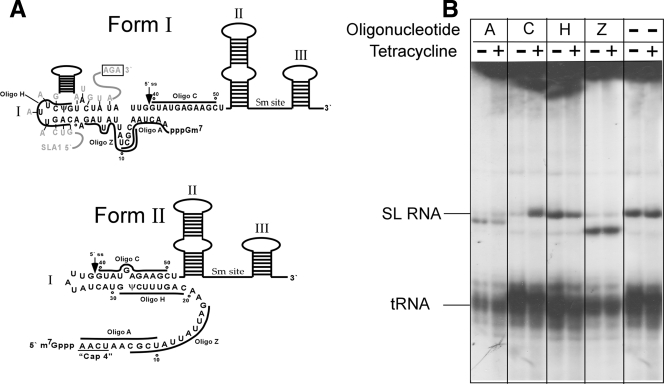
Since the lack of ψ on SL RNA had a very minor effect on trans splicing and since SLA1 was shown not to detach from the SL RNA unless Sm assembly took place, we next examined the function of the association of SLA1 with SL RNA during biogenesis. We suggested that the main function of SLA1 is to induce and preserve a distinct fold of the SL RNA that is necessary for interactions with proteins and possibly with the capping machinery. Support for such an effect was already obtained with cells depleted for CBF5, in which the SLA1 level is significantly reduced. In these cells, the capping of the SL RNA is perturbed (3). We therefore wished to examine the effect SLA1 might have on the folding of nascently transcribed SL RNA. The most suitable system to answer this question is the permeable-cell system. In this system, the SL RNA is transcribed efficiently and utilizes in trans splicing (34, 35). Furthermore, this system was previously used to demonstrate the structure of SL RNA by examining the accessibility of SL RNA to digestion with RNase H in the presence of cDNA oligonucleotides. In such experiments, the RNA secondary structure, another RNA, or a protein at or near the site of oligonucleotide annealing might block or partially inhibit digestion by RNase H. The accessibilities of various regions of the SL RNA during synthesis were determined previously (36). It was reasoned that, under these conditions, the oligonucleotides will be able to bind the regions not yet bound to proteins, since the oligonucleotide is added to permeable cells at the start of the incubation together with [α-32P]UTP. After a 10-min labeling in the presence of the oligonucleotide and RNase H, the SL RNA is analyzed by gel electrophoresis.
We therefore compared the patterns of oligonucleotide-mediated SL RNA digestion in permeable cells made from cells silenced for CBF5 before and after induction. In these silenced cells, the level of SLA1 is markedly reduced (3). In the presence of SLA1 (without tetracycline), we observed the same accessibility of the oligonucleotides to SL RNA as that previously described by Ullu and Tschudi (36) (the oligonucleotides used are shown in Fig. 5A). To eliminate the possibility that CBF5 silencing can affect the synthesis of SL RNA, the same experiment was performed with uninduced and induced cells in the absence of added oligonucleotide. Figure 5B shows that CBF5 silencing has no effect on SL RNA synthesis. Interestingly, the change in cleavage pattern between uninduced and CBF5-silenced cells is most prominent for oligonucleotide C. It was proposed previously that SL RNA can be found in two conformations: form II, which is prevalent in the steady state and is energetically favorable (11), and form I, which is present in minute amounts in the steady state but is the prevalent form detected in nascently transcribed SL RNA (11). The resistance to oligonucleotide C cleavage observed after significant reduction of SLA1 is best explained if under SLA1 depletion SL RNA is found in form II (Fig. 5B). Indeed, the domain covered by oligonucleotide C is engaged in intermolecular base pairing in form II but not in form I, which explains why cleavage with oligonucleotide C was observed in uninduced cells but not under silencing. Susceptibility to cleavage was observed to occur in the first 20 nt of the SL RNA in both induced and uninduced cells (Fig. 5B, oligonucleotides A and Z), suggesting that this domain is largely unprotected by proteins or RNAs in both form I and form II. Interestingly, the domain that is base paired with SLA1 (oligonucleotide H) is less protected in the absence of SLA1 but is not completely susceptible, most probably because, in form II, this domain is largely involved in intramolecular base pair interactions. Together, these data suggest that the main function of SLA1 RNA is to interact with the SL RNA and to maintain it in form I, which might be the preferred structure needed for interaction with the capping enzymes and Sm proteins. This interaction is required because form I is not energetically favorable. Indeed, the ΔG of form I (folding it from 1 to 50 nt, using the program mfold [http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi]) is −3 kcal/mole, whereas that of form II is −10 kcal/mole. SLA1 is thus needed to prevent the SL RNA from folding into its most energetically favorable form, form II.
