Abstract
Establishing the occurrence of endocytosis in filamentous fungi was elusive in the past mainly due to the lack of reliable indicators of endocytosis. Recently, however, it was shown that the fluorescent dye N-(3-triethylammoniumpropyl)-4-(p-diethyl-aminophenyl-hexatrienyl)pyridinium dibromide (FM4-64) and the plasma membrane protein AoUapC (Aspergillus oryzae UapC) fused to enhanced green fluorescent protein (EGFP) were internalized from the plasma membrane by endocytosis. Although the occurrence of endocytosis was clearly demonstrated, its physiological importance in filamentous fungi still remains largely unaddressed. We generated a strain in which A. oryzae end4 (Aoend4), the A. oryzae homolog of Saccharomyces cerevisiae END4/SLA2, was expressed from the Aoend4 locus under the control of a regulatable thiA promoter. The growth of this strain was severely impaired, and its hyphal morphology was altered in the Aoend4-repressed condition. Moreover, in the Aoend4-repressed condition, neither FM4-64 nor AoUapC-EGFP was internalized, indicating defective endocytosis. Furthermore, the localization of a secretory soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) was abnormal in the Aoend4-repressed condition. Aberrant accumulation of cell wall components was also observed by calcofluor white staining and transmission electron microscopy analysis, and several genes that encode cell wall-building enzymes were upregulated, indicating that the regulation of cell wall synthesis is abnormal in the Aoend4-repressed condition, whereas Aopil1 disruptants do not display the phenotype exhibited in the Aoend4-repressed condition. Our results strongly suggest that endocytosis is crucial for the hyphal tip growth in filamentous fungi.
The filamentous fungus Aspergillus oryzae has been used in industrial fermentation processes and is regarded to be safe for humans. A. oryzae can secrete several proteins, such as alpha-amylase, into the medium. Thus, A. oryzae is a potential host for heterologous protein production. Since the completion of A. oryzae genome sequencing (18) in recent years, many applied and basic studies have been conducted on A. oryzae using its genome sequencing data. In particular, studies on vesicular trafficking, including the secretory pathway, are of increasing importance because they are closely related to protein production. For example, endoplasmic reticulum and vacuole dynamics and systematic soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) protein analyses have been performed in A. oryzae (16, 19, 23, 30, 31, 32). However, endocytosis, an intracellular trafficking pathway, has not been studied as well in A. oryzae as in other filamentous fungi.
Endocytosis is an important cellular process that occurs, for example, in signal transduction and reconstruction of cell polarity and is conserved in eukaryotic cells. The detailed mechanism of endocytosis has been well studied in model organisms such as yeasts. Many proteins are involved in the endocytic process, which is regulated spatiotemporally (12). Saccharomyces cerevisiae END4/SLA2 (synthetic lethal with ABP1) is an endocytosis-associated gene that has been studied in detail (3, 6, 22, 27, 35, 43, 44). End4p/Sla2p is essential for fluid-phase and receptor-mediated endocytosis and actin assembly and polarization (27). The protein has the epsin N-terminal homology (ENTH) and the AP180 N-terminal homology (ANTH) domains, which bind to phosphatidylinositol-4,5-bisphosphate in the plasma membrane in the N-terminal region, and the I/LWEQ domain, which is proposed to be the actin-binding domain in the C-terminal region; it also functions as an adaptor that connects the invaginated plasma membrane and actin cytoskeleton, which plays an important role in endocytosis, to generate force for invaginating the plasma membrane into the intracellular space and forming endocytic pits (13, 33). Abp1p (actin-binding protein) forms actin patches by polymerization of the actin cytoskeleton. It is suggested that endocytosis occurs at the sites in which Abp1p localizes, i.e., cortical actin patches (21, 22). Hence, Abp1p has been used as a tool to investigate the subcellular space in which endocytosis occurs (21).
Establishing the existence of endocytosis in filamentous fungi was elusive in the past mainly due to the lack of reliable indicators of endocytosis (28). However, it has been confirmed that the fluorescent dye N-(3-triethylammoniumpropyl)-4-(p-diethyl-aminophenyl-hexatrienyl)pyridinium dibromide (FM4-64) and the plasma membrane protein AoUapC (Aspergillus oryzae UapC [uric acid-xanthine permease]) fused to enhanced green fluorescent protein (EGFP) were internalized from the plasma membrane by endocytosis (8, 25). Moreover, recently, in Aspergillus nidulans, the localization of components required for endocytosis has been analyzed in living hyphae (1, 37, 41). ActA and FimA, which are actin and fimbrin, respectively, are mostly localized in the hyphal tip region (41). Furthermore, AbpA, an actin-binding protein, is primarily localized in the apical region and is used as an endocytic site marker. AmpA, the amphiphysin homolog in A. nidulans, and SlaB, the End4p/Sla2p homolog, are also localized in sites in which AbpA is localized (1). These endocytic components are localized near the hyphal tip regions but slightly away from the apex where exocytosis preferentially occurs (37). Although the occurrence of endocytosis was clearly demonstrated and the localization of endocytic components was analyzed, the physiological importance of endocytosis in filamentous fungi still remains largely unaddressed.
In this report, we analyzed the physiological significance of endocytosis by generating strains that conditionally express A. oryzae end4 (Aoend4), the A. oryzae homolog of S. cerevisiae END4/SLA2. Hyphae grown in the Aoend4-repressed condition displayed aberrant morphology; endocytic defects in AoUapC-EGFP and FM4-64; abnormal apical recycling of EGFP-fused AoSnc1, which is a vesicle SNARE required for secretion; and abnormal cell wall synthesis. These results suggest that endocytosis plays crucial roles in the physiology of hyphal growth.
MATERIALS AND METHODS
A. oryzae strains and plasmids.
The A. oryzae strains used in this study are listed in Table 1. A. oryzae RIB40 is the wild-type strain that was used as the DNA donor. The cDNA was prepared as follows. Total RNA (1 μg) was treated with DNase (Clontech) and used as the template. The cDNA was amplified using oligo(dT)12-18 primers (Invitrogen, Tokyo, Japan) and Prime Script reverse transcriptase (TaKaRa, Kyoto, Japan). For the rapid amplification of cDNA 5′-end analysis of Aoend4, we used the Gene Racer kit (Invitrogen) according to the manufacturer's instructions. For DNA or cDNA cloning, the Pyrobest DNA polymerase (TaKaRa) was used. For Aoend4 cDNA cloning, the Aoend4 cDNA-F (5′-ATGAGTCGCACGGAG-3′) and Aoend4 cDNA-R (5′-GTCCTCCTGGTACGAGATCTT-3′; the stop codon is excluded for EGFP fusion to the C terminus of AoEnd4) primers were used. For Aoabp1 cDNA cloning, the Aoabp1 cDNA-F (5′-ATGGCATCCCTTAACCTTTC-3′) and Aoabp1 cDNA-R (5′-CTTTCGAAGTTCTACATAATTTGC-3′; the stop codon is excluded for mDsRed fusion to the C terminus of AoAbp1) primers were utilized. All plasmids used for A. oryzae transformation in this study were constructed by the MultiSite Gateway system (Invitrogen) (17). To generate strains that conditionally express Aoend4, we constructed the plasmids as follows. The Aoend4up-F (5′-TCCGATATCAGGTCGACATACCAGACGAG-3′ [the EcoRV site is underlined]) and Aoend4up-R (5′-TCCCCCGGGGCTTCGTCACCTCCTGAGTT-3′ [the SmaI site is underlined]) primers were used to clone the Aoend4 5′ untranslated region. Using these primers, a DNA fragment was amplified by PCR and inserted into a pg5′Pp vector digested with SmaI. The resultant plasmid was named pg5′e4up. The PthiA sequence from pBTHI II digested with XhoI was blunted and introduced into the pgEHH vector digested with SmaI. The resultant plasmid, named pgEPt, was digested with SmaI; subsequently, the adeA sequence was introduced from pAdeA that had been digested with EcoRI and PstI. The resultant plasmid was named pgEaAPt. The Aoend4 g-F (5′-ATGAGTCGGTAAGTGTTTTTGGGAC-3′) and Aoend4 g-R (5′-TCCctcgagGATATCGCTCTTCCAGGTCTTTCACAC-3′; lowercase and underlined characters indicate the XhoI and EcoRV sites, respectively) primers were utilized for cloning the 1.7-kb Aoend4 open reading frame from the start codon. The amplified DNA fragment was introduced into the pg3′HH vector digested with SmaI. The resultant plasmid was named pg3′e4. The pg5′e4up, pgEaAPt, and pg3′e4 plasmids were used for Gateway LR recombination, and the resultant plasmid was digested with EcoRV, and a 7.0-kb fragment was used as the DNA cassette for A. oryzae transformation. To generate Aopil1 disruptants, a DNA fragment amplified by PCR using the Aopil1 up-F (5′-CTGCAGCATGGCCTGCGCAATTTTCT-3′ [the PstI site is underlined]) and Aopil1 up-R (5′-GCTACGGTTTGTATGGGAAG-3′) primers was introduced into pg5′Pp digested with SmaI, and a DNA fragment amplified by PCR using the Aopil1 dw-F (5′-GCCAATTGCAGCCACAAACA-3′) and Aopil1 dw-R (5′-CTGCAGATCACACACAGGATCCAGGA-3′ [the PstI site is underlined]) primers was inserted into pg3′HH digested with SmaI; the resultant plasmids were named pg5′DP and pg3′DP, respectively. The pg5′DP, pgEaA, and pg3′DP plasmids were used by Gateway LR recombination, and the DNA cassette from the resultant plasmid digested with PstI was used for A. oryzae transformation. For A. oryzae transformation, the DNA fragments or plasmids were introduced into each host strain using a standard method (15).
