Abstract
Cryptococcus neoformans capsular polysaccharide is composed of at least two components, glucuronoxylomannan (GXM) and galactoxylomannans (GalXM). Although GXM has been extensively studied, little is known about the location of GalXM in the C. neoformans capsule, in part because there are no serological reagents specific to this antigen. To circumvent the poor immunogenicity of GalXM, this antigen was conjugated to protective antigen from Bacillus anthracis as a protein carrier. The resulting conjugate elicited antibodies that reacted with GalXM in mice and yielded an immune serum that proved useful for studying GalXM in the polysaccharide capsule. In acapsular cells, immune serum localized GalXM to the cell wall. In capsulated cells, immune serum localized GalXM to discrete pockets near the capsule edge. GalXM was abundant on the nascent capsules of budding daughter cells. The constituent sugars of GalXM were found in vesicle fractions consistent with vesicular transport for this polysaccharide. In addition, we generated a single-chain fraction variable fragment antibody with specificity to oxidized carbohydrates that also produced punctate immunofluorescence on encapsulated cells that partially colocalized with GalXM. The results are interpreted to mean that GalXM is a transient component of the polysaccharide capsule of mature cells during the process of secretion. Hence, the function of GalXM appears to be more consistent with that of an exopolysaccharide than a structural component of the cryptococcal capsule.
Cryptococcus neoformans is an encapsulated fungal pathogen that causes meningitis primarily in immunocompromised patients (22, 27). The incidence of cryptococcosis increased dramatically at the end of the 20th century in association with advanced human immunodeficiency virus infection. Other groups at risk are patients receiving immunosuppressive therapies for cancers and transplants (3, 8). C. neoformans has several well-defined virulence factors that include a polysaccharide capsule. Classically, the capsular polysaccharide was defined as being composed of glucuronoxylomannan (GXM), galactoxylomannan (GalXM), and mannoproteins (MPs) (17, 25, 32). However, this composition has been assumed based on analysis of exopolysaccharides. Although GXM has been extensively studied and is associated with many deleterious effects in the host, considerably less is known about GalXM. There is no direct evidence for a structural role of GalXM and MP in capsule assembly or architecture. In recent years, evidence has emerged that GalXM is a more potent immunomodulatory molecule than GXM (9, 28). Percolini et al. showed that GalXM inhibits T-cell proliferation and peripheral blood mononuclear cells. The study also revealed that GalXM increased the production of the cytokines gamma interferon and interleukin-10 (28). GalXM upregulates Fas and initiates apoptosis of T lymphocytes by DNA fragmentation through the activation of caspase 8 (28). GalXM also causes apoptosis in macrophages through a FasL-related mechanism (34).
GalXM constitutes about 8% of the shed polysaccharide found in cryptococcal culture supernatants (3, 32) and has an α1,6-galactan backbone containing four potential short oligosaccharide branch structures. The branches are 3-O linked to the backbone and consist of an α1,3-mannose, α1,4-mannose, β-galactosidase trisaccharide with variable amounts of β1,2- or β1,3-xylose side groups (3, 20, 32). The GalXM backbone consists of galactopyranose and a small amount of galactofuranose (32), unlike GXM, which contains only mannopyranose (3). The molar composition of GalXM components revealed xylose at 22%, mannose at 29%, and galactose at 50% (10, 32). Proton nuclear magnetic resonance (NMR) revealed the anomeric region to be between 5.4 and 4.3 ppm in a one-dimensional (1D) 1H spectrum recorded at 600 MHz and 56°C (10, 32). GalXMs from serotypes A, C, and D each contain galactose, mannose, and xylose, but the molar ratios of these sugars are not identical, suggesting structural variability. GalXMs are thought to be a group of complex closely related polysaccharides (16, 32).
GalXM, with an average mass of 1 × 105 Da (3, 20), is significantly smaller than GXM (1.7 × 106 Da). Since GalXM has a smaller molecular mass, GalXM is the most numerous polysaccharide in shed capsular polysaccharide preparations on a molar basis, with 2 to 3.5 mol of GalXM for each mol of GXM (20).