FIG. 5.
Secondary structure of nascently transcribed SL RNA in CBF5-silenced cells. (A) Schematic representation of the secondary structures of the two forms of SL RNA. The secondary-structure predictions are based on data from Harris et al. (11). The three stem-loops and the Sm site of the SL RNA are indicated. The locations of the oligonucleotides (Oligo) used for RNase H cleavage are depicted on the SL RNA structure. The interaction domain of SLA1 with SL RNA is shown in gray. (B) RNase H cleavage of nascently transcribed SL RNA in CBF5-silenced cells. CBF5-silenced cells, uninduced (Tetracycline −) and induced (Tetracycline +) (72 h), were permeabilized and incubated with different oligonucleotides in the presence of RNase H. RNA was extracted from the cells and separated on a denaturing 6% polyacrylamide gel. The designations of the oligonucleotides are shown (their positions are given in panel A). The positions of the SL RNA and tRNA are indicated.
DISCUSSION
In this study, the localization and association of SLA1 with SL RNA and the implications for trans splicing and SL RNA biogenesis were studied. We report that mutated SL RNAs, which cannot undergo pseudouridylation, are affected only slightly as substrates of trans splicing. Thus, we describe a dual role for SLA1 to direct the pseudouridylation on SL RNA but also to maintain the SL RNA in the form I structure, thereby preventing it from adopting the more energetically favorable form II structure. We demonstrate that SLA1 associates with the SL RNA even when SL RNA biogenesis is blocked in Sm-depleted cells, suggesting that the SLA1 interaction with SL RNA is an early event in SL RNA biogenesis. SLA1 associates with SL RNA in a compartment containing tSNAP42 and Sm proteins, where SL RNP particles are formed, but also in a domain at the nucleolus border.
The data presented in this study suggest that the lack of pseudouridylation at position −12 only mildly affects the ability of the SL RNA to undergo trans splicing. Our experimental system (19, 41) was previously used to study the roles of domains or individual nucleotides of the SL RNA in the trans-splicing reaction. Those studies identified domains and even single nucleotides that cannot tolerate mutations. For instance, by mutating each of the individual cap nucleotides (nt 1 to 3) we demonstrated that these residues are absolutely essential for trans splicing; in contrast, the cap +4 position can tolerate mutations, but its mutation still reduces trans splicing by almost 50% (20). Another single base substitution that affects trans splicing is mutation of the +2 position of the 5′ splice site, which completely eliminates splicing. The trans-splicing defect observed with −12 mutations is minor compared to the splicing defects observed with the mutations mentioned above, suggesting that ψ at the −12 position is not critical for the trans-splicing reaction.
Severe defects in splicing were observed when the pseudouridylation of snRNAs was perturbed. It was demonstrated that modified but not unmodified U2 snRNA can rescue splicing in extracts depleted of U2 snRNP (43). Indeed, the importance of ψ in the branch point recognition domain was demonstrated by elegant nuclear magnetic resonance studies. These findings provided the first direct evidence that ψ could induce a structural change in the RNA and suggested that Ψ35 of U2 snRNA may function to better position the branch site adenosine for recognition and activity during splicing (23, 24).
This seems to not be the case for the ψ at the −12 position of the SL RNA. Note that the −12 position is part of the extensive intramolecular base pairing network between the exon and the intron of SL RNA (forming stem I of the SL RNA). Interestingly, mutations that change the sequence of this stem can be tolerated as long as the stem structure is conserved (20). Indeed, a mutation introduced in L. tarentolae including the −12 position induced no splicing defect, suggesting, as observed in this study, that the ψ in this position is not essential for splicing (29). The Ψ at the −12 position, however, can play a role in stabilizing the intermolecular interactions between SL RNA and U5/U6 snRNAs. Indeed, the −12 position has the potential to interact by base pairing with the intron G at position +5. The +5 intron position was shown to interact by base pairing with both U5 and U6 snRNAs (41). The ψ at the −12 position may affect the potential to form these interactions by affecting the intramolecular/intermolecular base pairing in stem I of SL RNA. This effect, however, must be relatively minor.