TABLE 1.
A. oryzae strains used in this study
| Strain(s) | Relevant genotype | Reference or source |
|---|---|---|
| RIB40 | Wild type | |
| niaD300 | niaD− | 20 |
| NSR13 | niaD−sC−adeA− | 11 |
| NSRku70-1-1 | niaD−sC−adeA− ΔargB Δku70::argB | 38 |
| niaD300N-1 | niaD−niaD | This study |
| NS13 | niaD−sC−adeA−adeA | This study |
| NSRku70-1-1A | niaD−sC−adeA−adeA ΔargB Δku70::argB | This study |
| TE4-1 and TE4-2 | niaD−sC−adeA− ΔAoend4::(PthiA-Aoend4 adeA) | This study |
| AAD1 | niaD− (PamyB-Aoabp1-mdsred niaD) | This study |
| TEUE3 | niaD−sC− (PamyB-AouapC-egfp sC) adeA− ΔAoend4::(PthiA-Aoend4 adeA) | This study |
| TEUA1 | niaD− (PamyB-Aoabp1-mdsred niaD) sC− (PamyB-AouapC-egfp sC) adeA− ΔAoend4::(PthiA-Aoend4 adeA) | This study |
| TESn1 | niaD− (PamyB-egfp-Aosnc1 niaD) sC−adeA− ΔAoend4::(PthiA-Aoend4 adeA) | This study |
| TEN1 | niaD−niaD sC−adeA− ΔAoend4::(PthiA-Aoend4 adeA) | This study |
| TEAEN1 | niaD− (PamyB-Aoend4-egfp niaD) sC−adeA− ΔAoend4::(PthiA-Aoend4 adeA) | This study |
| DP1, DP2, and DP3 | niaD−sC−adeA− ΔAopil1::adeA ΔargB Δku70::argB | This study |
Southern blot analysis.
Southern blot analyses were performed using the ECL detection kit according to the manufacturer's instructions (Amersham, United Kingdom), and the bands were detected using a luminescent image analyzer LAS-4000miniEPUV (Fujifilm, Japan). Specific probes for Southern blot analyses were constructed as follows. A 1.2-kb DNA fragment from pg3′e4 digested with KpnI and PstI was exploited as the specific probe for confirmation of the TE4 strains. pg3′DP was digested with XhoI, and the 0.7-kb DNA fragment was exploited as the specific probe for confirming the Aopil1 disruptants.
Fluorescence microscopy, culture media, and staining.
For fluorescence microscopy, we used an Olympus System microscope model BX52 (Olympus, Tokyo, Japan) equipped with an UPlanApo 100× objective lens (1.35 numerical aperture) (Olympus). A GFP filter (495/520-nm excitation, 510-nm dichroic, 530/535-nm emission) (Chroma Technologies, Brattleboro, VT) was used for observing EGFP fluorescence. A DsRed filter (570/620-nm excitation, 590-nm dichroic, 630/660-nm emission) (Chroma Technologies) was used to observe the fluorescence of FM4-64 and DsRed. A BHDMU (330- to 385-nm excitation, 400-nm dichroic, >420-nm emission) UV excitation cube (Olympus) was used to observe the fluorescence of calcofluor white. The images were analyzed by using MetaMorph software (Molecular Devices Co., Sunnyvale, CA). Confocal microscopy was performed with an IX71 inverted microscope (Olympus) equipped with 100× and 40× Neofluor objective lenses (1.40 numerical aperture); 488-nm (Furukawa Electric, Japan) and 561-nm (Melles Griot) semiconductor lasers; GFP, DsRed, and DualView filters (Nippon Roper, Chiba, Japan); a CSU22 confocal scanning system (Yokogawa Electronics, Tokyo, Japan); and an Andor iXon cooled digital charge-coupled-device camera (Andor Technology PLC, Belfast, United Kingdom). Images were analyzed with the Andor iQ software (Andor Technology PLC). For fluorescence recovery after photobleaching (FRAP) analysis, the MicroPoint ablation laser system (Photonic Instruments Inc., Tokyo, Japan) was used, and the images were analyzed with MetaMorph software (Molecular Devices Co.).
Approximately 105 conidia were inoculated in 100 μl liquid medium and incubated on cover glasses for fluorescence microscopy of calcofluor white staining or in glass-based dishes (Asahi Techno Glass, Chiba, Japan) for other microscopic observations using confocal laser microscopy. They were grown at 30°C for approximately 20 h, and indirect immunofluorescence microscopy was performed as described below. Czapek-Dox (CD) medium (0.3% NaNO3, 0.2% KCl, 0.1% KH2PO4, 0.05% MgSO4·7H2O, 0.002% FeSO4·7H2O, 2% glucose [pH 5.5]), CDm medium (CD medium with 0.0015% methionine), M medium [0.2% NH4Cl, 0.1% (NH4)2SO4, 0.05% KCl, 0.05% NaCl, 0.1% KH2PO4, 0.05% MgSO4·7H2O, 0.002% FeSO4·7H2O, 2% glucose (pH 5.5)], and MM medium (M medium with 0.15% methionine) were used for cultivation to suit the auxotrophy of each strain. To lower the expression of fusion genes under PamyB, we used each medium containing glycerol (2% glucose was replaced with 1.95% glycerol and 0.05% glucose) as the carbon source (36).
The induction of AoUapC-EGFP internalization was performed as described previously (8). FM4-64 (Molecular Probes, Eugene, OR) was prepared as a 1.6 mM solution in dimethyl sulfoxide. Approximately 20 h after inoculation, the cultures were transferred into a medium containing 8 μM FM4-64 and incubated for 2 min at room temperature. After incubation, FM4-64-containing medium was replaced with fresh dye-free medium, and the samples were examined. For calcofluor white (Sigma) staining, the cultures were fixed with 0.5% glutaraldehyde, 3.7% formaldehyde, and 50 mM phosphate-buffered saline (PBS) (8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, 0.24 g of KH2PO4[all in 1 liter] [pH 7.4]) for 10 min, washed twice with PBS, and incubated in medium containing 0.1 mg/ml calcofluor white for 3 min at room temperature. After two washes with PBS, the samples were observed by fluorescence microscopy. For indirect immunofluorescence microscopy, the cultures that had been incubated for approximately 12 h were fixed with 3.7% formaldehyde, 5 mM MgSO4, and 2.5 mM EGTA in PBS for 15 min. The cultures were washed twice with PBS containing 0.05% Tween 20 (PBST) after each operation. After fixation, the cultures were digested with 3 mg/ml Yatalase (TaKaRa), 1 mg/ml lysing enzyme (Sigma), and 10 mg/ml egg white (Sigma) in PBST and thereafter incubated in methanol for 10 min at −20°C. Primary and secondary antibody reactions were performed for 1 h. The primary antibody was the rabbit anti-actin antibody (1:500 dilution; Sigma), while the secondary antibody was the fluorescein isothiocyanate-conjugated immunoglobulin G antibody (1:200 dilution; Sigma); both antibodies were diluted in PBS containing 0.1 mg/ml bovine serum albumin. The cultures were mounted in PBST and examined by microscopy.