The location of GalXM in the C. neoformans capsule is uncertain. In fact, it is not clear whether GalXM is a constituent of the capsule or an exopolysaccharide. An attempt at immunolocalization with the monoclonal antibody (MAb) CN6, which is no longer available, suggested that GalXM was located within the cytoplasm and the cell wall of the acapsular mutant cap67 (16, 32). Given the usefulness of antibodies in studying capsule (5, 13, 26), we have generated a serological reagent for the localization of GalXM. The results suggest that GalXM is a transient component of the capsule, is associated with newly formed capsules, and may be present in vesicular fractions.
MATERIALS AND METHODS
C. neoformans strains.
Several strains of C. neoformans were used in this study: 24067 (serotype D), acapsular mutant cap67 and its parental strain B3501 (serotype D), and NIH 34 (a serotype C reference strain typically used for the production of rabbit anti-C serum) (29). NYC1343, a clinical isolate of serotype C from New York (18), NIH 112, a serotype B strain (15), and serotype A/D strain MAS92-203 were also tested. We also used strains KN99α (serotype A parent strain of GalXM mutants), a ugt1Δ mutant (GalXM mutant in which the UGT gene encodes a putative UDP-galactose transporter), and a uge1Δ mutant (a GalXM mutant in which the UGE gene encodes a putative UDP-glucose epimerase) (23). The uge1Δ strain was also constructed in the serotype D background of Jec21, and the strains were designated as follows: Jec21 (MATα), NE570 (MATα uge1Δ::NAT1), and NE571 MATα uge1Δ::NAT1). To generate the uge1Δ strain (serotype D), the disruption cassette for the gene UGE1 was constructed by overlapping PCR as previously described (24). The primer sequences used are given in Table S1 in the supplemental material. The PCR-amplified fragment was used to transform strain Jec21 by biolistic DNA delivery, and transformants were selected on yeast extract-peptone-dextrose containing 100 μg/ml of nourseothricin (Werner BioAgents). They were then screened for homologous integration, first by PCR and then by Southern blotting, as previously described (24). Two mutant strains were isolated from two independent transformations and stored at −80°C for further studies. The tagged plasmid pNATSTM, used to amplify the selective marker, was kindly provided by Jennifer Lodge (St. Louis, MO).
GXM and GalXM isolation.
GXM was isolated as described by Cherniak et al. (6). GalXM was isolated from the culture supernatant by the method described in references 20 and 32. Briefly, a 400-ml culture of the C. neoformans acapsular mutant of strain cap67 was grown in peptone supplemented with 2% galactose for 7 days. The culture supernatant was separated from the cells by centrifugation at 900 × g for 15 min at room temperature and then concentrated using an Amicon centrifugal filter (Millipore, Bedford, MA) with a molecular-weight-cutoff of 10,000. The material was then dialyzed for 1 week against distilled water, and the 10-kDa retentate, containing GalXM and MPs, was passed through a 0.2-μm filter. The resulting filtrate was then lyophilized and stored at room temperature. The freeze-dried mixture was dissolved in 25 ml start buffer: 0.01 M Tris base and 0.5 M NaCl solution (pH 7.2), to which CaCl2 and Mn(II)Cl2 were sequentially added to final concentrations of 1 mM. To separate the GalXM and MP, solution was then continuously passed through a concanavalin A (ConA)-Sepharose 4B column (2.5 by 10 cm) (Sigma) for 16 h at 4°C using a peristaltic pump with a flow rate of 16 ml/h. The flowthrough and 11 column washes with start buffer were collected as 20-ml fractions (20). To identify carbohydrate-containing fractions, we tested these using the phenol-sulfuric acid assay (12). The fractions were combined, concentrated by ultrafiltration, and dialyzed against water for 7 days. GalXM was then recovered by lyophilization. Compositional analysis of GalXM was done by combined gas chromatography-mass spectrometry (GC-MS) of the per-O-trimethylsilyl derivatives of the monosaccharide methyl glycosides produced from the sample by acidic methanolysis (21). The possibility of ConA contamination was tested by Western blotting with an anti-ConA antibody (Vector Laboratories, Burlingame, CA). No ConA contamination was detected (not shown).
