Abstract
The sucrose nonfermenting 1 (SNF1) protein kinase of yeast plays a central role in the transcription of glucose-repressible genes in response to glucose starvation. In this study, we deleted an ortholog of SNF1 from Gibberella zeae to characterize its functions by using a gene replacement strategy. The mycelial growth of deletion mutants (ΔGzSNF1) was reduced by 21 to 74% on diverse carbon sources. The virulence of ΔGzSNF1 mutants on barley decreased, and the expression of genes encoding cell-wall-degrading enzymes was reduced. The most distinct phenotypic changes were in sexual and asexual development. ΔGzSNF1 mutants produced 30% fewer perithecia, which matured more slowly, and asci that contained one to eight abnormally shaped ascospores. Mutants in which only the GzSNF1 catalytic domain was deleted had the same phenotype changes as the ΔGzSNF1 strains, but the phenotype was less extreme in the mutants with the regulatory domain deleted. In outcrosses between the ΔGzSNF1 mutants, each perithecium contained ∼70% of the abnormal ascospores, and ∼50% of the asci showed unexpected segregation patterns in a single locus tested. The asexual spores of the ΔGzSNF1 mutants were shorter and had fewer septa than those of the wild-type strain. The germination and nucleation of both ascospores and conidia were delayed in ΔGzSNF1 mutants in comparison with those of the wild-type strain. GzSNF1 expression and localization depended on the developmental stage of the fungus. These results suggest that GzSNF1 is critical for normal sexual and asexual development in addition to virulence and the utilization of alternative carbon sources.
Gibberella zeae (anamorph Fusarium graminearum) is an important plant pathogen that is the major cause of Fusarium head blight (FHB) of cereal crops, such as wheat, barley, and rice. The fungus causes severe yield losses in many regions of the world and produces mycotoxins that are harmful to humans and animals (13). It produces sexual spores (ascospores) within perithecia and asexual spores (conidia) that are resistant to unfavorable environmental conditions and well suited for dispersal into susceptible host tissue. Both ascospores and conidia may serve as disease inocula, but ascospores may be more important than conidia in FHB epidemics because FHB inoculum requires aerial dispersal to the cereal heads (53) and ascospores are forcibly discharged into the air (56, 57). Spore dispersal gradients are consistent with the disease foci of G. zeae originating from airborne ascospores (14). Thus, sexual and asexual forms of reproduction of G. zeae are important developmental processes for disease outbreak and are regulated by specific pathways (16, 17, 36, 44, 52).
Mating type (MAT) loci control sexual development in many ascomycete fungi (12). In G. zeae, the MAT1-1 and MAT1-2 idiomorphs are tightly linked on the same chromosome and coexist within a single nucleus (65). The inactivation of either the MAT1-1 or MAT1-2 function results in self-sterility, but strains with MAT1-1 or MAT1-2 deleted can be outcrossed with wild-type strains and with each other (35). Some genes required for sexual development have been identified in G. zeae, but most have pleiotropic functions, such as virulence and other mycological traits (18, 19, 28, 33, 47, 51, 59, 63). In contrast, the molecular mechanisms underlying conidial development and germination in G. zeae remain largely unknown, although genes whose expression changes during conidial germination have recently been identified in microarray studies (52). The deletion of mes1 in G. zeae reduces sexual and asexual reproduction; in particular, mutants with mes1 deleted alter cell wall deposition and the shape of conidia and hyphae (47).
During the infection process, plant pathogenic fungi usually are nutrient deprived until they gain access to the living tissue. Thus, cell-wall-degrading enzymes may play an important role in nutrient acquisition. Fungi usually control the expression of cell-wall-degrading enzymes via protein phosphorylation catalyzed by protein kinases in the signal transduction pathway. Among protein kinases, sucrose nonfermenting 1 (SNF1) kinase is implicated in responses to nutritional and environmental stresses (1, 50).
SNF1 encodes a serine/threonine protein kinase that plays a central role in carbon catabolite repression (8) and is highly conserved in eukaryotes, including mammals, plants, and fungi (6, 23). SNF1 kinase complexes contain the α subunit encoded by SNF1; the β subunits encoded by GAL83, SIP1, and SIP2; and the γ subunit encoded by SNF4. SNF1 is phosphorylated and activated by upstream kinases, such as SAK1, TOS3, and ELM1, and is inactivated by the REG1-GLC7 protein phosphatase (49). When glucose starvation occurs, SNF1 induces the derepression of glucose-repressed genes, including SUC2, which encodes the invertase that hydrolyzes sucrose to glucose and fructose. In addition to enabling metabolic adaptation to different environmental conditions, SNF1 affects many developmental processes, such as sporulation (7), filamentation and invasive growth (10), survival, and life span (20, 50).
The role of SNF1 in fungal virulence has been examined in several plant pathogenic fungi. The SNF1 kinase gene of Cochliobolus carbonum (CcSNF1), a foliar pathogen of maize, is required for the expression of numerous cell-wall-degrading enzymes (55). CcSNF1 is also required for the fungal penetration of the host through its control of cell wall degradation and the metabolism of the resulting sugars. SNF1 mutations in Fusarium oxysporum, a vascular wilt fungus, do not produce cell-wall-degrading enzymes, cannot utilize some carbon sources, and reduce virulence (42). The SNF1 of Colletotrichum gloeosporioides f. sp. malvae (CgSNF1) is highly expressed during appressorium formation and is weakly expressed during subsequent necrotrophic growth in the host, suggesting that CgSNF1 regulates host penetration (15).
The importance of SNF1 in the virulence and reproduction of G. zeae is currently unknown. To address this question, we have functionally characterized the SNF1 ortholog from G. zeae. The mutant with GzSNF1 deleted (ΔGzSNF1) exhibited a dramatic decrease in virulence on barley and was severely impaired in terms of both sexual and asexual reproduction. Perithecium maturation and spore germination are considerably delayed in ΔGzSNF1 strains relative to the wild-type strain. Our results provide the first evidence for a central role for SNF1 during the reproduction process in G. zeae.
MATERIALS AND METHODS
Strain and culture conditions.
Gibberella zeae wild-type strain GZ3639 isolated from Kansas (3) and mutants derived from it were stored as frozen conidial suspensions in 20% glycerol at −80°C (see Table S1 in the supplemental material). The growth of the wild-type and mutants was measured on minimal medium (MM) (37) supplemented with 2% (wt/vol) arabinose, fructose, galactose, glucose, sucrose, trehalose, xylan, or xylose (Sigma-Aldrich Co., St. Louis, MO). For sexual reproduction, the strains were incubated on carrot agar as previously described (37). The number of perithecia on each plate was counted by defining an arbitrarily selected area (35 mm2). Eight areas were counted from each plate by using a grid layover in Adobe Photoshop. For conidial production, strains were inoculated in carboxymethyl cellulose (CMC) (5) and yeast malt agar (YMA) (21) were used as previously described. The Saccharomyces cerevisiae snf1D mutant strain [BY4742 snf1::kanMX4] obtained from EUROSCARF (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/) was used for the complementation studies.
Nucleic acid manipulations, primers, and PCR conditions.
Fungal genomic DNA and total RNA were prepared as previously described (18). Standard procedures were used for restriction endonuclease digestion, agarose gel electrophoresis, and Southern and Northern hybridizations (48). The PCR primers (see Table S2 in the supplemental material) used in this study were synthesized by the Bioneer oligonucleotide synthesis facility (Bioneer, Daejeon, South Korea). General PCRs were performed as previously described (32). Plasmid DNA was purified from Escherichia coli grown in 3 ml of LB medium (48) for 18 h at 37°C by using a plasmid purification kit (NucleoGen, Siheung, South Korea). DNA sequencing was performed at the National Instrumentation Center for Environmental Management (Seoul National University). Total RNA was extracted from mycelia or conidia by using an Easy-Spin total RNA extraction kit (Intron Biotech., Seongnam, South Korea), and the first strand cDNA was synthesized by using SuperScriptIII reverse transcriptase (Invitrogen, Carlsbad, CA). Reverse transcriptase PCR (RT-PCR) used AccuPowerRT/PCR PreMix (Bioneer). Quantitative real-time PCR (qRT-PCR) was performed with the Sybr green super mix (Bio-Rad, Hercules, CA) and a 7500 real-time PCR system (Applied Biosystems, Foster, CA). The PCRs were repeated three times, with three replicates per run.