TEM analysis.
Transmission electron microscopy (TEM) analysis was performed as described previously (10). After incubation for 27 h in a submerged culture, the mycelia were harvested and fixed for 4 h in 4% glutaraldehyde with 0.1 M phosphate buffer and thereafter for 3 h in 1% osmium tetroxide at 4°C. Ultrathin sections were stained with uranyl acetate for 30 min and then with lead citrate for 5 min, and these sections were observed with a JEOL transmission electron microscope (JEM-1010).
Real-time RT-PCR analysis.
Real-time reverse transcription-PCR (RT-PCR) analysis was performed as described previously (14). Template cDNA from A. oryzae TEUE3 cultured in Aoend4-expressed or Aoend4-repressed conditions was prepared as described above. The specific primers used for real-time RT-PCR analysis in this study are listed in Table 2. The expression of each gene was normalized to that of gpdA.
TABLE 2.
Primers for real-time RT-PCR analysis
| Gene | Gene IDa | Sequenceb |
|---|---|---|
| chsA | AO090012000084 | 5′-CGATCAACATCCTGATGGTG-3′ |
| 5′-CCCACAGAATGACTCGGAAA-3′ | ||
| chsB | AO090701000589 | 5′-TACCTGGACCCTTGGCATAT-3′ |
| 5′-ACTTGTCCATACCGTCACTG-3′ | ||
| chsC | AO090011000449 | 5′-TCACGGATCTGTTCACCAAC-3′ |
| 5′-CTTGTAGTAATCGGCCTGTC-3′ | ||
| chsY | AO090026000323 | 5′-CGAAAAAGAGATGGGAGGAG-3′ |
| 5′-CTTCGGTGGCTGAGTTGATA-3′ | ||
| chsZ | AO090026000321 | 5′-CTGTCCAGCTTTAGTCGGTA-3′ |
| 5′-GCCAATTCCTGGTCAATCTG-3′ | ||
| Aofks1 | AO090009000174 | 5′-TGGACTGGAAAGTGGTACAG-3′ |
| 5′-TTGTGTTTCTCGTCAAGGGG-3′ | ||
| gpdA | AO090003001322 | 5′-TCAGAACATCATCCCCAGCT-3′ |
| 5′-ATCGACCTTGGAGATGTAGG-3′ |
Gene ID, gene identification.
For each gene, the top sequence shows the sequence of the forward primer, and the bottom sequence is the sequence of the reverse primer.
Western blot analysis.
Mycelia grown in submerged culture at 30°C for 24 h were harvested, ground to a powder by liquid nitrogen, and suspended in a buffer (10 mM MgSO4, 100 mM Tris-HCl [pH 7.5], 1 mM phenylmethylsulfonyl fluoride, and 1:100 protease inhibitor cocktail [Sigma]). After 1 h at 4°C, the samples were centrifuged at 500 × g for 5 min to remove the debris, and the supernatant was removed and boiled with 4× sample buffer. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 6% acrylamide gel, the samples were transferred to a nitrocellulose membrane. Blocking was performed using 5% skim milk in TBS (2.42 g of Tris, 29.22 g NaCl, and 1 g of Tween 20 [all in 1 liter] [pH 7.5]) for 30 min. The anti-GFP mouse monoclonal antibody (1:5,000 dilution, Clontech) was used as the primary antibody, while the peroxidase-labeled mouse immunoglobulin G antibody (1:500 dilution; Vector) was used as the secondary antibody. Both antibody reactions were performed for 1 h. Detection was carried out using the ECL detection reagents (Pierce) and a luminescent image analyzer LAS-4000miniEPUV (Fujifilm, Japan).
RESULTS
Generation of strains that conditionally express Aoend4.
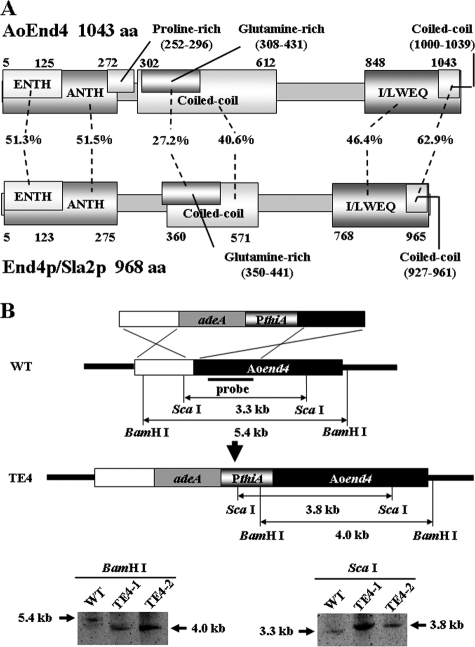
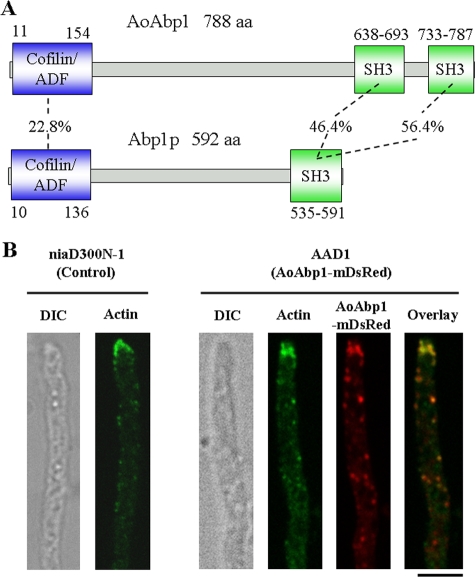
To study the physiological importance of endocytosis in fungal cells, we attempted to generate strains that had endocytic defects. S. cerevisiae cells lacking END4/SLA2 exhibited endocytic defects. Hence, we used the A. oryzae genome database (http://www.bio.nite.go.jp/dogan/MicroTop?GENOME_ID=ao) to clone the cDNA of Aoend4 (DDBJ accession no. AB430739), which is the A. oryzae homolog of S. cerevisiae END4/SLA2, from the A. oryzae RIB40 wild-type strain. We performed rapid amplification of cDNA 5′-end analysis and determined the Aoend4 start codon. The deduced AoEnd4 sequence consists of 1,043 amino acids and displays 34.6% amino acid identity to End4p/Sla2p (Fig. 1A). Based on Pfam (http://pfam.sanger.ac.uk/) motif analysis, AoEnd4 has the ENTH and ANTH domains in the N-terminal region and the I/LWEQ domain in the C-terminal region. Moreover, according to COILS program (http://www.ch.embnet.org/software/COILS_form.html) analysis, AoEnd4 has two coiled-coil regions, which may allow for interactions with other endocytic proteins. These sequence analyses suggested that AoEnd4 is similar to End4p/Sla2p in terms of endocytic functions. AoEnd4 is also similar to A. nidulans SlaB (84.6% amino acid identity) (1).
FIG. 1.
Generation of strains that conditionally express Aoend4. (A) Motif analysis of A. oryzae AoEnd4 and S. cerevisiae End4p/Sla2p. The number of amino acids (aa) and the sequence identity between the corresponding domains are shown. (B) Schematic diagram of the generation of strains that conditionally express Aoend4. Two strains expressing Aoend4 under the thiA promoter from the Aoend4 locus were generated and confirmed by Southern blot analysis using the BamHI and ScaI restriction endonucleases. WT, wild type.
Next, we tried to generate an Aoend4 disruptant. We obtained three heterokaryon strains; however, the Aoend4 disruptant was not obtained, probably because Aoend4 is essential for hyphal growth (data not shown). This result is consistent with that obtained from the A. nidulans slaB mutant (1). Thus, we generated strains in which Aoend4 was expressed under the thiA promoter (PthiA) from the Aoend4 locus. In this construct, PthiA was located just upstream of Aoend4, whereas the endogenous promoter region of Aoend4 was located upstream of PthiA. Thus, Aoend4 was regulated by PthiA and not by the endogenous Aoend4 promoter (Fig. 1B). Two strains were obtained, and these were named the TE4 strains (TE4-1 and TE4-2). For a control, NS13 strain was generated, in which Aoend4 is regulated under its own promoter, and has the same auxotrophy as TE4 strains. TE4 strains were confirmed to be homokaryon strains by Southern blot analysis (Fig. 1B). When cultured in the absence of thiamine, PthiA drives the downstream gene, whereas in the presence of thiamine, PthiA represses it. When cultured with more than 10 μM thiamine, PthiA almost completely represses the downstream gene (29). In the following experiments, we used 10 μM thiamine to shut off Aoend4 expression.