GalXM-Bacillus anthracis PA conjugate.
GalXM hydroxyl groups were activated with cyanogen bromide (Sigma, St. Louis, MO). Briefly, GalXM (10 mg/ml in 0.2 M NaCl) was activated with 10 mg/ml of cyanogen bromide with continuous pH monitoring such that the pH was maintained between 10.5 and 11.0 for 6 min at 4°C. The activated GalXM was then derivatized with the bifunctional linker adipic acid dihydrazide (ADH) (Sigma, St. Louis, MO). To do this, an equal volume of 0.5 M NaHCO3 at pH 8.5 containing 0.5 M ADH was added to the activated GalXM. The reaction mixture was then tumbled at 4°C for 18 h and dialyzed against 0.2 M NaCl for 7 days. The reaction mixture containing GalXM-ADH was then mixed with 5.0 mg/ml of protective antigen (PA) from Bacillus anthracis (Wadsworth Laboratories, New York, NY) and brought to pH 5.6 with 0.05 N HCl and 0.05 M of the water-soluble carbodiimidine, 1-ethyl-3-(3 dimethylaminopropyl)-carboiimide (EDAC) (Sigma, St.Louis, MO). The pH was continuously maintained at 5.6 for 1 h at 4°C. The reaction mixture was then dialyzed against 0.2 M NaCl for 24 h at 4°C (11). Bio-Rad Quick start protein assay was used according to the manufacturer's instructions to detect protein in the conjugate.
Animals.
Six- to 8-week-old BALB/c female mice were obtained from the National Cancer Institute. Long Evans rats (body weight, 200 to 250 g) were obtained from Charles River Laboratories (Wilmington, MA). All animal experiments were done according to institutional guidelines.
Immunizations with GalXM.
Mice and rats were given an intraperitoneal immunization with 0.5, 5, 50, and 500 μg of GalXM; saline; or 100 μl of GalXM-PA conjugate in complete Freund's adjuvant. Mice were then boosted with 0.1 μg of GalXM or 50 μl of the conjugate in incomplete Freund's adjuvant at day 14. The mice were bled, and serum titers were tested. Mice were sacrificed by asphyxiation with CO2.
Serum antibodies.
Blood was collected from mice, and serum was analyzed by enzyme-linked immunosorbent assay (ELISA). Costar plates were coated with 50 μg of GalXM, the plates were blocked with 2% bovine serum albumin (BSA) or Superblock (Pierce, Rockford, IL), and a 1:100 dilution of serum was serially diluted in a 1:3 ratio along the plate. A cocktail of alkaline phosphatase-conjugated anti-immunoglobulin M (IgM), -IgA, and -IgG (H+L) polyclonal antibodies (Southern Biotechnology, Birmingham, AL) at 1 μg/ml was used as the secondary antibody for the detection of bound antibodies. Reactions were developed with p-nitrophenyl phosphate (PNPP), and the absorbance was measured at 405 nm. BSA-coated plates were used as a negative control.
Biotinylated GXM and GalXM.
GalXM (1 mg/ml) was oxidized with 20 mM sodium periodate (Pierce, Rockford, IL) in dimethyl sulfoxide (DMSO) for 30 min at 4°C in the dark. To stop the oxidation reaction, 15 mM glycerol was added to the mixture for 5 min at 4°C. The samples was then passed through Zeba desalting columns (Pierce, Rockford, IL), previously equilibrated with 0.1 M sodium acetate (pH 5.5). A 5 mM solution of biotin hydrazide was added to the oxidized carbohydrate and incubated at room temperature for 2 h. To separate the biotinylated carbohydrates from the unreacted biotin, the samples were again passed through Zeba desalting columns. To test if GXM and GalXM had been biotinylated, an ELISA was done by coating the plate with the biotinylated sample by blocking with 1% BSA for 1 h. A 1:1,000 dilution of streptavidin conjugated to alkaline phosphatase was used as the secondary reagent (Southern Biotechnology, Birmingham, AL), and absorbance was measured at 405 nm after development with PNPP.