Elongation factor 1-beta (EF1b [FGSG_01008.3]) was used as an endogenous control for normalization. The changes in fluorescence of the Sybr green dye in each cycle were monitored by the system software, and the threshold cycle (CT) above the background for each reaction was calculated. The CT value of EF1b was subtracted from that of GzSNF1 to obtain a ΔCT value. The ΔCT value of an arbitrary calibrator was subtracted from the ΔCT value of each sample to obtain a ΔΔCT value. The GzSNF1 expression level relative to the calibrator was expressed as 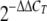 . Tukey's test using SPSS 12.0 software (SPSS, Inc., Chicago, IL) was performed to examine the significant differences (P < 0.05) of
. Tukey's test using SPSS 12.0 software (SPSS, Inc., Chicago, IL) was performed to examine the significant differences (P < 0.05) of 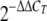 among the mean values of the samples.
among the mean values of the samples.
Targeted gene deletion and complementation.
Fusion PCR products used for the fungal transformation were constructed by using a double-joint PCR procedure (64) with slight modification. To delete GzSNF1, both the 5′ and 3′ flanking regions of GzSNF1 were amplified by PCR with primer pairs GzSNF1-F/GzSNF1-FGT and GzSNF1-RGT/GzSNF1-R (see Table S2 in the supplemental material), respectively. The geneticin-resistance gene cassette (gen; 1.8 kb), including the Aspergillus nidulans trpC promoter and terminator, was amplified from pII99 (40) with primers gen-for and gen-rev. The three amplicons were mixed in a 1:2:1 molar ratio and used as templates for the second-round PCR, which was followed by a nested PCR using the new primer pair GzSNF1-NF and GzSNF1-NR.
Two different approaches were used to complement the ΔGzSNF1 mutant. In the first, the entire GzSNF1 gene, including the native promoter and terminator, was amplified from G. zeae strain GZ3639 with the primer set GzSNF1-NF/GzSNF1-NR. This amplicon was cotransformed with pUCHI harboring the hygromycin phosphotransferase cassette (hygB) (58) into ΔGzSNF1 to generate ΔGzSNF1::GzSNF1. For the second approach, a 4.4-kb fusion PCR product, in which ScSNF1 was flanked by the GzSNF1 promoter and terminator, was amplified by primers GzSNF1-F and GzSNF-R, and was cotransformed with pUCHI into ΔGzSNF1 to generate ΔGzSNF1::ScSNF1. To make the 4.4-kb fusion PCR product, ScSNF1 (1.9 kb) was amplified from the genomic DNA of S. cerevisiae with primers ScSNF1-F and ScSNF1-R, and the 5′ flanking (1.4 kb) and 3′ flanking (1.3 kb) regions of GzSNF1 were amplified from GZ3639 with primers GzSNF1-F/GzSNF1-ScSNF1-FT and GzSNF1-ScSNF1-RT/GzSNF1-R, respectively. Finally, the three amplicons were fused and used as a template to construct the 4.4-kb final product (see Fig. S1 in the supplemental material).
Fungal transformation.
Fungal conidia produced in liquid CMC medium were inoculated into 50 ml of YPG liquid medium (3 g yeast extract, 10 g peptone, and 20 g glucose per liter) at 106 conidia per ml and grown for 12 h at 25°C in an orbital shaker (120 rpm). Mycelia harvested through filtration were incubated in 80 ml of 1 M NH4Cl containing Driselase (10 mg per ml; InterSpex Products, San Mateo, CA) for 2 h at 30°C to generate protoplasts. Further transformation steps were done as previously described (32). Each transformant was transferred to a fresh potato dextrose agar plate amended with the desired antibiotics and purified by isolating a single conidium. Standard methods were used for the transformation of the S. cerevisiae strains (4).
Virulence tests.
One milliliter of conidial suspension (106 conidia per ml) was sprayed onto the heads of the barley cultivar SangRok, which is susceptible to FHB, at the early anthesis stage. The plants were incubated in a growth chamber (25°C with 100% relative humidity) for 2 days and then transferred to a greenhouse until disease symptoms appeared.
Domain deletion and complementation.
The catalytic and regulatory domain were deleted independently by following a common strategy. The 5′ flanking region including the regulatory domain of GzSNF1 and the 3′ flanking region were amplified with primers GzSNF1-F/GzSNF1-Cat-FGT and GzSNF1-R/GzSNF1-Cat-RGT, respectively. The two amplicons were fused with gen in a second round of PCR, followed by a final round of PCR with primers GzSNF1-NF/GzSNF1-NR. The construct was introduced into GZ3639 to generate ΔGzSNF1-cat mutants. For the deletion of the regulatory domain, the 5′ and 3′ flanking regions including the catalytic domain of GzSNF1 were amplified with primers GzSNF1-F/GzSNF1-Reg-RGT and GzSNF1-Reg-RGT/GzSNF1-R, respectively. After the fusion of the two amplicons with gen, the final construct was amplified with primers GzSNF1-NF/GzSNF1-NR and was introduced to GZ3639 to generate ΔGzSNF1-reg (see Fig. S2 in the supplemental material). For the complementation of the catalytic domain in ΔGzSNF1, the terminator region of GzSNF1 (primers GzSNF1-F and GzSNF1-Cat-RT) was fused with the catalytic region including the promoter of GzSNF1 (primers GzSNF1-Cat-F and GzSNF1-R). This fusion construct was cloned into the pGEM-T-Hyg vector, and the clone was transformed into ΔGzSNF1 to generate ΔGzSNF1::GzSNF-cat. To complement the regulatory domain in ΔGzSNF1, the regulatory domain including the terminator of GzSNF1 (primers GzSNF1-F and GzSNF1-Cat-RT) was fused with the promoter region of GzSNF1 (primers GzSNF1-Cat-F and GzSNF1-R). Using the same procedure as described above, the ΔGzSNF1::GzSNF-reg mutant was generated.
Outcrosses.
Three different crosses were performed to characterize the outcrossing ability of ΔGzSNF1 by using the method described previously (35). In the first cross, a mutant with MAT1-1 deleted (ΔGzMAT1) served as the female parent and was fertilized with 1 ml of a conidial suspension (105 conidia per ml in aqueous 2.5% Tween 60) of ΔGzSNF1-pIGPAPA, which is a ΔGzSNF1 mutant tagged with green fluorescent protein (GFP), serving as the male parent. A double mutant, ΔGzMAT1 ΔGzSNF1, was generated through outcrossing between ΔGzMAT1 and ΔGzSNF1. The double mutant served as the female for the wild-type and the ΔGzSNF1 mutant for the second and third crosses, respectively. The formation of perithecia and ascospores was observed 9 days after fertilization.
Microscopy and TEM.
ΔGzSNF1-pIGPAPA and ΔGzSNF1::GzSNF1-GFP were observed with a confocal laser microscope (Radiance 2000 MP; Bio-Rad) and a DE/Axio Imager A1 microscope (Carl Zeiss, Oberkochen, Germany) with excitation and emission wavelengths of 488 and 515/530 nm, respectively. Spores and mycelia were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen, Carlsbad, CA) and 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt (DID) (Invitrogen, Carlsbad, CA) to observe nuclei and plasma membranes, respectively. For the transmission electron microscopy (TEM), ascospores and conidia were fixed as previously described (34). After dehydration in a graded ethanol series (30, 50, 70, and 80%), the specimens were embedded in London Resin White. Ultrathin sections were cut with a diamond knife in an ultramicrotome (MT-X; RMC, Tucson, AZ). The ultrathin sections were mounted on copper grids and stained with 2% uranyl acetate and Reynolds' lead citrate (46), each for 7 min. The sections were examined with an energy-filtering transmission electron microscope (LIBRA 120; Carl Zeiss) operated at an accelerating voltage of 120 kV. Zero-loss energy-filtered images were recorded with a 4 K slow-scan charge-coupled device camera (4000 SP; Gatan, Pleasanton, CA).
Germination test.
Each strain was incubated in 50-ml potato dextrose broth (PDB) for 72 h at 25°C on a rotary shaker (150 rpm), and mycelia were harvested and then washed twice with 50 ml sterile distilled water. Mycelia were spread on YMA medium to induce conidiation. After 48 h, conidia from the YMA culture were collected in distilled water, filtered through gauze, washed twice with distilled water, and centrifuged (1,000 × g, 24°C, 5 min). One milliliter of conidial suspension (106 conidia per ml) was incubated in 50 ml MM and PDB to allow germination. Two hundred conidia were observed in each examination with light microscopy. The total number of conidia germinated was counted after incubation for 0, 2, 4, 8, 12, and 24 h. Conidium germination was defined as the point at which the length of the germ tube is the same as the width of the conidium. Ascospores were collected by placing the plate containing the carrot agar upside down and allowing the mature perithecia to discharge ascospores onto petri dish covers. Ascospores were collected from the petri dish lids in distilled water, washed twice with distilled water, and centrifuged under the same conditions as described above. Ascospores were tested for germination as described above for conidia. The experiment was performed twice with three replicates, and Tukey's test using SPSS 12.0 software (SPSS, Inc., Chicago, IL) was performed to examine the significant differences (P < 0.05) of the germination percentages of spores among the mean values of the samples.