Initially, it was proposed that snoRNAs may have a chaperone-like activity during rRNA maturation via base pairing with the rRNA domains, thereby avoiding unwanted intramolecular base pairing and assisting and directing the folding of the huge precursor rRNA molecule through a highly constrained productive pathway (2). Since the discovery that these RNAs can actually guide chemical modification, their role as chaperones has been disregarded (32, 40). However, in the case presented here, the role of SLA1 as a chaperone might be more critical than the pseudouridylation that it guides. An important function of SLA1 is to keep the SL RNA in a distinct conformation (form I) described previously by Harris et al. (11), a conformation which could be favored by the capping machinery. The accessibility to cleavage by oligonucleotides suggests a major difference in the secondary structures of the SL RNA in cells carrying or depleted for SLA1. In the absence of SLA1, the SL RNA is found mainly in form II (11). As noted by us previously, in the absence of SLA1, defects in cap 4 formation are observed (3). After cap 4 formation and until Sm assembly, SLA1 associates with the SL RNA; it then detaches, and the SL RNA is folded into its more energetically favored form, form II. We suggest that SLA1 has a dual function: it guides the modification at −12 and acts as a chaperone during early steps of SL RNA biogenesis. Note that although the lack of this modification did not dramatically affect trans splicing (Fig. 1), we cannot exclude the possibility that this modification is important for the fidelity of splicing or that it may be important under ambient stresses imposed on the parasites in the host or in cycling between the hosts.
The pioneering studies performed by Tschudi and Ullu to determine the structure of SL RNA in vivo using the powerful and elegant permeable-cell system are fully supported by our results. For instance, the domain covered by oligonucleotide H (Fig. 5) is resistant to degradation in form I, and U24 to U26 display the unexpected behavior of being completely protected from chemical attack in vivo. These results were unexpected, and the reason for these observations was not immediately apparent. Our results explain these observations. Domain H is either covered by SLA1 (in form I) or engaged in extensive base pairing (in form II). The almost complete protection from chemical modification seen in the entire domain of G23 to U33 stems from the same effect; this domain is covered with SLA1 in form I or is engaged in base pairing in form II. It was also not clear why domain C21 to U33 is conserved in 16 trypanosomatid species examined, whereas the sequences that base pair with it vary. We can now interpret this sequence conservation as reflecting the fact that domain C21 to U33 is the interaction domain with SLA1, which is highly conserved among the trypanosomatids (17). In addition, the potential to form the two conformations (I and II) is highly conserved.
If the interaction of SLA1 is so important, why in its absence is the defect in trans splicing (as examined by reduction of the Y-structure intermediate) only ∼30% (3)? Note that in the absence of SLA1, the SL RNA is already folded in form II, which is the prevalent form in which the SL RNA is found in the cell and is most probably the form which is utilized in trans splicing. The absence of SLA1 may mainly affect the efficiency of capping. However, capping of the SL RNA can tolerate major perturbation without severely affecting trans splicing. Indeed, the knockdown of individual capping enzymes does not result in major splicing defects or a lethal phenotype (1, 44).
In this study, we also describe a novel nuclear compartment for SLA1, which is distinct from its localization with SL RNA and the nucleolar RNAs. This domain resembles the Cajal bodies, which are nuclear sites essential for the maturation of snRNA and snoRNAs (21). It was proposed that C/D and H/ACA snoRNAs move quickly from the Cajal bodies to the nucleolus. However, several RNAs and especially small Cajal body RNAs, which guide modification on snRNAs, are retained in this compartment. Other RNAs, such as snRNAs, C/D and H/ACA snoRNAs, and telomerase RNA, traverse these bodies but then migrate to their final destination (21). So far, there is no evidence for the existence of small Cajal body RNAs in trypanosomes (16), though these RNAs are expected to exist, since trypanosomes carry conserved modifications on their snRNAs (3). Trypanosomes may not possess Cajal bodies identical to those found in metazoa, because their genome lacks coilin, the protein that specifies these bodies in higher eukaryotes (26). However, yeasts also lack Cajal bodies but do contain nuclear bodies that might have similar functions (38). The nuclear localization of SLA1 outside the site of colocalization with SL RNA might be such a domain through which snoRNP/snRNA traverse on their way to their final destination.
We also present evidence in this study that SLA1 exists in the SL RNP factory, where SL RNA transcription and modification and Sm assembly take place. Indeed, clear colocalization among tSNAP42, the Sm protein, and SLA1 was observed. The finding of a distinct single focus of SL RNA transcription was reported initially for T. cruzi (7) and then for T. brucei (5, 31). The same observation was also recently reported for L. tarentolae (13). However, two or even four foci (in cells before division) were also observed in T. brucei (37). The two foci may represent transcription for the two alleles carrying the SL RNA genes. Interestingly, we found that SLA1 exists mostly in a single locus, even when we observed two loci in the nucleus, suggesting that mostly one locus is fully active as the SL RNP factory.