Phage display peptide library screening.
The phage display library was synthesized with mRNA extracted from chickens immunized with fluorescein isothiocyanate (FITC)-coupled BSA (1). The light and heavy chain variable regions were amplified by reverse transcription-PCR, cloned into the pComb3X vector, and transformed into Escherichia coli cells following protocols in reference 2. The selection was made in four rounds: rounds 1 to 3 used biotinylated GalXM, cap67 cells, and biotinylated GalXM, respectively. The 4th round was made initially with intact cells, which yielded no phages. We then repeated this round with biotinylated GalXM. Positive clones were selected by ELISA after the final round of selection. These clones were sequenced, and the encoded single-chain fragment variable regions (scFvs) were purified from cell lysates by affinity chromatography in nickel-nitrilotriacetic acid columns (the vector encodes a C-terminal His6 tag). GalXM-specific binding was assessed initially by ELISA and then by immunofluorescence with uge1Δ and ugt1Δ mutants.
Panning with the antigens.
Phage-biotinylated carbohydrate complexes were captured by streptavidin-coated magnetic beads (Dynal Biotech, Lake Success, NY). Beads were washed five times with 1 ml of Tris-buffered saline-Tween 20 (TBST) and pelleted using a magnetic rack each time. Antigen-specific phages were eluted from the beads using 0.1 M glycine at pH 2.2 for 10 min at room temperature. The eluate was then neutralized and transferred to 2 ml of XL-2 cells at an optical density at 600 nm (OD600) of 1 and incubated at room temperature for 15 min.
Selection with intact cells.
Because GalXM binds irregularly to ELISA plates, we did the panning using biotinylated soluble antigen. To avoid selection of irrelevant antibiotin or antistreptavidin bead phages, we decided to alternate rounds of selection with whole cells. The reasoning was that the cells express GalXM in their surface and would not select for the same irrelevant antibodies. Since the two antigen forms could result in the isolation of antibodies with a different range of specificities, this design aimed to isolate scFvs which bind GalXM in solution and in the surface of intact cells. Briefly, cap67 cells were washed twice with TBST, counted and diluted in 1% BSA-TBS to108 cells/ml. They were then incubated with the phage as with the beads above. Phage precipitation and amplification was also done as described above. Input and output titers were measured after each round of panning in ampicillin-supplemented LB (LB-AMP) plates.
Fluorescence microscopy.
C. neoformans strains were grown in Sabouraud dextrose broth (Difco Laboratories, Detroit, MI) for 3 days at 30°C. The cells were washed three times with sterile phosphate-buffered saline (PBS [pH 7.4]) and then counted with a hemocytometer. Strains were normalized to a suspension of 2 × 106 cells/ml and incubated with a 1:100 dilution of GalXM-PA polyclonal serum or nonimmune serum as a control in IF buffer (2% BSA and 0.05% goat serum in PBS). Cells were washed three times with IF buffer and incubated with 4 μg/ml of goat anti-mouse IgM-FITC. Alternatively, to determine cell age, cells were grown in Sabouraud dextrose broth for 2 days. The cell wall was labeled first in a solution of 4-mg/ml EZ-Link sulfo-NHS-LC-biotin [sulfosuccinimidyl-6-(biotinamido)hexanoate] in PBS (Pierce, Rockford, IL) for 30 min at room temperature at a cell density of 2 × 107 cells/ml. Cells were extensively washed with PBS and then placed in the capsule-inducing medium for 2 days. Biotinylated cells were detected using 20 μg/ml of streptavidin-Texas red (Jackson Immunoresearch, West Grove, PA) (19). For staining with wheat germ agglutinin (WGA), 106 yeast cells were suspended in 100 μl of a 5-μg/ml solution of the Alexa Fluor 594 conjugate of WGA (Molecular Probes, Eugene, OR) and incubated for 30 min at 37°C after incubation with GalXM-PA polyclonal serum at same dilution as mentioned earlier (30). For staining with the lipophilic carbocyanine dye Vybrant DiI (Molecular Probes, Eugene, OR), we used 1:100 dilution of GalXM-PA polyclonal serum for 1 h at room temperature and 1:100 goat anti-mouse (GAM)-IgM-FITC and 5 μM Vybrant DiI overnight at 4°C. Cells were suspended in mounting medium (50% glycerol and 50 mM N-propyl gallate in PBS) and analyzed under an Olympus AX70 microscope. Images were acquired using a QImaging Retiga 1300 digital camera and processed using QCapture suite V2.46 software (QImaging, Burnaby, British Columbia, Canada).