Expression and localization of GzSNF1 during the sporulation and germination stages.
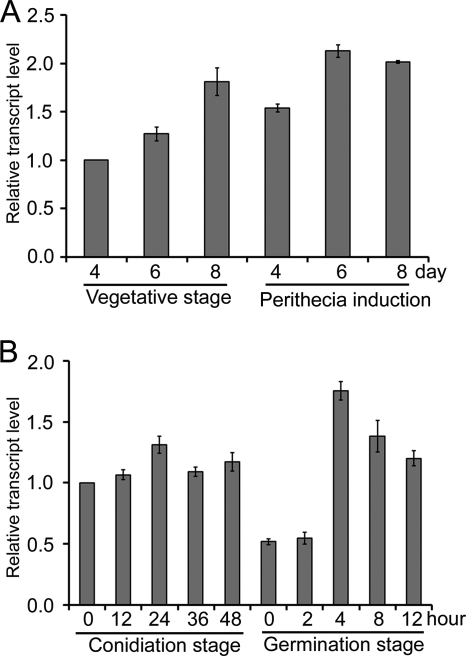
We defined the conidiation and germination stages to evaluate the expression and localization of GzSNF1: the conidiation stage is the stage in which conidiation is induced on YMA, and the germination stage is the stage in which conidia are allowed to germinate in PDB as described above. Total RNA was extracted 0, 12, 24, 36, and 48 h after conidiation began and 0, 2, 4, 8, and 12 h after the germination process began. To measure the expression of GzSNF1 during the vegetative and perithecia induction stages, total RNA was extracted from cultures growing for 4, 6, and 8 days on carrot media before and after induction, and qRT-PCR was performed.
For cellular localization studies, GFP was fused to the C terminus of GzSNF1. GFP was amplified from pIGPAPA (27) with primers pIGPAPA-gfpF and pIGPAPA-gfpR. GzSNF1, including the native promoter, was amplified from GZ3639 with primers GzSNF1-pIGPAPA-tail-F and GzSNF1-R, and the 3′ flanking region (1.3 kb) of GzSNF1 was amplified with primers pIGPAPA-gfpNF and GzSNF1-NR. Three amplicons were mixed for the final round of PCR, and a 4.3-kb final product was cloned into pGEM-T-Hyg. The clone was transformed into ΔGzSNF1 to generate ΔGzSNF1::GzSNF1-GFP. The transformants were observed under a fluorescent microscope.
RESULTS
Sequence similarity.
The ortholog of ScSNF1 was identified in the Fusarium graminearum genome database (http://www.broad.mit.edu/annotation/genome/fusarium_group/). The putative open reading frame (ORF) of GzSNF1 is annotated as FGSG_09897.3 and is 2,136 bp long with three putative introns. The predicted GzSNF1 protein contains 712 amino acids with a protein kinase domain at the N terminus. The deduced GzSNF1 amino acid sequence is similar to those for the SNF1 proteins of other fungal species, such as F. oxysporum (96% identity; FoSNF1, GenBank accession no. AF420488) (42), Hypocrea jecorina (79% identity; HjSNF1, GenBank accession no. AF291845) (11), C. carbonum (48% identity; CcSNF1, GenBank accession no. AF159293) (55), and S. cerevisiae (55% identity; ScSNF1, GenBank accession no. M13971) (8). The similarity of GzSNF1 to its homologs was high in the catalytic domain (approximately amino acid residues 1 to 404) but was only weakly conserved in the regulatory domain (approximately amino acid residues 405 to 712) (see Fig. S3 in the supplemental material). The catalytic domain region contains an activation segment (approximately amino acid residues 205 to 233) that was identical for all of the filamentous fungi (20, 31). The C terminus of GzSNF1 includes three putative nuclear localization signals (PPKKTKP [∼524 to ∼530], PKKTKPV [∼525 to ∼531], and PKKRYNL [∼600 to ∼606]) which are also found in the other fungal homologs (42, 55, 60).
Complementation of an S. cerevisiae ΔScSNF1 mutant with GzSNF1.
The p426GPD vector containing either the GzSNF1 ORF or the ScSNF1 ORF was transformed into the ΔScSNF1 mutant. All of the transformants grew well on synthetic complete (SC) medium containing glucose, whereas only the transformants carrying GzSNF1 or ScSNF1 grew on SC medium containing 2% glycerol/3% ethanol as the carbon source, although the growth of ΔScSNF1::GzSNF1 cells was reduced in comparison with that of ΔScSNF1::ScSNF1 cells (see Fig. S4 in the supplemental material).
Targeted deletion of the GzSNF1 gene and complementation.
We deleted the GzSNF1 ORF from wild-type strain GZ3639 and replaced it with the gen cassette to generate mutants with GzSNF1 deleted (ΔGzSNF1) (see Fig. S1A in the supplemental material). EcoRI-digested genomic DNA from eight independent ΔGzSNF1 transformants had a single 7.9-kb hybridizing DNA fragment instead of the 3.2-kb fragment found in the wild-type strain. This pattern confirmed that the 2.1-kb GzSNF1 locus in G. zeae had been replaced with the gen cassette (see Fig. S1D, left, in the supplemental material). ΔGzSNF1 was complemented by introducing GzSNF1 or ScSNF1 into the ΔGzSNF1 strain through cotransformation (see Fig. S1B and C in the supplemental material). The entire GzSNF1, including the native promoter and terminator from GZ3639, was introduced into the ΔGzSNF1 strain along with pUCH1. ScSNF1 flanked by the promoter and terminator of GzSNF1 was introduced into the ΔGzSNF1 strain along with pUCH1. Three of the ΔGzSNF1::GzSNF1 and two of the ΔGzSNF1::ScSNF1 mutants carried the newly introduced GzSNF1 and ScSNF1 sequences, respectively. The presence of these sequences was confirmed by Southern analysis, in which the transformants had either a 3.2-kb or a 7.8-kb hybridizing fragment for GzSNF1 or ScSNF1, respectively (see Fig. S1D in the supplemental material).
Growth characteristics of ΔGzSNF1 mutants on different carbon sources.
The ΔGzSNF1 mutant grew like its wild-type parent when supplied with arabinose, fructose, or xylose as the sole carbon source (data not shown). However, growth was reduced by 21 to 74% when galactose, sucrose, trehalose, or xylan served as the sole carbon source. The introduction of GzSNF1 into ΔGzSNF1 (ΔGzSNF1::GzSNF1) restored the wild-type growth rate level, whereas growth was only partially restored by the introduction of ScSNF1 (ΔGzSNF1::ScSNF1) (see Fig. S5 in the supplemental material).
GzSNF1 mutants can alter virulence and the expression of cell-wall-degrading enzymes.
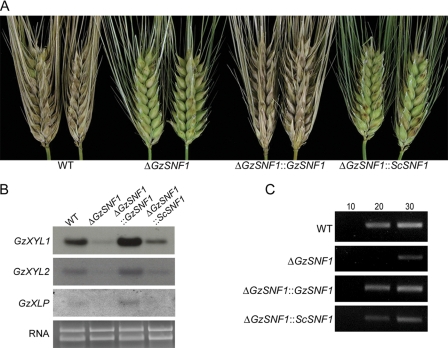
ΔGzSNF1 mutants produced small necrotic spots on spikelets 10 days after inoculation, whereas the wild-type strain caused typical head blight symptoms (Fig. 1A). ΔGzSNF1::GzSNF1 had wild-type virulence, suggesting that GzSNF1 is required for virulence by G. zeae. The virulence of ΔGzSNF1::ScSNF1 was not the same as that of the wild type, although the percentage of infected spikelets was slightly higher than in ΔGzSNF1 deletion mutants (Fig. 1A).
FIG. 1.
Virulence and expression of cell-wall-degrading enzymes. (A) Virulence assay on barley heads. Photograph was taken 10 days after inoculation. (B) Expression of cell-wall-degrading enzymes. Northern analyses were performed with β-1,4-xylanase I (GzXYL1), β-1,4-xylanase II (GzXYL2), and putative pectate lyase (GzXLP) as probes. Total RNAs were extracted from 3-day-old minimal liquid cultures supplemented with 2% xylan. Ethidium-bromide-stained gel is shown as a loading control. (C) Expression of a putative pectate lyase (GzPL). For RT-PCR, total RNA was extracted from 3-day-old minimal liquid culture supplemented with 2% polygalacturonic acid. The numbers above the panel indicate cycles of amplification. WT, G. zeae wild-type strain GZ3639.