During Sm depletion, normal SL RNA biogenesis is blocked, and after massive accumulation in the nucleus, the SL RNA in its SL RNP-C complex migrates to the cytoplasm (5) together with SLA1, as demonstrated here. SL RNP-C is a disposal mechanism that removes defective SL RNA from the nucleus. It therefore was and remains puzzling why such defective SL RNA is not degraded like the U snRNAs in the same cells (5). One possibility is that SLA1 protects the SL RNA from degradation, since it remains associated with SL RNA in Sm-depleted cells. Indeed, the domain that engages with SLA1 was shown to protect the SL RNA from cleavage with oligonucleotide H (11, 33). We recently argued that degradation of the SL RNA might be deleterious to trypanosomes (5). The association of SLA1 with SL RNA might protect SL RNA in the SL RNP-C complex. We have recently purified the SL RNP-C complex to homogeneity and, among its binding proteins, identified helicase, ATPase, and a specific nuclear protein that migrates with the RNA from the nucleus to the cytoplasm (our unpublished data). SL RNP-C and its accompanying SLA1 may represent a novel approach to discarding RNA from the nucleus to the cytoplasm without degrading it.
This study dissects the function of a very special guide RNA, which possesses a chaperone function in addition to the modification it guides. Additional RNAs with similar properties may exist in trypanosomes and in other eukaryotes. This study emphasizes that the function of snoRNAs as chaperones should be revisited and that snoRNAs should be considered potential noncoding RNAs that regulate mRNA fate.
Acknowledgments
This study was supported by grants from the United States-Israel Binational Science Foundation and the Israel Science Foundation and by an International Research Scholar grant from the Howard Hughes Medical Institute to S.M. S.M. holds the David and Inez Myers Chair in RNA silencing of diseases.
Footnotes
Published ahead of print on 21 November 2008.
REFERENCES
- 1.Arhin, G. K., H. Li, E. Ullu, and C. Tschudi. 2006. A protein related to the vaccinia virus cap-specific methyltransferase VP39 is involved in cap 4 modification in Trypanosoma brucei. RNA 1253-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachellerie, J. P., J. Cavaillé, and A. Hüttenhofer. 2002. The expanding snoRNA world. Biochimie 84775-790. [DOI] [PubMed] [Google Scholar]
- 3.Barth, S., A. Hury, X. H. Liang, and S. Michaeli. 2005. Elucidating the role of H/ACA-like RNAs in trans-splicing and rRNA processing via RNA interference silencing of the Trypanosoma brucei CBF5 pseudouridine synthase. J. Biol. Chem. 28034558-34568. [DOI] [PubMed] [Google Scholar]
- 4.Bell, M., and A. Bindereif. 1999. Cloning and mutational analysis of the Leptomonas seymouri U5 snRNA gene: function of the Sm site in core RNP formation and nuclear localization. Nucleic Acids Res. 273986-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biton, M., M. Mandelboim, G. Arvatz, and S. Michaeli. 2006. RNAi interference of XPO1 and Sm genes and their effect on the spliced leader RNA in Trypanosoma brucei. Mol. Biochem. Parasitol. 150132-143. [DOI] [PubMed] [Google Scholar]
- 6.Darzacq, X., B. E. Jády, C. Verheggen, A. M. Kiss, E. Bertrand, and T. Kiss. 2002. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 212746-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dossin, F. M., and S. Schenkman. 2005. Actively transcribing RNA polymerase II concentrates on spliced leader genes in the nucleus of Trypanosoma cruzi. Eukaryot. Cell 4960-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dungan, J. M., K. P. Watkins, and N. Agabian. 1996. Evidence for the presence of a small U5-like RNA in active trans-spliceosomes of Trypanosoma brucei. EMBO J. 154016-4029. [PMC free article] [PubMed] [Google Scholar]
- 9.Ganot, P., M. L. Bortolin, and T. Kiss. 1997. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89799-809. [DOI] [PubMed] [Google Scholar]