Confocal microscopy.
For GalXM staining using the GalXM-PA polyclonal serum, 2 × 106 C. neoformans cells in 100 μl of a buffer (2% BSA with 0.5% goat serum) were incubated with 2 μl of serum overnight at 4°C. The cells were washed three times with buffer and incubated with 4 μg/ml of anti-IgM-FITC as the secondary antibody for 1 h at room temperature. For staining of C. neoformans using the scFv, 2 × 106 cells in 100 μl of IF buffer were incubated with 100 μg/ml of the purified scFv clone D8 overnight at 4°C. The cells were washed three times with buffer and incubated with a 1:100 dilution of antihemagglutinin (anti-HA)-Alexa Fluor 488 as the secondary antibody for 1 h at room temperature. Stained cells were mounted on glass slides as described above and imaged with a Leica SP2 laser scanning confocal microscope. 3D reconstructions of the captured Z-stacks were made using ImageJ (NIH; http://rsb.info.nih.gov/ij/) and Voxx (Indiana University; www.nephrology.iupui.edu/imaging/voxx/) software.
Vesicle isolation and carbohydrate analysis.
Isolation of extracellular vesicles was done using the protocol described by Rodrigues et al. (31). Briefly, cell-free culture supernatants of cap67 cells were obtained by sequential centrifugation at 4,000 and 15,000 × g (15 min at 4°C). These supernatants contained vesicles and were concentrated by approximately 20-fold using an Amicon ultrafiltration system (cutoff of 100 kDa). The concentrate was again centrifuged at 4,000 and 15,000 × g (15 min at 4°C) for cell and debris removal and then at 100,000 × g for 1 h at 4°C. The supernatants were discarded, and pellets were washed by five sequential suspension and centrifugation steps, each consisting of 100,000 × g for 1 h at 4°C with 0.1 M TBS. The 100,000 × g pellets were then suspended in 75% cold ethanol (1 ml). A white precipitate was immediately formed that was collected by centrifuging this suspension for 5,000 × g for 5 min. The supernatant was then discarded, and the pellet was dried by vacuum centrifugation. The dried pellet (1 mg) was then dissolved in methanol-1 M HCl at 80°C (18 to 22 h) and then per-O-trimethylsilylated by treatment with Tri-Sil (Pierce) at 80°C (0.5 h). Sugar composition was then determined by GC-MS analysis of the resulting per-O-trimethylsilyl-derivatized monosaccharides on an HP 5890 gas chromatograph interfaced to a 5970 MSD mass spectrometer, using a Supelco DB-1 fused-silica capillary column (30 m by 0.25 mm inside diameter). The carbohydrate standards used were arabinose, rhamnose, fucose, xylose, glucuronic acid, galacturonic acid, mannose, galactose, glucose, mannitol, dulcitol, and sorbitol.
Modification of C. neoformans cells for epitope analysis. (i) Proteinase K digestion.