In Northern analyses with the putative genes for endo-1,4-β-xylanase 1 precursor (GzXYL1), endo-1,4-β-xylanase 2 precursor (GzXYL2), extracellular β-xylosidase (GzXLP), and hypothetical protein similar to pectate lyase (GzPL1), the transcript levels of GzXYL1, GzXYL2, and GzXLP were decreased in the ΔGzSNF1 mutants and fully or partially restored in ΔGzSNF1::GzSNF1 and ΔGzSNF1::ScSNF1, respectively (Fig. 1B). No transcripts for GzPL1 were detected by Northern analysis. In an RT-PCR analysis, the transcription of GzPL1 also decreased in ΔGzSNF1 mutants and was fully or partially restored by complementation with GzSNF1 or ScSNF1, respectively (Fig. 1C).
ΔGzSNF1 and sexual reproduction.
In self-fertilization, the ΔGzSNF1 mutants produced fewer perithecia than the wild-type parent. Thirteen days after induction, perithecia production by the ΔGzSNF1 mutants was ∼30% of that by the wild type and the maturation of perithecia was delayed (see Table S3 in the supplemental material). Three days after induction, ascospores were not seen in most of the perithecia produced by either the wild type or the ΔGzSNF1 mutant. On the 5th day, ∼65% of the wild-type perithecia contained ascospores, whereas ∼20% of the ΔGzSNF1 perithecia contained ascospores. On the 13th day, perithecia maturation was maintained at ∼90% in the wild type, and it had increased up to ∼65% in the ΔGzSNF1 mutants (see Table S3 in the supplemental material).
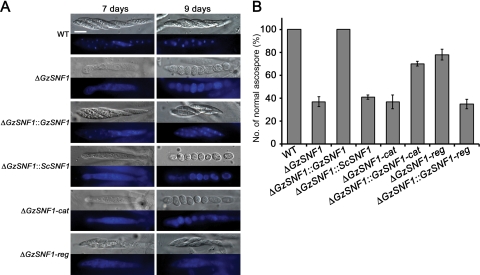
GzSNF1 also affects ascospore formation. The wild-type strain produced asci containing eight normal spindle-shaped ascospores. Approximately 70% of the asci produced by the ΔGzSNF1 mutants contained one to eight ascospores that were round or oval with a single cell. The remaining ∼30% of the asci contained eight normally shaped ascospores (Fig. 2). We observed ascospores in the wild-type asci 7 days after induction, while ascospores in asci of the ΔGzSNF1 mutants were observed 9 days after induction (Fig. 2A). Staining of the wild-type asci with DAPI showed intact nuclei, whereas DAPI staining was dispersed across the entire cytoplasm of the ΔGzSNF1 ascospores (Fig. 2A). All of the phenotypes, perithecium production, ascospore formation, and DAPI staining were fully restored by reintroducing GzSNF1 but not by introducing ScSNF1 (Fig. 2; see also Table S3 in the supplemental material).
FIG. 2.
Morphology and formation of ascospores. (A) Microscopic observation of ascospores in asci and nuclei stained with DAPI. Ascospores were sampled at 7 days and 9 days after induction. Differential interference contrast and staining with DAPI are shown. The scale bar represents 10 μm. (B) Percentages of normally shaped ascospores. WT, G. zeae wild-type strain GZ3639.
Relative importance of catalytic and regulatory domains of GzSNF1 for sexual development.
The catalytic and regulatory domains of GzSNF1 were deleted separately with the same procedure used for targeted gene deletion (see Fig. S2A and B in the supplemental material). These deletions were confirmed by the presence of a 9-kb or a 3.9-kb hybridizing band in a Southern blot of EcoRI-digested genomic DNA, instead of the 3.2-kb band found for the wild-type strain (see Fig. S2C in the supplemental material). These deletions were complemented in the ΔGzSNF1 strain with either the terminator region of GzSNF1 fused to the catalytic region, including the promoter of GzSNF1, or with the regulatory domain, including the terminator of GzSNF1 fused with the promoter region of GzSNF1. Each fusion construct was cloned into a pGEM-T-Hyg vector and then transformed into the ΔGzSNF1 mutant. Eight ΔGzSNF1::GzSNF-cat and six ΔGzSNF1::GzSNF-reg transformants were generated and confirmed by Southern analysis (see Fig. S2D in the supplemental material). The ΔGzSNF1-cat had the same phenotype as ΔGzSNF1 in both perithecium production and maturation (see Table S3 in the supplemental material). The ΔGzSNF1-reg mutants had a phenotype that in terms of the number of perithecia produced was intermediate between the ΔGzSNF1-cat mutant and the wild type (see Table S3 in the supplemental material). The domain deletion mutants also produced abnormal ascospores when self-fertilized. The ratios of the abnormal to normal ascospores were 7 to 3 and 3 to 7 in the ΔGzSNF1-cat and ΔGzSNF1-reg mutants, respectively (Fig. 2B). Similar results were obtained in the strains in which the individual domains were introduced into the ΔGzSNF1 mutant (Fig. 2; see also Table S3 in the supplemental material).
Effects of GzSNF1 in outcrossing.
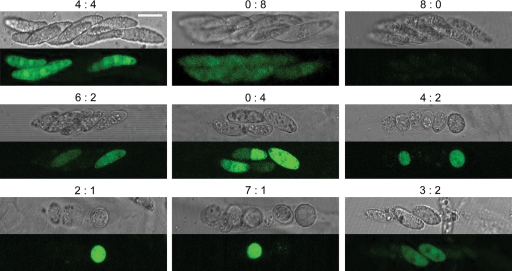
We fertilized the female ΔGzMAT1 strain with the male ΔGzSNF1 strain tagged with GFP and fertilized the ΔGzMAT1 ΔGzSNF1 double mutant with the wild type and the ΔGzSNF1 mutant tagged with GFP. All of the perithecia produced should be from outcrosses since the female strains were self-sterile (35). All asci from the first set of crosses contained eight normally shaped ascospores per ascus, the same as the self-fertilized wild-type strain. The ratio between the GFP and non-GFP ascospores within an ascus was always 1 to 1 (data not shown). When the double mutant was fertilized by the wild-type strain, ∼95% of the ascospores had the normal morphology shape, as in the first set, but ∼5% of the ascospores were round or oval single cells. Asci containing abnormal ascospores did not have a 1:1 ratio of GFP to non-GFP ascospores (data not shown). When both parents carried ΔGzSNF1, the shape of the ascospores was the same as that of the self-fertilized ΔGzSNF1 mutant. Each perithecium contained ∼70% abnormal ascospores, and the number of ascospores per ascus ranged from one to eight. The segregation ratio between the GFP and non-GFP ascospores within asci was remarkably irregular (Fig. 3). Less than 50% of the asci had a 1:1 ratio, and asci with all of the possible segregation ratios were observed.
FIG. 3.
Outcross between the ΔGzMAT1 ΔGzSNF1 and ΔGzSNF1-pIGPAPA strains. The ΔGzSNF1-pIGPAPA strain served as the male for the female ΔGzMAT1 ΔGzSNF1, double deletion mutants of GzSNF1 and GzMAT1-1, respectively. Ascospores were viewed under a Bio-Rad Radiance 2000 confocal microscope. The numbers above each panel indicate the ratio of non-GFP- to GFP-producing ascospores. The scale bar represents 10 μm.
GzSNF1 and conidiation.
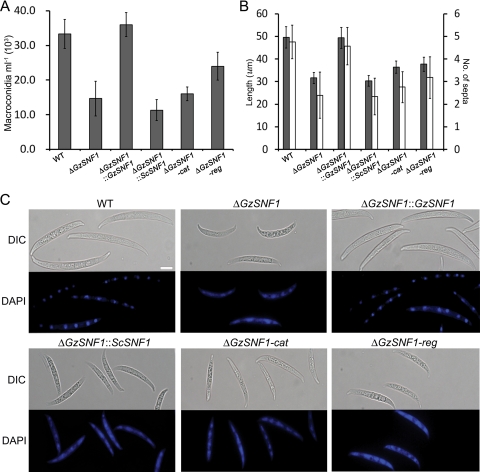
GzSNF1 mutants produced ∼55% of the number of conidia produced by the wild-type parent in CMC liquid medium (Fig. 4A). Conidia produced by the ΔGzSNF1 mutant on YMA medium were shorter than those of the wild-type parent (Fig. 4B and C). The number of septa in those conidia on YMA medium also was reduced (Fig. 4B and C). The shape of the conidia produced by the ΔGzSNF1 mutants also was abnormal. The wild-type conidia were moderately curved on the dorsal side and straight on the ventral surface with distinct septa. Mutant conidia were shorter and curved on both sides with poorly defined septa (Fig. 4C). The wild-type conidia contained distinct nuclei that could be clearly stained with DAPI that were not seen in DAPI-stained ΔGzSNF1 conidia (Fig. 4C). ΔGzSNF1-cat mutants produced a similar number of conidia with the same morphology as that of ΔGzSNF1 mutants. Conidia produced by ΔGzSNF1-reg mutants also had this morphological abnormality but were produced in greater numbers than were conidia from ΔGzSNF1-cat or the ΔGzSNF1-cat mutants (Fig. 4).