- 10.Goncharov, I., Y. X. Xu, Y. Zimmer, K. Sherman, and S. Michaeli. 1998. Structure-function analysis of the trypanosomatid spliced leader RNA. Nucleic Acids Res. 262200-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris, K. A., Jr., D. M. Crothers, and E. Ullu. 1995. In vivo structural analysis of spliced leader RNAs in Trypanosoma brucei and Leptomonas collosoma: a flexible structure that is independent of cap4 methylations. RNA 1351-362. [PMC free article] [PubMed] [Google Scholar]
- 12.Hausner, T. P., L. M. Giglio, and A. M. Weiner. 1990. Evidence for base-pairing between mammalian U2 and U6 small nuclear ribonucleoprotein particles. Genes Dev. 42146-2156. [DOI] [PubMed] [Google Scholar]
- 13.Hitchcock, R. A., S. Thomas, D. A. Campbell, and N. R. Sturm. 2007. The promoter and transcribed regions of the Leishmania tarentolae spliced leader RNA gene array are devoid of nucleosomes. BMC Microbiol. 744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly, S., J. Reed, S. Kramer, L. Ellis, H. Webb, J. Sunter, J. Salje, N. Marinsek, K. Gull, B. Wickstead, and M. Carrington. 2007. Functional genomics in Trypanosoma brucei: a collection of vectors for the expression of tagged proteins from endogenous and ectopic gene loci. Mol. Biochem. Parasitol. 154103-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang, X. H., L. Liu, and S. Michaeli. 2001. Identification of the first trypanosome H/ACA RNA that guides pseudouridine formation on rRNA. J. Biol. Chem. 27640313-40318. [DOI] [PubMed] [Google Scholar]
- 16.Liang, X. H., S. Uliel, A. Hury, S. Barth, T. Doniger, R. Unger, and S. Michaeli. 2005. A genome-wide analysis of C/D and H/ACA-like small nucleolar RNAs in Trypanosoma brucei reveals a trypanosome-specific pattern of rRNA modification. RNA 11619-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang, X. H., Y. X. Xu, and S. Michaeli. 2002. The spliced leader-associated RNA is a trypanosome-specific sn(o) RNA that has the potential to guide pseudouridine formation on the SL RNA. RNA 8237-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, L., H. Ben Shlomo, Y. X. Xu, M. Z. Stern, I. Goncharov, Y. Zhang, and S. Michaeli. 2003. The trypanosomatid signal recognition particle consists of two RNA molecules, a 7SL RNA homologue and a novel tRNA-like molecule. J. Biol. Chem. 27818271-18280. [DOI] [PubMed] [Google Scholar]
- 19.Mandelboim, M., S. Barth, M. Biton, X. H. Liang, and S. Michaeli. 2003. Silencing of Sm proteins in Trypanosoma brucei by RNA interference captured a novel cytoplasmic intermediate in spliced leader RNA biogenesis. J. Biol. Chem. 27851469-51478. [DOI] [PubMed] [Google Scholar]
- 20.Mandelboim, M., C. L. Estrano, C. Tschudi, E. Ullu, and S. Michaeli. 2002. On the role of exon and intron sequences in trans-splicing utilization and cap 4 modification of the trypanosomatid Leptomonas collosoma SL RNA. J. Biol. Chem. 27735210-35218. [DOI] [PubMed] [Google Scholar]
- 21.Matera, A. G., R. M. Terns, and M. P. Terns. 2007. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8209-220. [DOI] [PubMed] [Google Scholar]
- 22.Mittra, B., J. R. Zamudio, J. M. Bujnicki, J. Stepinski, E. Darzynkiewicz, D. A. Campbell, and N. R. Sturm. 2008. The TbMTr1 spliced leader RNA cap 1 2′-O-ribose methyltransferase from Trypanosoma brucei acts with substrate specificity. J. Biol. Chem. 2833161-3172. [DOI] [PubMed] [Google Scholar]
- 23.Newby, M. I., and N. L. Greenbaum. 2001. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA 7833-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newby, M. I., and N. L. Greenbaum. 2002. Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc. Natl. Acad. Sci. USA 9912697-12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ni, J., A. L. Tien, and M. J. Fournier. 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89565-573. [DOI] [PubMed] [Google Scholar]
- 26.Ogg, S. C., and A. I. Lamond. 2002. Cajal bodies and coilin—moving towards function. J. Cell Biol. 15917-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palfi, Z., G. L. Xu, and A. Bindereif. 1994. Spliced leader-associated RNA of trypanosomes. Sequence conservation and association with protein components common to trans-spliceosomal ribonucleoproteins. J. Biol. Chem. 26930620-30625. [PubMed] [Google Scholar]