Strains 24067, B3501, and cap67 were incubated with 1 mg/ml proteinase K (Roche, Nutley, NJ) in 100 mM Tris-HCl (pH 8.0) for 3 h at 37°C. Proteinase K was heat inactivated by boiling samples for 5 min. As a control, cells not treated with proteinase K were boiled for 5 min.
(ii) Sodium metaperiodate oxidation.
Strains 24067, B3501, and cap67 were incubated with 1 mM or 11 mM sodium metaperiodate (Pierce, Rockford, IL) overnight at 4°C. The reaction was stopped with ethylene glycol, and the cells were washed several times in PBS and IF buffer. Periodate concentrations greater than 10 mM oxidize many sugar residues, such as galactose and mannose (http://www.piercenet.com/files/1161dh5.pdf).
(iii) DMSO.
DMSO removes capsule polysaccharides (4, 14). Strains 24067, B3501, and cap67 were incubated with in pure DMSO (Sigma, St. Louis, MO) for 1, 3 and 24 h. DMSO was washed with PBS, and cells were prepared for immunofluorescence.
Proton NMR of GalXM.
GalXM was dissolved in D2O, and all NMR experiments were performed at 329 K on a Bruker DRX 600MHz spectrometer equipped with a 5-mm inverse-triple-resonance probe. 1D proton spectra were collected with 512 scans of 64 K points over 20 ppm and a total recycle delay of 4 s. The residual water in the spectrum was removed using presaturation of the HOD signal. Spectra were processed with an exponential line broadening of 1 Hz, and the proton chemical shifts were referenced to internal 3-(trimethylsilyl) propionate.
Nucleotide sequence accession number.
The sequence from scFv D8 has been submitted to GenBank under accession no. FJ233885.
RESULTS
Generation of serological reagents for the study of GalXM.
Immunization of mice or rats with GalXM elicited no significant antibody response (data not shown). To circumvent this problem we synthesized a GalXM-protein conjugate using the PA protein from B. anthracis. We then immunized mice and rats with the conjugate and attempted to make MAbs. Although the PA-polysaccharide conjugate was immunogenic, we were unable to generate MAbs using either mice or rats because the hybridomas were unstable. Hence, we hyperimmunized mice and rats and generated polyclonal serum to GalXM. Specific antibodies in this serum were mostly IgM. This serum was used for immunofluorescence on C. neoformans strains cap67, B3501, and 24067. Immunofluorescence revealed a punctate pattern around the cell wall in acapsular strains, a pattern consistent with prior reports that GalXM is mostly cell wall associated (32, 33). Encapsulated strains showed a punctate pattern near or on the edge of the capsule. 3D reconstructions of confocal images produced the same pattern (Fig. 1).
FIG. 1.
Indirect immunofluorescence microscopy of C. neoformans after staining with GalXM-PA polyclonal sera. GalXM hyperimmune serum was used to analyze the strains 24067, cap67, and its parent strain, B3501. (Top panels) Merged bright-field and fluorescence images for strains 24067 and B3501 cells. Punctate fluorescence is observed for GalXM (green). The scale bar is 10 μm. (Lower left panel) Merged images of bright-field and fluorescence images for strain cap67. (Lower right panel) Three-dimensional reconstructions from Z-series stacks of cap67 cells. Green, GalXM; red, GXM; blue, calcofluor white, which stains cell wall chitins. The scale bar is 5 μm.
GalXM-deficient mutants confirm specificity of polyclonal antibody.
To ascertain that the polyclonal serum obtained from GalXM-PA-immunized mice was specific for GalXM, we tested its reactivity for C. neoformans mutants that made no GalXM. First, we tested the uge1Δ and ugt1Δ mutants that were constructed in the serotype A background since these were already available (23). There was no binding to the mutants or to the parental strain KN99α. Earlier work demonstrated that GalXM is not a single molecular entity in serotypes A, C, and D (16). Hence, we hypothesized that our polyclonal reagent was GalXM serotype specific since we had raised the antibody against GalXM from serotype D cap67. Consequently, a GalXM-deficient uge1Δ mutant in the serotype D (Jec21) background was constructed and tested. The results revealed no fluorescence for the uge1Δ mutant and a positive signal for the parental strain Jec21, demonstrating the specificity for GalXM (data not shown).