FIG. 4.
Production and morphology of conidia. (A) Number of conidia in 3-day-old CMC cultures. (B) Length (gray) and number of septa (white) of conidia on YMA medium. Error bars indicate the standard deviations. (C) Morphology of conidia and nuclei stained with DAPI. Differential interference contrast (DIC) and staining with DAPI are shown. The scale bar represents 10 μm. WT, wild-type strain GZ3639.
Deletion of GzSNF1 delays spore germination.
Spores produced by the ΔGzSNF1 mutants were as viable as the wild-type spores, but spore germination was delayed on both MM and PDB. More than 90% of the wild-type spores germinated within 12 h on MM or PDB (Table 1). On MM, the germination of ΔGzSNF1 spores reached 90% by 48 h, and on PDB, 90% of the spores had germinated by 36 h (data not shown).
TABLE 1.
Germination percentages of spores in MM and PDB
| Straina | % Germinationb
|
|||||||
|---|---|---|---|---|---|---|---|---|
| MM
|
PDB
|
|||||||
| 4c | 8 | 12 | 24 | 4 | 8 | 12 | 24 | |
| Ascospores | ||||||||
| Wild type | 1.3 A | 52.3 A | 90.3 A | 99.0 A | 0.3 A | 58.0 A | 90.7 A | 99.0 A |
| ΔGzSNF1 | 0 A | 0 B | 5.0 C | 33.0 D | 0 A | 0.3 C | 8.3 C | 68.3 C |
| ΔGzSNF1::GzSNF1 | 0 A | 53.3 A | 93.0 A | 99.0 A | 0.7 A | 58.3 A | 93.3 A | 99.0 A |
| ΔGzSNF1::ScSNF1 | 0 A | 0 B | 8.3 C | 36.7 D | 0 A | 0.3 C | 9.3 C | 68.7 C |
| ΔGzSNF1-cat | 0 A | 0 B | 7.7 C | 50.7 C | 0 A | 0.3 C | 10.3 C | 74.7 C |
| ΔGzSNF1-reg | 0 A | 4.0 B | 19.3 B | 60.7 B | 0 A | 4.7 B | 34.3 B | 90.7 B |
| Conidia | ||||||||
| Wild type | 27.7 A | 85.0 A | 96.0 A | 99.0 A | 28.0 A | 84.3 A | 96.7 A | 99.0 A |
| ΔGzSNF1 | 0 B | 1.0 C | 6.7 D | 35.0 D | 0 B | 2.0 C | 8.3 D | 76.3 B |
| ΔGzSNF1::GzSNF1 | 27.4 A | 86.3 A | 96.3 A | 99.0 A | 24.3 A | 85.3 A | 96.3 A | 99.0 A |
| ΔGzSNF1::ScSNF1 | 0 B | 3.3 C | 12.3 CD | 42.7 CD | 0 B | 2.7 C | 15.7 C | 81.3 B |
| ΔGzSNF1-cat | 0 B | 2.3 C | 15.0 C | 45.0 C | 0.3 B | 4.7 C | 20.7 C | 78.0 B |
| ΔGzSNF1-reg | 4.0 B | 14.3 B | 31.0 B | 73.7 B | 3.0 B | 18.0 B | 39.7 B | 91.3 A |
A spore suspension of each strain was incubated in 50 ml MM And PDB at 25°C on a rotary shaker (150 rpm). Two hundred spores were observed in each examination with light microscopy, and the number of conidia germinated was counted.
Values within a column not sharing a letter are significantly different according to Tukey's test (P < 0.05).
Incubation time (hour).
GzSNF1 is necessary for proper nucleation for germination.
We followed the germination of ascospores and conidia microscopically in PDB. Nuclei in the wild-type spores could be seen when stained with DAPI at 0 h (see Fig. S6 in the supplemental material). When the spores were swollen at 2 h, DAPI diffused in the cytoplasm of the spores, indicative of nucleus replication. At 4 h, spores began to germinate and germ tubes containing nuclei appeared, although DAPI was still diffused in the cytoplasm. At 8 h, germ tubes were elongated and nuclei were condensed within them. In the ΔGzSNF1 mutants, spores did not contain clear nuclei until 2 h. Nuclei were visualized at 4 h, but spores did not germinate. At 8 h, spores initiated germination and the nuclei were diffused in the cytoplasm (see Fig. S6 in the supplemental material).
Ascospores from the wild type and the ΔGzSNF1 mutants also were observed with TEM. The ΔGzSNF1 ascospores were structurally different from the wild-type ascospores, which contained well-developed organelles such as nuclei, nucleoli, and mitochondria (Fig. 5). Neither a nucleus, nuclear membrane, nor septum was observed in the abnormal ascospores of ΔGzSNF1. The cell wall and cell membrane of the wild type were thick and clear, while those of the ΔGzSNF1 mutants were thin and unclear. Chromatin materials were absent in the wild type but were irregularly spread throughout the cells of the ΔGzSNF1 ascospores (Fig. 5). The ΔGzSNF1 conidia were similar to the wild-type conidia except for the nuclear membrane. The ΔGzSNF1 conidia contained cellular organelles, cell membranes, and cell walls, but the nuclear membranes of the mutants were thinner and less clear than that of the wild type. In addition, there were additional unidentified electron dense materials in the cytoplasm of the ΔGzSNF1 conidia (Fig. 5).
FIG. 5.
Transmission electron micrographs of ascospores and conidia of the G. zeae wild-type strain and the ΔGzSNF1 mutant. WT, wild-type strain GZ3639; AC, ascus; AS, ascospore; C, chromatin materials; E, electron dense materials; L, lipid globule; M, mitochondrion; N, nucleus; NM, nuclear membrane; NO, nucleolus; P, peroxisome; V, vacuole. The scale bar represents 0.5 μm.
GzSNF1 expression and localization during sporulation and germination.
GFP was fused to the C terminus of GzSNF1 by single-joint PCR and then cloned into a pGEM-T-Hyg vector carrying hygB. This plasmid was introduced into ΔGzSNF1 to generate ΔGzSNF1::GzSNF1-GFP. Four transformants restored the wild-type phenotype and were confirmed by Southern analysis (data not shown).
The GFP in fresh ascospores was commonly associated with the plasma membrane and septa, but GFP also was associated with the cytoplasm after the ascospores were swollen for germination (Fig. 6A). The GFP signal weakened 72 h after germination but was associated with both the cytoplasm and nuclei. During conidiation, GFP was found in the cytoplasm of hyphae, conidiophores with phialides, and young conidia. It was transported to the plasma membrane by vesicles during conidium maturation (data not shown). Finally, GFP was also localized on the plasma membrane and septa in mature conidia (Fig. 6B). The localization of GFP in conidia was similar to its localization in ascospores. When conidia were swollen to germinate at 2 h after incubation in PDB, GFP was localized in the cytoplasm of spores. After 8 h of incubation in PDB, GFP was localized in the cytoplasm of hyphae. Not until 72 h postgermination was most of the GFP localized in the cytoplasm and nuclei (Fig. 6B). Spores stained with DID showed that the GFP signal was associated with the plasma membrane and septa in ascospores and conidia (see Fig. S7 in the supplemental material).
FIG. 6.
Localization of GzSNF1 in ascospores (A) and conidia (B). GFP was fused to the C terminus of GzSNF1. Nuclei were stained with DAPI. Differential interference contrast (DIC), staining with DAPI, and GFP fluorescence are shown. The scale bar represents 10 μm.
Based on qRT-PCR, GzSNF1 was constitutively expressed throughout the entire fungal life cycle. On carrot medium, GzSNF1 expression increased gradually during vegetative growth (Fig. 7A). During the perithecium induction stage, GzSNF1 expression decreased slightly 4 days after induction but then increased as perithecia developed and matured (Fig. 7A). After 72 h of incubation in PDB, the mycelia strongly expressed GzSNF1. When mycelia were spread on YMA to induce conidiation, GzSNF1 expression increased slightly during the conidiation stage (Fig. 7B). Fresh conidia (0-h germination stage) harvested from 48-h-old YMA to induce germination expressed GzSNF1 less than mycelia from the conidiation stage, but expression increased by 4 h of incubation. After 4 h, expression decreased to the level of conidiation (Fig. 7B).