Growth defect in the Aoend4-repressed condition.
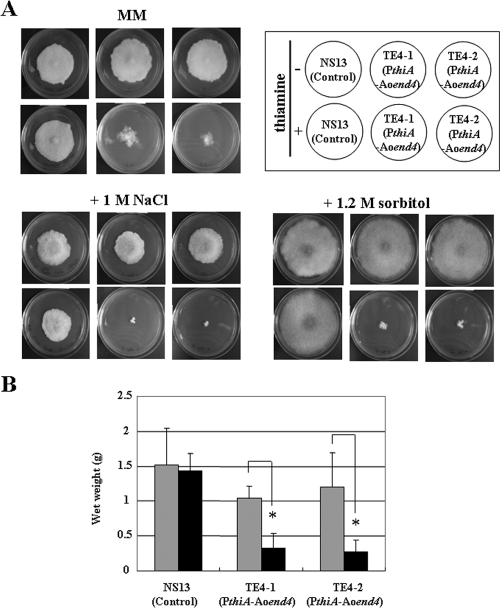
The TE4 strains displayed a remarkable growth defect and formed irregular-shaped colonies on the culture plates as a result of apical growth defects that occurred when thiamine was added to the medium by inoculation with conidia (Fig. 2A). We confirmed that the TE4 strains in the Aoend4-repressed condition also showed a similar growth defect in liquid culture (Fig. 2B). Moreover, a more severe growth defect was observed when the TE4 strains were cultured on plates containing thiamine and 1 M NaCl or 1.2 M sorbitol, suggesting that the TE4 strains in the Aoend4-repressed condition exhibited higher salt and osmotic stress sensitivities (Fig. 2A).
FIG. 2.
Mycelia in the Aoend4-repressed condition exhibit severe growth defects. (A) Approximately 102 conidia of the A. oryzae NS13 (control), TE4-1, and TE4-2 strains were inoculated and incubated on several culture plates at 30°C for 15 days. The strains were cultured in the absence (−) or presence (+) of thiamine on MM alone or supplemented with NaCl and sorbitol. (B) The wet weight of each strain that was grown in the MM liquid medium for 4 days was measured. The gray and black bars indicate culturing in the absence or presence of thiamine, respectively. Five independent experiments were performed. The error bars represent the standard deviations. Values that were significantly different (P < 0.05) by Student's t test are indicated (*).
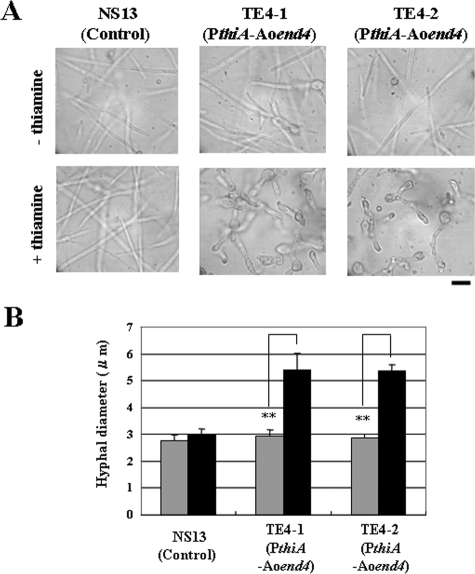
Next, we studied the hyphal morphology of the TE4 strains by microscopy. The hyphal morphology in the Aoend4-repressed condition was abnormal in comparison with that in the Aoend4-expressed condition (Fig. 3A). Moreover, the hyphal diameter in the Aoend4-repressed condition was larger than that in the Aoend4-expressed condition, suggesting defective apical cell polarity in the Aoend4-repressed condition (Fig. 3B). Since the two TE4 strains displayed almost the same growth defects when thiamine was added to the medium, we used the TE4-1 strain as the host strain in subsequent experiments. Furthermore, there was no significant difference in the growth, hyphal diameter, and morphology of strains NS13 and TE4-1 grown in the absence of thiamine; thus, we concluded that TE4-1 grown in the absence of thiamine could be used as a control. Therefore, for the control experiments, we used the phenotype of the TE4-1 strain when it was cultured in the absence of thiamine.
FIG. 3.
Hyphae in the Aoend4-repressed condition display abnormal morphology. (A) Hyphae that were grown in the MM liquid medium in the absence (−) or presence (+) of thiamine at 30°C for 18 h were observed by microscopy. Bar, 10 μm. (B) After 20 h of incubation at 30°C, the maximum width of hyphae within 10 μm from the hyphal tip was measured using the Andor iQ software. For each strain and condition, 20 hyphal tips were measured, and the average diameters are shown. The gray and black bars indicate the hyphal diameters (in microns) cultured in the absence or presence of thiamine, respectively. Four independent experiments were performed. The error bars represent standard deviations. Values that were significantly different (P < 0.01) by Student's t test are indicated (**).
Endocytic defects in the Aoend4-repressed condition.
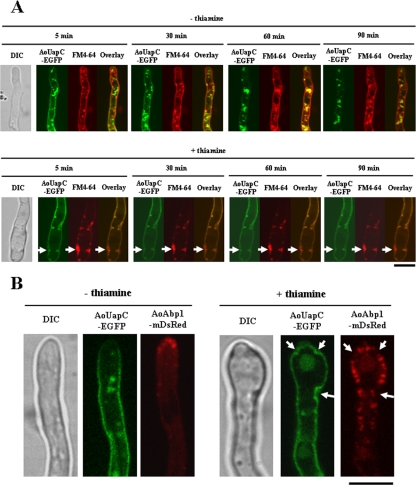
To test whether the hyphae grown in the Aoend4-repressed condition exhibited endocytic defects, the TEUE3 strain, which expresses the AouapC-egfp fusion gene in the conditional Aoend4 background, was generated. AoUapC is a putative plasma membrane purine transporter, and AoUapC-EGFP is internalized from the plasma membrane by endocytosis after ammonium addition to the medium (8). Together with the endocytic induction of AoUapC-EGFP, we applied FM4-64, an amphiphilic styryl endocytic marker (Fig. 4A). Under the Aoend4-expressed condition, AoUapC-EGFP was internalized from the plasma membrane 90 min after ammonium addition. Similarly, FM4-64 was also internalized and observed in the endosomal structures (Fig. 4A, top panels). Moreover, moving endosomal structures labeled with both AoUapC-EGFP and FM4-64 were observed, and these were found to mostly colocalize in the hyphae (Fig. 4A, top panels). When the TEUE3 strain was cultured in the Aoend4-repressed conditions, its hyphal growth was remarkably inhibited as in the case of the host strain TE4-1. In the Aoend4-repressed condition, neither AoUapC-EGFP nor FM4-64 was internalized, and certain parts of the plasma membrane were observed as large invagination structures labeled with AoUapC-EGFP, indicating that endocytosis of both the membrane protein and its lipid cargo was defective in the Aoend4-repressed condition (Fig. 4A, bottom panels, arrows). Furthermore, in contrast to the Aoend4-expressed condition, in the Aoend4-repressed condition, moving endosomal structures labeled with AoUapC-EGFP and FM4-64 were not observed in the hyphae. This result also strongly suggested endocytic deficiencies in the Aoend4-repressed condition.
FIG. 4.
Hyphae in the Aoend4-repressed condition show endocytic deficiency. (A) Endocytosis of AoUapC-EGFP and the FM4-64 stain was induced simultaneously in cells cultured in the absence (−) or presence (+) of thiamine. Arrows show FM4-64 accumulation in the large invagination structures labeled with AoUapC-EGFP. The time after endocytic induction of AoUapC-EGFP and simultaneous FM4-64 staining is shown. The image of a hypha by differential interference contrast (DIC) is shown to the left. Bar, 5 μm. (B) AoAbp1-mDsRed was localized in the hyphal tip region in the absence (−) or presence (+) of thiamine. In the absence of thiamine, AoAbp1-mDsRed was localized not only in the tip but dispersed in the plasma membrane in patches. Note that AoAbp1-mDsRed was hardly localized in the invagination structures labeled with AoUapC-EGFP (arrows). Bar, 5 μm.