GalXM is consistent with being vesicle associated.
The punctate fluorescence patterns observed led us to address whether these pockets were transiently secreted or were bound to the capsule. Although GXM was found in extracellular vesicles (31), no information is available with respect to the secretion of other capsular components. Due to the apparent diversity of vesicle morphology, composition, and functionality, we hypothesized that the punctate pattern observed by immunofluorescence for GalXM suggests that it is exopolysaccharide and thus may be secreted in vesicles. To investigate this possibility, vesicles were prepared from culture supernatants of cap67 cells as described previously (31) and analyzed for monosaccharide components by GC-MS. The most abundant monosaccharide detected was glucose, which is present in high concentrations in culture medium. Except for glucose, the only other monosaccharides detected were galactose, mannose, and xylose (Table 1), the components of GalXM.
TABLE 1.
Glycosyl composition analysis of extracellular vesicles produced by cap67 cellsa
| Glycosyl residue | Mass (μg) |
|---|---|
| Arabinose | NDb |
| Ribose | ND |
| Rhamnose | ND |
| Fucose | ND |
| Xylose | + |
| Glucuronic acid | ND |
| Galacturonic acid | ND |
| Mannose | + |
| Galactose | + |
| Glucose | +++ |
| N-Acetylgalactosamine | ND |
| N-Acetylglucosamine | ND |
| Heptose | ND |
| 3-Deoxy-2-manno-2-octulsonic acid | ND |
Positive results (+) are expressed as function of the relative intensity of the peaks in the chromatograms. The identification of the sugars detected by GC (xylose, mannose, galactose, and glucose) was confirmed by MS analysis.
ND, none detected.
Phage display antibodies against GalXM.
Since we were unable to generate MAbs to GalXM due to hybridoma instability, we sought an alternative approach employing a phage display. We first screened a library that expressed scFv antibody fragments from chickens immunized with FITC-BSA, naïve against cryptococcal antigens. After several alternating rounds of panning using biotinylated immunobilized GalXM and C. neoformans cells, we obtained several scFvs that produced punctate immunofluorescence patterns similar to that observed with GalXM-PA polyclonal serum. However, unlike the abundant puncta observed with GalXM-PA serum on the capsule, the scFvs bound at only a few points along the capsule. As all scFvs appeared to have the same binding pattern, we concentrated further studies in one single clone, which we named scFv D8. To investigate whether the polyclonal serum to GalXM and scFv D8 were binding to the same epitope, we simultaneously bound both probes to the capsule and found that they rarely colocalized, implying reactivity with different epitopes (Fig. 2). To investigate the epitope specificity of scFv D8, we studied its binding to biochemically modified C. neoformans cells using proteinase K, DMSO, and sodium metaperiodate. Proteinase K, which degrades all proteins, or DMSO, which partially removes the capsule, had no effect on the binding of serum or the binding of scFv D8. However, sodium metaperiodate abrogated the binding of polyclonal serum, while a stronger punctate pattern was observed using scFv D8. These results suggest that periodate oxidation is disrupting the GalXM epitopes recognized by the polyclonal serum but allows increases in the number of binding sites to scFv D8 (Fig. 3).
FIG. 2.
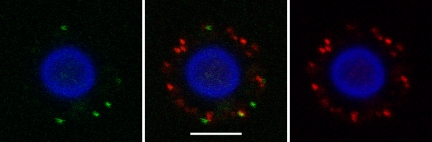
Indirect immunofluorescence microscopy after binding with GalXM-PA polyclonal sera and scFv D8 to C. neoformans cells reveals little colocalization. Indirect immunofluorescence with GalXM-PA polyclonal antibody (red) and phage display protein scFv D8 (green) reveals that both stain the capsule of C. neoformans strain B3501, but only occasional areas suggest colocalization, as implied by the presence of the yellow area. The cell body was stained with Uvitex 2B, a probe that stains cell wall chitins (blue). The scale bar is 5 μm.