FIG. 7.
Expression profiles of GzSNF1. Transcription of GzSNF1 was analyzed by quantitative real time-PCR (qRT-PCR). (A) Expression of GzSNF1 in the vegetative growth and perithecium induction stages. (B) Expression of GzSNF1 gene in the conidiation and germination stages.
DISCUSSION
Organisms encounter a variety of nutritional conditions during their life cycle, and must turn on or off metabolic pathways to survive under different conditions. One of the best understood pathways is glucose repression, for which the key regulator is SNF1 (8). When glucose is limited, SNF1 regulates the expression of metabolic genes by controlling transcriptional activators and repressors (6). We found that the SNF1 gene of G. zeae affects a broader spectrum of functions than is known for its homologs in other filamentous fungi. SNF1 in G. zeae affects fungal virulence; developmental processes, such as sexual and asexual reproduction; spore maturation and germination; and the utilization of certain carbon sources in G. zeae.
GzSNF1 contains two major domains, a catalytic and a regulatory domain, that are common to all fungal SNF1 sequences (30). GzSNF1 enables the utilization of alternative carbon sources in SNF1-defective yeast cells (see Fig. S4 in the supplemental material). Similarly in G. zeae strains lacking GzSNF1 function, SNF1 partially complements the deficiency and enables these cells to use alternative carbon sources. These results suggest that the GzSNF1 gene is an ortholog of S. cerevisiae SNF1.
The deletion of GzSNF1 in G. zeae reduces growth on some carbon sources, but this list of regulated carbon sources differs slightly from those in other filamentous fungi, e.g., F. oxysporum and C. carbonum (42, 55). For example, the ΔFoSNF1 strain grows similarly on xylan and xylose, while ΔGzSNF1 strains grow differently on xylan than they do on xylose. ΔFoSNF1 mutants also use pectin efficiently, whereas ΔCcSNF1 mutants could not use either pectin or xylan as a carbon source (42, 55). Xylan and pectin are major components of the plant cell wall (22, 43, 62). The differences in the carbon source utilization pattern among the different fungal species could result from differences in the activity of cell-wall-degrading enzymes depolymerized by genes regulated by SNF1.
The expression of cell-wall-degrading enzymes is an important virulence factor in plant pathogenic fungi. Cytological studies have provided indirect evidence that G. zeae produces cellulose, xylanase, and pectinase during plant penetration and colonization (62). In addition, G. zeae grown on the plant cell wall in vitro expresses more than 40 putative genes related to polysaccharide degradation or carbohydrate catabolism (43), and at least 30 putative xylanases are expressed in the plant cell wall medium (22). ΔGzSNF1 mutants had reduced virulence capabilities and expression of cell-wall-degrading enzymes, as has been found in similar mutants in other plant pathogenic fungi (42, 55). The lower virulence in ΔGzSNF1 mutants would be due to a reduced ability to use sucrose, which is one of the major plant sugars, and a reduced ability to degrade cell wall tissues.
We expected that GzSNF1 would have a role in the sexual reproduction of G. zeae because the ability of S. cerevisiae to reproduce sexually was abolished when the SNF1 gene was deleted (7). However, the sexual function of ΔGzSNF1 mutants was not as severe since these mutants still produced perithecia containing viable ascospores, although the number of perithecia was reduced and the ascospores formed were morphologically abnormal. In S. cerevisiae, glucose starvation triggers the derepression of SNF1, which induces metabolic pathways that use alternative carbon sources, and sexual reproduction, with acetate being required to complete sexual reproduction (26). To induce sexual reproduction, G. zeae may also require a certain nutrient(s), and GzSNF1 could enable the utilization of an alternative nutrient(s) that is required to complete perithecium formation.
Outcrosses with ΔGzSNF1, ΔGzMAT1 ΔGzSNF1, and the wild-type parent exhibited different segregation patterns. The ΔGzSNF1 strain was fertile as a normal male when crossed with ΔGzMAT1 as the female. When a ΔGzMAT1 ΔGzSNF1 strain was used as the female and the wild-type strain as the male, ∼5% of the ascospores produced were abnormal. Thus, there was some maternal effect in the phenotype. In crosses between the female ΔGzMAT1 ΔGzSNF1 and the male ΔGzSNF1 strains, unexpected ascus segregation patterns were observed (Fig. 3). In ΔScSNF1 strains of S. cerevisiae, chromosome replication is retarded (26). The retardation of chromosome replication during meiosis or mitosis could result in incomplete segregation and the formation of aneuploid nuclei that were not viable. Depending on when/where the aberrant segregation occurred, asci with any number of spores could be formed (45). Aberrations in meiosis I could result in nonviable ascospores if both of the daughter cells are aneuploid. It could also result in all of the ascospores having the same SNF1 genotype if only one of the daughter cells survives the process with a viable number of chromosomes. The production of more than four spores with the same SNF1 genotype could result from traditional gene conversion or from irregular meiotic and/or mitotic divisions. Distinguishing these possibilities is not possible with the strains described in this paper, which were all derived from the same parent and differ only at the deleted markers and not at the numerous loci on multiple chromosomes that would be required to differentiate between these two hypotheses.
Spore germination of the ΔGzSNF1 mutants also was delayed, although most spores did germinate given sufficient time (Table 1). Spore germination in filamentous fungi is controlled by multiple factors and pathways. Nutrients, including amino acids, sugars, and inorganic salts, stimulate germination in saprophytic fungi such as A. nidulans and Neurospora crassa (41). The spores of plant pathogenic fungi such as Magnaporthe grisea and Colletotrichum spp. are able to germinate in the absence of nutrients (25, 54). Although the spore germination of G. zeae has been studied (21), at least two factors could play a role in retarding spore germination in the ΔGzSNF1 mutants. First, spores of the ΔGzSNF1 mutants may not have accumulated sufficient nutrients for germination, while the wild-type spores contain enough of all nutrients required for germination. GzSNF1 is expressed constitutively during sporulation and is localized at the plasma membrane of mature spores. The slower maturation of these spores and their aberrant shapes could result from inadequate or unbalanced nutrient accumulation during spore formation. GzSNF1 also could trigger the expression of cell-wall-depolymerizing enzymes necessary for spore germination. Accumulating the necessary nutrients or sufficiently degrading the cell wall could both delay spore germination. Alternatively, delays in spore germination could result from delays in nucleus formation due to delayed chromosome condensation and nucleus organization. Nuclei in ΔGzSNF1 spores were neither condensed nor organized immediately following sporulation, although the nuclei in the wild-type spores clearly were (see Fig. S6 in the supplemental material). Nuclei became diffuse and then recondensed during the germination of both the ΔGzSNF1 and wild-type G. zeae spores, but the process in the ΔGzSNF1 spores was slower than in the wild-type spores.
The germination delay may be responsible for the decreased virulence that we observed as well. Spore germination needs to occur rapidly under favorable environmental conditions for successful infection, as the time that the plant host is susceptible to infection is quite short. Freshly discharged wild-type ascospores germinate within 4 h in 100% relative humidity (2), and wild-type conidia germinate within 6 to 12 h after inoculation when the relative humidity is high (62). Thus, delayed spore germination even in a favorable environment could make it more difficult for the fungus to infect the plant.
The expression and localization of GzSNF1 are dependent on the developmental stage. GzSNF1 was constitutively expressed during sporulation and is localized in the cytoplasm of immature spores of G. zeae. As spores mature, GzSNF1 is strongly expressed, and GzSNF1 is transported via vesicle trafficking to the plasma membrane, suggesting that GzSNF1 is targeted to the plasma membrane through a typical eukaryotic secretory pathway (38) even though the GzSNF1 protein does not have a signal peptide at its N terminus. GzSNF1 also is found in both the cytoplasm and the nuclei of vegetative hyphae. The regulatory domain of GzSNF1 has three nuclear localization signals (NLS) recognized by importin (39), which could help localize GzSNF1 in the nuclei. In addition, the increase in GzSNF1 expression during germination also supports the functional requirement of GzSNF1 for spore germination. This result is consistent with a recent microarray study (52), in which GzSNF1 was upregulated more than twofold during the initiation of conidium germination.