S. cerevisiae Abp1p is localized in cortical actin patches in which endocytosis occurs. To detect actin patches in the Aoend4-repressed condition, we cloned Aoabp1 (DDBJ accession no. AB430740), the A. oryzae homolog of S. cerevisiae ABP1. AoAbp1 has 788 residues and shows 19.6% amino acid identity to Abp1p (Fig. 5A). AoAbp1 contains the cofilin/actin depolymerizing factor domain, which interacts with actin in the N-terminal region, and two SH3 (Src homology-3) domains, which are believed to interact with other endocytic proteins in the C-terminal region (Fig. 5A). The important Abp1p motif is conserved in AoAbp1, and AoAbp1 is similar to A. nidulans AbpA (55.7% amino acid identity) (1). Both AoAbp1 and A. nidulans AbpA have two SH3 domains, whereas Abp1p has one SH3 domain. Next, we generated a strain named AAD1 that expresses the Aoabp1-mdsred fusion gene from the niaD locus under the amyB promoter (PamyB). This is a highly inducible promoter that can be induced if the carbon source of the culture medium is changed. Under the culture condition in which Aoabp1-mdsred was expressed at a low level by PamyB (see Materials and Methods), AoAbp1-mDsRed was localized in patches of the plasma membrane, mainly in the hyphal tip region (Fig. 5B). However, it was possible that the AoAbp1-mDsRed localization was artificial and resulted from PamyB overexpression and Aoabp1-mdsred fusion gene ectopic expression. To exclude this possibility, we performed indirect immunofluorescence microscopy using the anti-actin antibody (Fig. 5B). Actin localization in AAD1 was similar to that in the control strain niaD300N-1. AoAbp1-mDsRed and actin were observed as patches and were largely colocalized throughout the hyphae, particularly in the hyphal tip, similar to the localization of A. nidulans AbpA (1). This suggested that actin patches were present wherever AoAbp1-mDsRed localized (Fig. 5B). Next, we generated TEUA1, a strain that expresses AouapC-egfp and Aoabp1-mdsred in the conditional Aoend4 background. When strain TEUA1 was examined in the Aoend4-expressed condition, AoAbp1-mDsRed was mainly localized in the tip region (Fig. 4B, left). In contrast, in the Aoend4-repressed condition, AoAbp1-mDsRed was dispersed not only in the hyphal tip but also in the plasma membrane (Fig. 4B, right). Moreover, the number of AoAbp1-mDsRed patches increased, and these were hardly observed in the aberrant invagination structures labeled with AoUapC-EGFP. These results suggest that AoAbp1 localization is dependent on AoEnd4 and is abnormal in the Aoend4-repressed condition.
FIG. 5.
Motif analysis and localization of AoAbp1. (A) Motif analysis of A. oryzae AoAbp1 and S. cerevisiae Abp1p. The number of amino acids (aa) and the sequence identity between the corresponding domains are shown. Note that AoAbp1 has an extra SH3 domain in comparison with Abp1p at the C terminus. ADF, actin depolymerizing factor. (B) Actin was detected by indirect immunofluorescence microscopy using the anti-actin antibody. AoAbp1-mDsRed was mostly colocalized with actin in the hyphal tip region in AAD1. Actin localization in strain AAD1 was almost the same as that in strain niaD300N-1, which is a control strain, indicating that AoAbp1-mDsRed expression in AAD1 did not alter actin localization. The image of a hypha by differential interference contrast (DIC) is shown to the left. Bar, 5 μm.
Abnormality of apical recycling and secretion in the Aoend4-repressed condition.
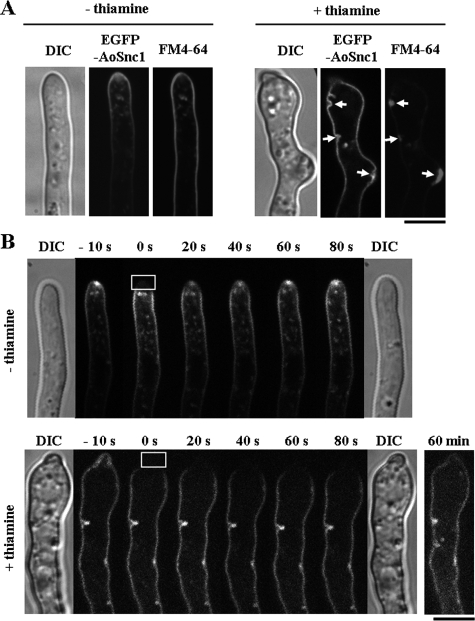
Hyphal tip elongation requires many components, some of which are possibly recycled to the tip region by endocytosis. AoSnc1, a vesicle SNARE protein localized both in the plasma membrane and endosome, is a candidate component for endocytic recycling in the tip region (16). In the Aoend4-expressed condition, EGFP-AoSnc1 was mainly localized in the tip region of the plasma membrane and Spitzenkörper-like structure (Fig. 6A, left). Membrane compartments labeled with EGFP-AoSnc1 were also stained with FM4-64 (Fig. 6A, left). In contrast, in the Aoend4-repressed condition, EGFP-AoSnc1 was not localized in the tip region and Spitzenkörper-like structure but accumulated throughout the plasma membrane. This suggested that EGFP-AoSnc1 is not recycled from the plasma membrane to the tip region by apical recycling of endocytosis and that the defects in SNARE protein recycling result in abnormal secretion to the tip, leading to defective apical polarity (Fig. 6A, right). Moreover, in the Aoend4-repressed condition, FM4-64 accumulated in the aberrant invagination structures on the plasma membrane that were labeled with EGFP-AoSnc1 (Fig. 6A, right, arrows).
FIG. 6.
Apical recycling and secretory defects in the Aoend4-repressed condition. (A) In the absence (−) of thiamine, EGFP-AoSnc1 was localized mainly in the tip region and was also stained with FM4-64. In contrast, in the presence (+) of thiamine, EGFP-AoSnc1 localization was altered, and it was localized throughout the plasma membrane. FM4-64 accumulated in the large invagination structures labeled with EGFP-AoSnc1 (arrows). The image of a hypha by differential interference contrast (DIC) is shown to the left. Bar, 5 μm. (B) The FRAP analysis was performed in the hyphal tip regions. The time after photobleaching is shown. Photobleaching was carried out in the squares outlined in white of the photographs taken at 0 s; these were taken just after photobleaching. DIC images were taken before (left) and after (right) photobleaching, and these revealed that there was no damage to the hyphae by laser irradiation. The fluorescence recovery of EGFP-AoSnc1 in the hyphal tip was observed in the presence of thiamine at 60 min after photobleaching. Bar, 5 μm.
To investigate the dynamics of EGFP-AoSnc1 in the hyphal tip in the Aoend4-repressed condition, we performed FRAP analysis. In the Aoend4-expressed condition, EGFP-AoSnc1 fluorescence was recovered in the Spitzenkörper-like structure in the tip region approximately 1 min after photobleaching (Fig. 6B, top). However, in the Aoend4-repressed condition, EGFP-AoSnc1 fluorescence was not recovered in the tip region at more than 1 min after photobleaching (Fig. 6B, bottom), although it was observed in this region 60 min after photobleaching, possibly as a result of membrane flux (Fig. 6B, bottom, right). These FRAP analyses demonstrated that in the Aoend4-repressed condition, secretion in the tip region is abnormal.
Regulation of cell wall components is abnormal in the Aoend4-repressed condition.
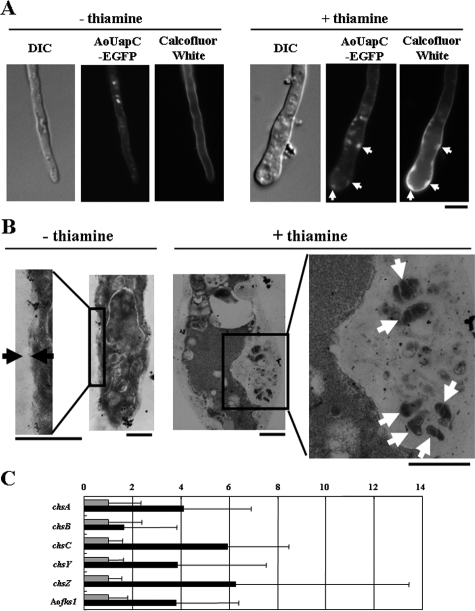
The TEM analysis showed that the cell wall of Δend4/sla2 mutants is thicker than that of the wild-type strains of S. cerevisiae and Schizosaccharomyces pombe (5, 6). Thus, we stained hyphae grown in the Aoend4-repressed condition with calcofluor white, which stains chitin, the major cell wall component in filamentous fungi (Fig. 7A). In the Aoend4-repressed condition, abnormal accumulation of the cell wall labeled with calcofluor white was observed, indicating that the cell wall in the Aoend4-repressed condition was abnormal in comparison with that in the Aoend4-expressed condition. However, in contrast to budding and fission yeasts, the cell wall in the Aoend4-repressed condition was not uniformly thicker at the cell surface but was thicker at specific sites on the cell surface. Moreover, the accumulation of the cell wall coincided with the presence of large invagination structures labeled with AoUapC-EGFP (Fig. 7A, right, arrows).