- 28.Ruan, J. P., S. Shen, E. Ullu, and C. Tschudi. 2007. Evidence for a capping enzyme with specificity for the trypanosome spliced leader RNA. Mol. Biochem. Parasitol. 156246-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sturm, N. R., J. Fleischmann, and D. A. Campbell. 1998. Efficient trans-splicing of mutated spliced leader exons in Leishmania tarentolae. J. Biol. Chem. 27318689-18692. [DOI] [PubMed] [Google Scholar]
- 30.Tkacz, I. D., S. Cohen, M. Salmon-Divon, and S. Michaeli. 2008. Identification of the complete set of Lsm proteins that bind U6 snRNA in Trypanosoma brucei. Mol. Biochem. Parasitol. 16022-31. [DOI] [PubMed] [Google Scholar]
- 31.Tkacz, I. D., Y. Lustig, M. Z. Stern, M. Biton, M. Salmon-Divon, A. Das, V. Bellofatto, and S. Michaeli. 2007. Identification of novel snRNA-specific Sm proteins that bind selectively to U2 and U4 snRNAs in Trypanosoma brucei. RNA 1330-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tollervey, D., and T. Kiss. 1997. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 9337-342. [DOI] [PubMed] [Google Scholar]
- 33.Tschudi, C., A. Djikeng, H. Shi, and E. Ullu. 2003. In vivo analysis of the RNA interference mechanism in Trypanosoma brucei. Methods 30304-312. [DOI] [PubMed] [Google Scholar]
- 34.Tschudi, C., and E. Ullu. 1990. Destruction of U2, U4, or U6 small nuclear RNA blocks trans splicing in trypanosome cells. Cell 61459-466. [DOI] [PubMed] [Google Scholar]
- 35.Ullu, E., and C. Tschudi. 1990. Permeable trypanosome cells as a model system for transcription and trans-splicing. Nucleic Acids Res. 183319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ullu, E., and C. Tschudi. 1993. 2′-O-methyl RNA oligonucleotides identify two functional elements in the trypanosome spliced leader ribonucleoprotein particle. J. Biol. Chem. 26813068-13073. [PubMed] [Google Scholar]
- 37.Uzureau, P., J. P. Daniels, D. Walgraffe, B. Wickstead, E. Pays, K. Gull, and L. Vanhamme. 2008. Identification and characterization of two trypanosome TFIIS proteins exhibiting particular domain architectures and differential nuclear localizations. Mol. Microbiol. 691121-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verheggen, C., J. Mouaikel, M. Thiry, J. M. Blanchard, D. Tollervey, R. Bordonné, D. L. Lafontaine, and E. Bertrand. 2001. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J. 205480-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watkins, K. P., J. M. Dungan, and N. Agabian. 1994. Identification of a small RNA that interacts with the 5′ splice site of the Trypanosoma brucei spliced leader RNA in vivo. Cell T6171-182. [DOI] [PubMed] [Google Scholar]
- 40.Weinstein, L. B., and J. A. Steitz. 1999. Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol. 11378-384. [DOI] [PubMed] [Google Scholar]
- 41.Xu, Y., L. Liu, and S. Michaeli. 2000. Functional analyses of positions across the 5′ splice site of the trypanosomatid spliced leader RNA. Implications for base-pair interaction with U5 and U6 snRNAs. J. Biol. Chem. 27527883-27892. [DOI] [PubMed] [Google Scholar]
- 42.Xu, Y. X., H. BenShlomo, and S. Michaeli. 1997. The U5 RNA of trypanosomes deviates from the canonical U5 RNA: the Leptomonas collosoma U5 RNA and its coding gene. Proc. Natl. Acad. Sci. USA 948473-8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, Y. T., M. D. Shu, and J. A. Steitz. 1998. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 175783-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zamudio, J. R., B. Mittra, S. Foldynova-Trantírková, G. M. Zeiner, J. Lukes, J. M. Bujnicki, N. R. Sturm, and D. A. Campbell. 2007. The 2′-O-ribose methyltransferase for cap 1 of spliced leader RNA and U1 small nuclear RNA in Trypanosoma brucei. Mol. Cell. Biol. 276084-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeiner, G. M., S. Foldynova, N. R. Sturm, J. Lukes, and D. A. Campbell. 2004. SmD1 is required for spliced leader RNA biogenesis. Eukaryot. Cell 3241-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeiner, G. M., N. R. Sturm, and D. A. Campbell. 2003. Exportin 1 mediates nuclear export of the kinetoplastid spliced leader RNA. Eukaryot. Cell 2222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]