Although GzSNF1 retains the function involved in a glucose repression, it has other functions that are quite different from the function of SNF1 in S. cerevisiae. First, the expression and localization of GzSNF1 are independent of the presence of glucose and are dependent on the developmental stage. In S. cerevisiae, SNF1 is localized in either the cytoplasm or the nucleus, depending on the presence of glucose (24, 61). The localization of SNF1 in S. cerevisiae is regulated by three β subunits encoded by SIP1, SIP2, and GAL83 (61). The genome of G. zeae has only one homolog (FGSG_07407.3) of the S. cerevisiae β subunits, and the deletion of the homolog in G. zeae did not result in any detectable phenotypic changes (S. Lee et al., unpublished data), which suggests that GzSNF1 localization is independent of the β subunits. Second, the regulation of GzSNF1 is different from that of ScSNF1. In S. cerevisiae, the regulatory domain binds the catalytic domain to autoinhibit the catalytic function of SNF1 in the presence of glucose and binds the SNF4 protein to activate the function in the absence of glucose (29). Thus, snf4 mutants constitutively inactivate ScSNF1 and ScSNF4, and snf1 mutants of S. cerevisiae have similar phenotypes (9). The catalytic function of GzSNF1 is independent from the interaction between the regulatory domain and GzSNF4 (FGSG_08998.3), because ΔGzSNF4 mutants have no detectable phenotype (Lee et al., unpublished). We do not know whether the phenotype changes in ΔGzSNF1-reg mutants result from the loss of the regulatory domain itself or from changes in the expression level of the catalytic domain. To test this hypothesis, the functions of chimeric constructs with ScSNF1 and GzSNF1 domains need to be characterized. Taking these functional differences together, GzSNF1 may undergo more evolutionary change than the yeast SNF1, which may allow GzSNF1 to carry more diverse roles for filamentous growth and/or virulence in G. zeae.
In conclusion, GzSNF1 has important roles in such nutrient-sensitive cellular processes as pathogenicity and reproduction in G. zeae. We have focused on dissecting the roles of GzSNF1 in G. zeae sexual and asexual development, which are neither well known nor well described for other filamentous fungi. This study is the first report to account for the functions of the SNF1 ortholog in sexual and asexual development of the filamentous fungus in detail. It will therefore be our main objective in future work to identify and characterize the interaction partners of GzSNF1 in the developmental stages as a prelude to a better understanding of multicellular development in G. zeae.
Supplementary Material
Acknowledgments
This work was supported by grant CG 1411 from the Crop Functional Genomics Center of the 21st Century Frontier Research Program, funded by the Korean Ministry of Education, Science and Technology, and by a Korea Research Foundation grant funded by the Korean government (MOEHRD) (KRF-2006-005-J04701). S.-H.L. and S.L. were supported by graduate fellowships from the Korean Ministry of Education, Science and Technology through the Brain Korea 21 project.
Footnotes
Published ahead of print on 21 November 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alepuz, P. M., K. W. Cunningham, and F. Estruch. 1997. Glucose repression affects ion homeostasis in yeast through the regulation of the stress-activated ENA1 gene. Mol. Microbiol. 2691-98. [DOI] [PubMed] [Google Scholar]
- 2.Beyer, M., and J. A. Verreet. 2005. Germination of Gibberella zeae ascospores as affected by age of spores after discharge and environmental factors. Eur. J. Plant Pathol. 111381-389. [Google Scholar]
- 3.Bowden, R. L., and J. F. Leslie. 1992. Nitate-nonutilizing mutants of Gibberella zeae (Fusarium graminearum) and their use in determining vegetative compatibility. Exp. Mycol. 16308-315. [Google Scholar]
- 4.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics, laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 5.Capellini, R. A., and J. L. Peterson. 1965. Macroconidium formation in submerged cultures by a nonsporulating strain of Gibberella zeae. Mycologia 57962-966. [Google Scholar]
- 6.Carlson, M. 1999. Glucose repression in yeast. Curr. Opin. Microbiol. 2202-207. [DOI] [PubMed] [Google Scholar]
- 7.Carlson, M., B. C. Osmond, and D. Botstein. 1981. Mutants of yeast defective in sucrose utilization. Genetics 9825-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Celenza, J. L., and M. Carlson. 1986. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science 2331175-1180. [DOI] [PubMed] [Google Scholar]
- 9.Celenza, J. L., and M. Carlson. 1989. Mutational analysis of the Saccharomyces cerevisiae SNF1 protein kinase and for functional interaction with the SNF4 protein. Mol. Cell. Biol. 95034-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cullen, P. J., and G. F. Sprague. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 9713619-13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cziferszky, A., B. Seiboth, and C. P. Kubicek. 2003. The SNF1 kinase of the filamentous fungus Hypocrea jecorina phosphorylates regulation-relevant serine residues in the yeast carbon catabolite repressor Mig1 but not in the filamentous fungal counterpart Cre1. Fungal Genet. Biol. 40166-175. [DOI] [PubMed] [Google Scholar]
- 12.Debuchy, R., and B. G. Turgeon. 2006. Mating-type structure, evolution and function in Euascomycetes, p. 293-321. In K. Esser, U. Kues, and R. Fischer (ed.), The mycota I: growth, differentiation and sexuality. Springer, Berlin, Germany.
- 13.Desjardins, A. E. 2006. Fusarium mycotoxins: chemistry, genetics and biology. APS Press, St. Paul, MN.
- 14.Fernando, W. G. D., J. D. Miller, W. L. Seaman, K. Seifert, and T. C. Paulitz. 2000. Daily and seasonal dynamics of airborne spores of Fusarium graminearum and other Fusarium species samples over wheat pots. Can. J. Bot. 78497-505. [Google Scholar]
- 15.Goodwin, P. H., and G. Y. J. Chen. 2002. High expression of a sucrose non-fermenting (SNF-1)-related protein kinase from Collectotrichum gloeosprioides f.sp. malvae is associated with penetration of Malva pusilla. FEMS Microbiol. Lett. 215169-174. [DOI] [PubMed] [Google Scholar]
- 16.Güldener, U., K.-Y. Seong, J. Boddu, S. Cho, F. Trail, J.-R. Xu, et al. 2006. Development of a Fusarium graminearum Affymetrix GeneChip for profiling fungal gene expression in vitro and in planta. Fungal Genet. Biol. 43316-325. [DOI] [PubMed] [Google Scholar]
- 17.Hallen, H. E., M. Huebner, S. H. Shiu, U. Güldener, and F. Trail. 2007. Gene expression shifts during perithecium development in Gibberella zeae (anamorph Fusarium graminearum), with particular emphasis on ion transport proteins. Fungal Genet. Biol. 441146-1156. [DOI] [PubMed] [Google Scholar]
- 18.Han, Y.-K., T. Lee, K.-H. Han, S.-H. Yun, and Y.-W. Lee. 2004. Functional analysis of the homoserine O-acetyltransferase gene and its identification as a selectable marker in Gibberella zeae. Curr. Genet. 46205-212. [DOI] [PubMed] [Google Scholar]
- 19.Han, Y.-K., M.-D. Kim, S.-H. Lee, S.-H. Yun, and Y.-W. Lee. 2007. A novel F-box protein involved in sexual development and pathogenesis in Gibberella zeae. Mol. Microbiol. 63768-779. [DOI] [PubMed] [Google Scholar]
- 20.Hardie, D. G., D. Carling, and M. Carlson. 1998. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67821-855. [DOI] [PubMed] [Google Scholar]
- 21.Harris, S. D. 2005. Morphogenesis in germinating Fusarium graminearum macroconidia. Mycologia 97880-887. [DOI] [PubMed] [Google Scholar]
- 22.Hatsch, D., V. Phalip, E. Petkovske, and J. M. Jeltsch. 2006. Fusarium graminearum on plant cell wall: no fewer than 30 xylanase genes transcribed. Biochem. Biophys. Res. Commun. 345959-966. [DOI] [PubMed] [Google Scholar]
- 23.Hedbacker, K., and M. Carlson. 2008. SNF1/AMPK pathway in yeast. Front. Biosci. 132408-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hedbacker, K., R. Townley, and M. Carlson. 2004. Cyclic AMP-dependent protein kinase regulates the subcellular localization of SNF1-SIP1 protein kinase. Mol. Cell. Biol. 241836-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hedge, Y., and P. E. Kolattukudy. 1997. Cuticular waxes relieve self-inhibition of germination and appressorium formation by the conidia of Magnaporthe grisea. Physiol. Mol. Plant Pathol. 5175-84. [Google Scholar]
- 26.Honigberg, S. M., and R. H. Lee. 1998. SNF1 kinase connects nutritional pathways controlling meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 184548-4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horwitz, B. A., A. Sharon, S. W. Lu, V. Ritter, T. M. Sandrock, O. C. Yoder, and B. G. Turgeon. 1999. A G protein alpha subunit from Cochliobolus heterostrophus involved in mating and appressorium formation. Fungal Genet. Biol. 2619-32. [DOI] [PubMed] [Google Scholar]
- 28.Hou, Z., C. Xue, Y. Peng, T. Katan, H. C. Kistler, and J.-R. Xu. 2002. A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant-Microbe Interact. 151119-1127. [DOI] [PubMed] [Google Scholar]
- 29.Jiang, R., and M. Carlson. 1996. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 103105-3115. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, R., and M. Carlson. 1997. The Snf1 protein kinase and its activating subunit, SNF4, interact with distinct domains of the SIP1/SIP2/GAL83 component in the kinase complex. Mol. Cell. Biol. 172099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson, L. N., M. E. M. Noble, and D. J. Owen. 1996. Active and inactive protein kinases: structural basis for regulation. Cell 85149-158. [DOI] [PubMed] [Google Scholar]
- 32.Kim, J.-E., J. Jin, H. Kim, J.-C. Kim, S.-H. Yun, and Y.-W. Lee. 2006. GIP2, a putative transcription factor regulates the aurofusarin biosynthetic gene cluster in Gibberella zeae. Appl. Environ. Microbiol. 721645-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim, J.-E., M.-D. Kim, W.-B. Shim, S.-H. Yun, and Y.-W. Lee. 2007. Functional characterization of acetylglutamate synthase and phosphoribosylamine-glycine ligase genes in Gibberella zeae. Curr. Genet. 5199-108. [DOI] [PubMed] [Google Scholar]
- 34.Kim, K.-W., and J.-W. Hyun. 2007. Nonhost-associated proliferation of intrahyphal hyphae of citrus scab fungus Elsinoe fawcettii: refining the perception of cell-within-a-cell organization. Micron 38565-571. [DOI] [PubMed] [Google Scholar]
- 35.Lee, J., T. Lee, Y.-W. Lee, S.-H. Yun, and B. G. Turgeon. 2003. Shifting fungal reproductive mode by manipulation of mating type genes: obligatory heterothallism of Gibberella zeae. Mol. Microbiol. 50145-152. [DOI] [PubMed] [Google Scholar]
- 36.Lee, S.-H., S. Lee, D. Choi, Y.-W. Lee, and S.-H. Yun. 2006. Identification of the down-regulated genes in a mat1-2-deleted strain of Gibberella zeae, using cDNA subtraction and microarray analyses. Fungal Genet. Biol. 43295-310. [DOI] [PubMed] [Google Scholar]
- 37.Leslie, J. F., and B. A. Summerell. 2006. The Fusarium laboratory manual. Blackwell Professional, Ames, IA.