FIG. 7.
Abnormality in cell wall regulation in the Aoend4-repressed condition. (A) Chitin was stained with calcofluor white. The TEUE3 strain was grown in the absence (−) or presence (+) of thiamine. In the presence of thiamine, chitin accumulated in the large invagination structures labeled with AoUapC-EGFP (arrows). The image of a hypha by differential interference contrast (DIC) is shown to the left. Bar, 5 μm. (B) In the absence of thiamine, the cell wall could be seen between the black arrows. In contrast, in the presence of thiamine, the cell wall became thicker, and non-cell wall components were observed as white arrows. Bars, 1 μm. (C) Real-time RT-PCR analysis was performed. The gray and black bars indicate culturing in the absence or presence of thiamine, respectively. The expression values of each gene were normalized to those of gpdA, and those in the absence of thiamine were fixed at 1. For each gene, the expression value in the presence of thiamine is indicated as the change (0-fold change to 14-fold change) relative to those in the absence of thiamine. Five independent experiments were performed. The error bars represent the standard deviations.
For a more precise analysis of the sites where the cell wall was thicker, we performed TEM analysis (Fig. 7B). In the Aoend4-expressed condition, the width of the cell wall was uniform (Fig. 7B, left). In the Aoend4-repressed condition, it was confirmed that the cell wall was thicker, and a non-cell wall component was observed in the large invagination structures (Fig. 7B, right, arrows), suggesting that cell wall regulation is abnormal in this condition.
On the basis of our observation that the cell wall is abnormal in the Aoend4-repressed condition, we further analyzed the expression of cell wall synthases by real-time RT-PCR in the Aoend4-expressed and Aoend4-repressed conditions (Fig. 7C). Five chitin synthases (chsA, chsB, chsC, chsY, and chsZ) and one 1,3-beta-glucan synthase (Aofks1), which display comparatively high expression under normal culture conditions, were selected for expression analyses. These six cell wall synthases tended to be expressed at higher levels in the Aoend4-repressed condition than in the Aoend4-expressed condition, suggesting that the upregulation of cell wall synthases is due to endocytic defects.
Complementation analysis of Aoend4.
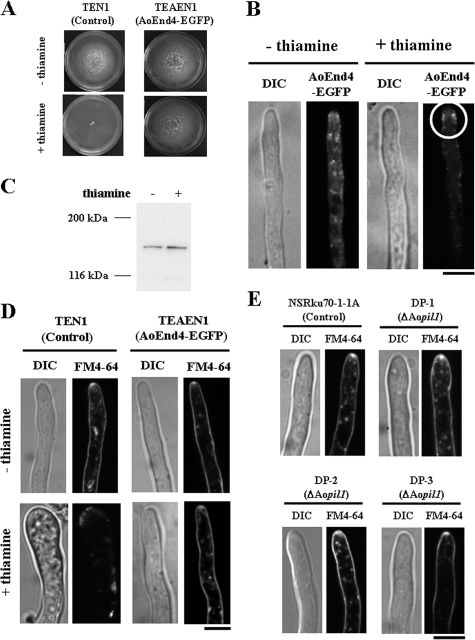
To complement the phenotype in the Aoend4-repressed condition and analyze AoEnd4 localization, we generated a strain that expressed the Aoend4-egfp fusion gene under PamyB at the niaD locus in the conditional Aoend4 background. When the control strain TEN1 was cultured in the endogenous Aoend4-repressed condition, mycelial growth was more severely inhibited than in the host strain, presumably due to differences in auxotrophy (Fig. 8A). In contrast, TEAEN1, a strain that expressed AoEnd4-EGFP in the conditional Aoend4 background, formed normal-sized and round colonies in the endogenous Aoend4-repressed condition; these colonies were similar to those formed in the endogenous Aoend4-expressed condition, even when Aoend4-egfp expression was low (Fig. 8A). This indicated that TEAEN1 growth is complemented in the endogenous Aoend4-repressed condition, and the AoEnd4-EGFP fusion protein is functional.
FIG. 8.
Complementation analysis of Aoend4. (A) Growth of the TEN1 and TEAEN1 strains on plates with (+) or without (−) thiamine. During culture in the presence of thiamine, the growth of the TEN1 mycelium was remarkably inhibited, whereas that of TEAEN1 was rescued. (B) AoEnd4-EGFP localization in the presence or absence of thiamine. In the absence of thiamine, AoEnd4-EGFP was scattered in the cytoplasm. In contrast, in the presence of thiamine, AoEnd4-EGFP was localized in the plasma membrane as patches, mostly in the apical region, slightly away from the apex (circle). The image of a hypha by differential interference contrast (DIC) is shown to the left. Bar, 5 μm. (C) Western blot analysis for detecting AoEnd4-EGFP in the presence or absence of thiamine using the anti-GFP antibody. The predicted molecular mass of AoEnd4-EGFP was approximately 145 kDa, and its expression was ascertained in the presence or absence of thiamine. (D) In the presence of thiamine, FM4-64 was not internalized in TEN1 hyphae but was internalized in those of TEAEN1. Photographs were taken approximately 30 min after FM4-64 staining. Bar, 5 μm. (E) Endocytic compartments, including Spitzenkörper-like structures, were stained with FM4-64 in the hyphal tip region of each strain. Bar, 5 μm.
We next analyzed AoEnd4-EGFP localization by using confocal microscopy. When hyphae were cultured in the endogenous Aoend4-expressed condition, AoEnd4-EGFP was localized not at specific sites but in the cytoplasm, suggesting that AoEnd4-EGFP could not localize to the appropriate region by competing with endogenous AoEnd4. On the other hand, in the endogenous Aoend4-repressed condition, AoEnd4-EGFP was mainly localized on the sides of the hyphal tip and partly in patches of the plasma membrane (Fig. 8B), which is consistent with A. nidulans SlaB localization (1). Western blot analysis was performed to confirm the expression of the AoEnd4-EGFP fusion protein in the endogenous Aoend4-expressed or Aoend4-repressed condition (Fig. 8C).
To verify the rescue of endocytosis in the endogenous Aoend4-repressed condition, we performed FM4-64 staining (Fig. 8D). When the TEN1 strain was cultured in the endogenous Aoend4-repressed condition, FM4-64 was not internalized, and instead, it accumulated in the plasma membrane. In contrast, when the TEAEN1 strain was cultured in both the endogenous Aoend4-expressed and Aoend4-repressed conditions, FM4-64 was internalized from the plasma membrane, and Spitzenkörper-like structures and endocytic compartments, including the vacuolar membrane, were stained. These results demonstrated that Aoend4 repression caused the phenotypes described above.
S. cerevisiae Pil1p functions as a component of the eisosome, which is a plasma membrane domain in which endocytosis occurs, and disruption of its gene results in the formation of aberrant invagination structures (42). To investigate whether Aopil1 (DDBJ accession no. AB430741), the sole A. oryzae homolog of PIL1, functions in endocytosis, we generated DP strains, Aopil1 disruptants. To investigate whether endocytosis occurred in the DP strains, we stained the samples with FM4-64. In the control and DP strains, FM4-64 was internalized, and Spitzenkörper-like structures and endocytic compartments were stained, indicating that normal endocytosis occurs in the DP strains (Fig. 8E). This result suggested that unlike AoEnd4, AoPil1 does not play a crucial role in endocytosis.
DISCUSSION
Recently, in filamentous fungi, the localization of endocytic components, such as A. nidulans AbpA, AmpA, and SlaB, was analyzed in living hyphae (1). However, there are few reports on the physiological importance of endocytosis in filamentous fungi, mainly due to the lack of reliable indicators of endocytosis. In this study, we investigated the physiological roles of endocytosis in filamentous fungi by generating strains that conditionally express Aoend4 and analyzed their phenotypes in the Aoend4-repressed condition.
On the basis of motif analysis, AoEnd4 is the homolog of S. cerevisiae End4p/Sla2p, suggesting that the function of AoEnd4 is similar to that of End4p/Sla2p in endocytosis. The A. nidulans fimA disruptants, fimA being the homolog of S. cerevisiae SAC6, also displayed deficiency in FM4-64 internalization, which was similar to that observed in the Aoend4-repressed condition (41). In the Aoend4-repressed condition, AoAbp1 was not localized at the hyphal tip but was dispersed in the plasma membrane as cortical patches. AoAbp1 was capable of localizing in the plasma membrane but not only in the tip region, indicating that AoEnd4 was not required for the formation of cortical actin patches and AoAbp1 localization was dependent on AoEnd4. In S. cerevisiae Δend4/sla2 cells, actin comet tails are observed, and the amount of actin increases, suggesting that End4p/Sla2p negatively regulates actin polymerization (9, 13). In the Aoend4-repressed condition, the number of AoAbp1 patches in the plasma membrane increased, which is consistent with the result from S. cerevisiae Δend4/sla2 cells. However, the AoAbp1 patches were hardly localized in the invagination structures. This result suggests that the components required for AoAbp1 recruitment do not localize in the invagination structures. However, further analyses of other endocytic components in filamentous fungi are required.
S. cerevisiae and C. albicans Δend4/sla2 cells display defects in filamentous growth (2, 44). In A. nidulans, SlaB is not required for hyphal germination but is essential for hyphal growth (1); this is consistent with the results obtained in the Aoend4-repressed condition in this study, which suggest that endocytosis is not required for hyphal germination but is required for polarized growth. Hyphae that showed the endocytic defect displayed aberrant hyphal morphology, probably due to the lack of endocytosis, which is the counterpart of exocytosis, which is required for hyphal growth in the tip region. In the Aoend4-repressed condition, high sensitivity to salt and osmotic stress was probably caused by endocytic defects in channels or receptors that sense the outer environment. In A. nidulans, ambient pH signaling might be regulated by the endocytosis of a seven-transmembrane protein PalH (7, 26).
Due to continuous tip elongation in filamentous fungi, it is thought that these organisms need to recycle certain components, such as cell wall-building enzymes, to the tip region (34, 41). Calcofluor white staining revealed that chitin, a major cell wall component in filamentous fungi, is accumulated in aberrant invagination structures in the Aoend4-repressed condition. In addition, TEM analysis revealed the presence of non-cell wall components in the aberrant invagination structures. These results predict that proteins involved in cell wall synthesis, such as chitin synthases, probably could not be recycled to the tip region and therefore accumulated in the aberrant invagination structures. Moreover, we found that the expression of cell wall synthases increased in the Aoend4-repressed condition. One possible explanation for the high expression of these genes is that regulatory component(s) of these cell wall synthases cannot be internalized by endocytosis. Of the cell wall synthases analyzed in this study, A. oryzae ChsY and ChsZ are the homologs of A. nidulans CsmA and CsmB, respectively (4, 39, 40). These chitin synthases have a myosin motor-like domain (MMD) and directly interact with actin, which has its most important functions in endocytosis. Thus, chitin synthases with MMD are thought to be possibly associated with endocytic recycling. Although MMD does not function like the myosin motor, which uses ATP, but interacts only with actin (39, 40), there is a possibility that chitin synthases with MMD bind to actin through this domain and undergo endocytic recycling to the tip region.
Filamentous fungi are important and have been examined in both basic and applied studies. Filamentous fungi, including A. oryzae, are potential hosts for heterologous protein production. However, there are many obstacles for protein production in these organisms. Further clarification and a better understanding of intracellular trafficking involving the endocytic pathway are required because endocytosis and exocytosis are closely related (34, 37). Further, filamentous fungi are now regarded as model organisms in studies on apical growth, and their machinery in the apical region, in which both exocytosis and endocytosis are vital, is being investigated (1, 34, 37, 41). Our results strongly indicate that endocytosis has a crucial role in hyphal growth by recycling of secretory SNARE and components of the plasma membrane, such as cell wall synthases in the tip region. However, endocytosis in filamentous fungi has not been investigated in detail so far. Endocytosis is a part of several physiological processes in many eukaryotic cells, and better understanding of filamentous fungi requires elucidation of its physiological roles in the organisms.
Acknowledgments
We thank S. Yamashita for TEM analysis. Y.H. was supported by research fellowships for young scientists from the Japan Society for the Promotion of Science.
This study was supported by Grant-in-Aid for Scientific Research (S) and Grant-in-Aid for Scientific Research on Priority Areas “Applied Genomics” from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 21 November 2008.
REFERENCES
- 1.Araujo-Bazán, L., M. A. Peñalva, and E. A. Espeso. 2008. Preferential localization of the endocytic internalization machinery to hyphal tips underlies polarization of the actin cytoskeleton in Aspergillus nidulans. Mol. Microbiol. 67891-905. [DOI] [PubMed] [Google Scholar]
- 2.Asleson, C. M., E. S. Bensen, C. A. Gale, A. S. Melms, C. Kurischko, and J. Berman. 2001. Candida albicans INT1-induced filamentation in Saccharomyces cerevisiae depends on Sla2p. Mol. Cell. Biol. 211272-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baggett, J. J., K. E. D'Aquino, and B. Wendland. 2003. The Sla2p talin domain plays a role in endocytosis in Saccharomyces cerevisiae. Genetics 1651661-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chigira, Y., K. Abe, K. Gomi, and T. Nakajima. 2002. chsZ, a gene for a novel class of chitin synthase from Aspergillus oryzae. Curr. Genet. 41261-267. [DOI] [PubMed] [Google Scholar]
- 5.Ge, W., T. G. Chew, V. Wachtler, S. N. Naqvi, and M. K. Balasubramanian. 2005. The novel fission yeast protein Pal1p interacts with Hip1-related Sla2p/End4p and is involved in cellular morphogenesis. Mol. Biol. Cell 164124-4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gourlay, C. W., H. Dewar, D. T. Warren, R. Costa, N. Satish, and K. R. Ayscough. 2003. An interaction between Sla1p and Sla2p plays a role in regulating actin dynamics and endocytosis in budding yeast. J. Cell Sci. 1162551-2564. [DOI] [PubMed] [Google Scholar]
- 7.Herranz, S., J. M. Rodriguez, H. J. Bussink, J. C. Sanchez-Ferrero, H. N. Arst, Jr., M. A. Peñalva, and O. Vincent. 2005. Arrestin-related proteins mediate pH signaling in fungi. Proc. Natl. Acad. Sci. USA 10212141-12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higuchi, Y., T. Nakahama, J. Y. Shoji, M. Arioka, and K. Kitamoto. 2006. Visualization of the endocytic pathway in the filamentous fungus Aspergillus oryzae using an EGFP-fused plasma membrane protein. Biochem. Biophys. Res. Commun. 340784-791. [DOI] [PubMed] [Google Scholar]
- 9.Holtzman, D. A., S. Yang, and D. G. Drubin. 1993. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J. Cell Biol. 122635-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horiuchi, H., M. Fujiwara, S. Yamashita, A. Ohta, and M. Takagi. 1999. Proliferation of intrahyphal hyphae caused by disruption of csmA, which encodes a class V chitin synthase with a myosin motor-like domain in Aspergillus nidulans. J. Bacteriol. 1813721-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin, F. J., J. Maruyama, P. R. Juvvadi, M. Arioka, and K. Kitamoto. 2004. Adenine auxotrophic mutants of Aspergillus oryzae: development of a novel transformation system with triple auxotrophic hosts. Biosci. Biotechnol. Biochem. 68656-662. [DOI] [PubMed] [Google Scholar]