- 38.Lowe, M., and F. A. Barr. 2007. Inheritance and biogenesis of organelles in the secretory pathway. Nat. Rev. Mol. Cell Biol. 8429-439. [DOI] [PubMed] [Google Scholar]
- 39.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67265-306. [DOI] [PubMed] [Google Scholar]
- 40.Namiki, F., M. Matsunaga, M. Okuda, I. Inoue, K. Nishi, Y. Fujita, and T. Tsuge. 2001. Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. sp. melonis. Mol. Plant-Microbe Interact. 14580-584. [DOI] [PubMed] [Google Scholar]
- 41.Osherov, N., and G. S. May. 2001. The molecular mechanisms of conidial germination. FEMS Microbiol. Lett. 199153-160. [DOI] [PubMed] [Google Scholar]
- 42.Ospina-Giraldo, M. D., E. Mullins, and S. Kang. 2003. Loss of function of the Fusarium oxysporum SNF1 gene reduces virulence on cabbage and Arabidopsis. Curr. Genet. 4449-57. [DOI] [PubMed] [Google Scholar]
- 43.Phalip, V., F. Delalande, C. Carapito, F. Goubet, D. Hatsch, E. Leize-Wagner, P. Dupree, A. V. Dorsselaer, and J. M. Jeltsch. 2005. Diversity of the exoproteome of Fusarium graminearum grown on plant cell wall. Curr. Genet. 48366-379. [DOI] [PubMed] [Google Scholar]
- 44.Qi, W., C. Kwon, and F. Trail. 2006. Microarray analysis of transcript accumulation during perithecium development in the filamentous fungus Gibberella zeae (anamorph Fusarium graminearum). Mol. Genet. Genomics 27687-100. [DOI] [PubMed] [Google Scholar]
- 45.Raju, N. B., and J. F. Leslie. 1992. Cytology of recessive sexual-phase mutants from wild strains of Neurospora crassa. Genome 35815-826. [DOI] [PubMed] [Google Scholar]
- 46.Reynolds, E. S. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17208-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rittenour, W. R., and S. D. Harris. 2008. Characterization of Fusarium graminearum Mes1 reveals roles in cell-surface organization and virulence. Fungal Genet. Biol. 45933-946. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Santangelo, G. M. 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70253-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanz, P. 2003. SNF1 protein kinase: a key player in the response to cellular stress in yeast. Biochem. Soc. Trans. 31178-181. [DOI] [PubMed] [Google Scholar]
- 51.Seo, B.-W., H.-K. Kim, Y.-W. Lee, and S.-H. Yun. 2007. Functional analysis of a histidine auxotrophic mutation in Gibberella zeae. Plant Pathol. J. 2351-56. [Google Scholar]
- 52.Seong, K.-Y., X. Zha, J.-R. Xu, U. Guldener, and H. C. Kistler. 2008. Conidial germination in the filamentous fungus Fusarium graminearum. Fungal Genet. Biol. 45389-399. [DOI] [PubMed] [Google Scholar]
- 53.Sutton, J. C. 1982. Epidemiology of wheat head blight and maize ear rot caused by Fusarium graminearum. Can. J. Plant Pathol. 4195-209. [Google Scholar]
- 54.Thines, E., H. Anke, and R. W. S. Weber. 2004. Fungal secondary metabolites as inhibitors of infection-related morphogenesis in phytopathogenic fungi. Mycol. Res. 10814-25. [DOI] [PubMed] [Google Scholar]
- 55.Tonukari, N. J., J. S. Scott-Craig, and J. D. Walton. 2000. The Cochliobolus carbonum SNF1 gene is required for cell wall-degrading enzyme expression and virulence on maize. Plant Cell 12237-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trail, F. 2007. Fungal cannons: explosive spore discharge in the Ascomycota. FEMS Microbiol. Lett. 27612-18. [DOI] [PubMed] [Google Scholar]
- 57.Trail, F., H. Xu, R. Loranger, and D. Gadoury. 2002. Physiological and environmental aspects of ascospore discharge in Gibberella zeae (anamorph Fusarium graminearum). Mycologia 94181-189. [PubMed] [Google Scholar]
- 58.Turgeon, B. G., R. C. Garber, and O. C. Yoder. 1987. Development of a fungal transformation system based on selection of sequences with promoter activity. Mol. Cell. Biol. 73297-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Urban, M., E. Mott, T. Farley, and K. Hammond-Kosack. 2003. The Fusarium graminearum MAP1 gene is essential for pathogenicity and development of perithecia. Mol. Plant Pathol. 4347-359. [DOI] [PubMed] [Google Scholar]
- 60.Vacher, S., P. Cotton, and M. Fèvre. 2003. Characterization of a SNF1 homolog from the phytopathogenic fungus Sclerotinia sclerotiorum. Gene 22113-121. [DOI] [PubMed] [Google Scholar]
- 61.Vincent, O., R. Townley, S. Kuchin, and M. Carlson. 2001. Subcellular localization of the SNF1 kinase is regulated by specific β subunits and a novel glucose signaling mechanism. Genes Dev. 151104-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wanjiru, W. M., K. Zhensheng, and H. Buchenauer. 2002. Importance of cell wall degrading enzymes produced by Fusarium graminearum during infection of wheat heads. Eur. J. Plant Pathol. 108803-810. [Google Scholar]
- 63.Yu, H.-Y., J.-A. Seo, J.-E. Kim, K.-H. Han, W.-B. Shim, S.-H. Yun, and Y.-W. Lee. 2008. Functional analyses of heterotrimeric G protein Gα and Gβ subunits in Gibberella zeae. Microbiology 154392-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu, J. H., Z. Hamari, K.-H. Han, J.-A. Seo, Y. Reyes-Dominguez, and C. Scazzocchio. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulation in filamentous fungi. Fungal Genet. Biol. 41973-981. [DOI] [PubMed] [Google Scholar]
- 65.Yun, S. H., T. Arie, I. Kaneko, O. C. Yoder, and B. G. Turgeon. 2000. Molecular organization of mating type loci in heterothallic, homothallic, and asexual Gibberella/Fusarium species. Fungal Genet. Biol. 317-20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.