- 12.Kaksonen, M., C. P. Toret, and D. G. Drubin. 2005. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell 123305-320. [DOI] [PubMed] [Google Scholar]
- 13.Kaksonen, M., Y. Sun, and D. G. Drubin. 2003. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115475-487. [DOI] [PubMed] [Google Scholar]
- 14.Kimura, S., J. Maruyama, M. Takeuchi, and K. Kitamoto. 2008. Monitoring global gene expression of proteases and improvement of human lysozyme production in the nptB gene disruptant of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 72499-505. [DOI] [PubMed] [Google Scholar]
- 15.Kitamoto, K. 2002. Molecular biology of the koji molds. Adv. Appl. Microbiol. 51129-153. [DOI] [PubMed] [Google Scholar]
- 16.Kuratsu, M., A. Taura, J. Y. Shoji, S. Kikuchi, M. Arioka, and K. Kitamoto. 2007. Systematic analysis of SNARE localization in the filamentous fungus Aspergillus oryzae. Fungal Genet. Biol. 441310-1323. [DOI] [PubMed] [Google Scholar]
- 17.Mabashi, Y., T. Kikuma, J. Maruyama, M. Arioka, and K. Kitamoto. 2006. Development of a versatile expression plasmid construction system for Aspergillus oryzae and its application to visualization of mitochondria. Biosci. Biotechnol. Biochem. 701882-1889. [DOI] [PubMed] [Google Scholar]
- 18.Machida, M., K. Asai, M. Sano, T. Tanaka, T. Kumagai, G. Terai, K. Kusumoto, T. Arima, O. Akita, Y. Kashiwagi, K. Abe, K. Gomi, H. Horiuchi, K. Kitamoto, T. Kobayashi, M. Takeuchi, D. W. Denning, J. E. Galagan, W. C. Nierman, J. Yu, D. B. Archer, J. W. Bennett, D. Bhatnagar, T. E. Cleveland, N. D. Fedorova, O. Gotoh, H. Horikawa, A. Hosoyama, M. Ichinomiya, R. Igarashi, K. Iwashita, P. R. Juvvadi, M. Kato, Y. Kato, T. Kin, A. Kokubun, H. Maeda, N. Maeyama, J. Maruyama, H. Nagasaki, T. Nakajima, K. Oda, K. Okada, I. Paulsen, K. Sakamoto, T. Sawano, M. Takahashi, K. Takase, Y. Terabayashi, J. R. Wortman, O. Yamada, Y. Yamagata, H. Anazawa, Y. Hata, Y. Koide, T. Komori, Y. Koyama, T. Minetoki, S. Suharnan, A. Tanaka, K. Isono, S. Kuhara, N. Ogasawara, and H. Kikuchi. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 4381157-1161. [DOI] [PubMed] [Google Scholar]
- 19.Maruyama, J., S. Kikuchi, and K. Kitamoto. 2006. Differential distribution of the endoplasmic reticulum network as visualized by the BipA-EGFP fusion protein in hyphal compartments across the septum of the filamentous fungus, Aspergillus oryzae. Fungal Genet. Biol. 43642-654. [DOI] [PubMed] [Google Scholar]
- 20.Minetoki, T., Y. Nunokawa, K. Gomi, K. Kitamoto, C. Kumagai, and G. Tamura. 1996. Deletion analysis of promoter elements of the Aspergillus oryzae agdA gene encoding alpha-glucosidase. Curr. Genet. 30432-438. [DOI] [PubMed] [Google Scholar]
- 21.Newpher, T. M., R. P. Smith, V. Lemmon, and S. K. Lemmon. 2005. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev. Cell 987-98. [DOI] [PubMed] [Google Scholar]
- 22.Newpher, T. M., and S. K. Lemmon. 2006. Clathrin is important for normal actin dynamics and progression of Sla2p-containing patches during endocytosis in yeast. Traffic 7574-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohneda, M., M. Arioka, H. Nakajima, and K. Kitamoto. 2002. Visualization of vacuoles in Aspergillus oryzae by expression of CPY-EGFP. Fungal Genet. Biol. 3729-38. [DOI] [PubMed] [Google Scholar]
- 24.Reference deleted.
- 25.Peñalva, M. A. 2005. Tracing the endocytic pathway of Aspergillus nidulans with FM4-64. Fungal Genet. Biol. 42963-975. [DOI] [PubMed] [Google Scholar]
- 26.Peñalva, M. A., J. Tilburn, E. Bignell, and H. N. Arst, Jr. 2008. Ambient pH gene regulation in fungi: making connections. Trends Microbiol. 16291-300. [DOI] [PubMed] [Google Scholar]
- 27.Raths, S., J. Rohrer, F. Crausaz, and H. Riezman. 1993. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J. Cell Biol. 12055-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Read, N. D., and E. R. Kalkman. 2003. Does endocytosis occur in fungal hyphae? Fungal Genet. Biol. 39199-203. [DOI] [PubMed] [Google Scholar]
- 29.Shoji, J. Y., J. Maruyama, M. Arioka, and K. Kitamoto. 2005. Development of Aspergillus oryzae thiA promoter as a tool for molecular biological studies. FEMS Microbiol. Lett. 24441-46. [DOI] [PubMed] [Google Scholar]
- 30.Shoji, J. Y., M. Arioka, and K. Kitamoto. 2008. Dissecting cellular components of the secretory pathway in filamentous fungi: insights into their application for protein production. Biotechnol. Lett. 307-14. [DOI] [PubMed] [Google Scholar]
- 31.Shoji, J. Y., M. Arioka, and K. Kitamoto. 2006. Vacuolar membrane dynamics in the filamentous fungus Aspergillus oryzae. Eukaryot. Cell 5411-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shoji, J. Y., Y. Higuchi, J. Maruyama, and K. Kitamoto. 2008. Polarized distribution of intracellular organelles involved in vesicular trafficking in filamentous fungal cells. Tanpakushitsu Kakusan Koso 53753-759. [PubMed] [Google Scholar]
- 33.Smythe, E., and K. R. Ayscough. 2006. Actin regulation in endocytosis. J. Cell Sci. 1194589-4598. [DOI] [PubMed] [Google Scholar]
- 34.Steinberg, G. 2007. On the move: endosomes in fungal growth and pathogenicity. Nat. Rev. Microbiol. 5309-316. [DOI] [PubMed] [Google Scholar]
- 35.Sun, Y., M. Kaksonen, D. T. Madden, R. Schekman, and D. G. Drubin. 2005. Interaction of Sla2p's ANTH domain with PtdIns(4,5)P2 is important for actin-dependent endocytic internalization. Mol. Biol. Cell 16717-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tada, S., K. Gomi, K. Kitamoto, K. Takahashi, G. Tamura, and S. Hara. 1991. Construction of a fusion gene comprising the Taka-amylase A promoter and the Escherichia coli beta-glucuronidase gene and analysis of its expression in Aspergillus oryzae. Mol. Gen. Genet. 229301-306. [DOI] [PubMed] [Google Scholar]
- 37.Taheri-Talesh, N., T. Horio, L. Araujo-Bazán, X. Dou, E. A. Espeso, M. A. Peñalva, S. A. Osmani, and B. R. Oakley. 2008. The tip growth apparatus of Aspergillus nidulans. Mol. Biol. Cell 191439-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahashi, T., T. Masuda, and Y. Koyama. 2006. Identification and analysis of Ku70 and Ku80 homologs in the koji molds Aspergillus sojae and Aspergillus oryzae. Biosci. Biotechnol. Biochem. 70135-143. [DOI] [PubMed] [Google Scholar]
- 39.Takeshita, N., A. Ohta, and H. Horiuchi. 2005. CsmA, a class V chitin synthase with a myosin motor-like domain, is localized through direct interaction with the actin cytoskeleton in Aspergillus nidulans. Mol. Biol. Cell 161961-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeshita, N., S. Yamashita, A. Ohta, and H. Horiuchi. 2006. Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor-like domain, perform compensatory functions that are essential for hyphal tip growth. Mol. Microbiol. 591380-1394. [DOI] [PubMed] [Google Scholar]
- 41.Upadhyay, S., and B. D. Shaw. 2008. The role of actin, fimbrin, and endocytosis in growth of hyphae in Aspergillus nidulans. Mol. Microbiol. 68690-705. [DOI] [PubMed] [Google Scholar]
- 42.Walther, T. C., J. H. Brickner, P. S. Aguilar, S. Bernales, C. Pantoja, and P. Walter. 2006. Eisosomes mark static sites of endocytosis. Nature 439998-1003. [DOI] [PubMed] [Google Scholar]
- 43.Wesp, A., L. Hicke, J. Palecek, R. Lombardi, T. Aust, A. L. Munn, and H. Riezman. 1997. End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell 82291-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, S., M. J. Cope, and D. G. Drubin. 1999. Sla2p is associated with the yeast cortical actin cytoskeleton via redundant localization signals. Mol. Biol. Cell 102265-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]