FIG. 3.
Epitope specificity of GalXM and scFv D8 shown by indirect immunofluorescence microscopy after binding of GalXM-PA sera and scFv D8 to modified C. neoformans cells. The y axis shows strains B3501 and cap67, which were indirectly stained with GalXM hyperimmune sera and scFv D8. Along the x axis, cells were chemically modified as follows: strains were treated with proteinase K treatment for 24 h, DMSO for 24 h, or 11 mM of sodium metaperiodate for 24 h. The scale bar is 10 μm.
GalXM is not part of the cell wall since it is not covalently bound to cell wall glucans (16). Unpublished results by van de Moer et al. suggested that like MPs GalXM diffuses slowly through the cell wall, cell membrane, and capsule (16). Three-dimensional reconstructions of confocal images using scFv clone D8 in C. neoformans H99 cells revealed a vesiclelike fluorescence structure projecting from the cell wall. Upon further magnification, it appears as if this vesicle-like projection has pushed through the GXM component and resides on the surface of capsule (Fig. 4). We also evaluated whether the scFv D8 reactivity was serotype specific. We tested the reagent with serotypes A, B, C, D, and A/D and found that the scFv binds serotypes A and D but not B and C and A/D (Table 2).
FIG. 4.
Indirect immunofluorescence microscopy after binding of MAb to GXM and scFv D8 to C. neoformans strain H99. scFv D8 (green) shows a vesicle-like structure emerging from the capsule. GXM was detected with MAb 18B7 (red), and the cell wall was detected with calcofluor (blue). The right panel is a magnification of the emerging vesicle. The scale bar is 5 μm. The images are 3D reconstructions using Voxx.
TABLE 2.
Serotype reactivity with scFv and polyclonal serum
| Serotype | Strain | Punctate pattern fora:
|
|
|---|---|---|---|
| GalXM-PA | scFv D8 | ||
| A | H99 | + | + |
| KN99α | − | − | |
| uge1Δ | − | + | |
| ugt1Δ | − | + | |
| B | NIH 112 | − | − |
| C | NIH 34 | − | − |
| NIH 1343 | − | − | |
| D | B3501 | +++ | + |
| cap67 | +++ | + | |
| 24067 | ++ | + | |
| Jec21 | +++ | + | |
| uge1Δ | − | + | |
| A/D | MAS92-203 | ++ | − |
GalXM-PA polyclonal serum and scFv D8 were tested with different serotypes of C. neoformans. +, positive reactivity by immunofluorescence; −, no fluorescence detected.
Proton NMR reveals GalXM variability.
Since the polyclonal serum was serotype specific, we investigated the possibility of serotype-related structural differences in GalXM by using NMR. Earlier studies had suggested that that GalXM may be structurally heterogeneous since its galactose, mannose, and xylose components are linked in 15 different ways (16). The biochemical complexity of that data suggested that the purified GalXM fractions could be part of a composite of several closely related antigens (16). Since our polyclonal antibody did not bind to the serotype A strains, we isolated GalXM from cap67 (serotype D) and compared it to GalXM extracted from two cap59 mutants derived from the serotype A background. Proton NMR revealed subtle differences among the different GalXM molecules on the anomeric region of the located between 5.4 and 4.3 ppm (Fig. 5). The results are consistent with fine structural differences in the GalXM molecule that could translate into antigenic variability.
FIG. 5.
Proton NMR of GalXM. (A) Proton NMR for GalXM of C. neoformans strains cap59 and cap67. (B) The lower panel shows peak differences when the cap67 spectrum is subtracted from the cap59 spectra. Upward peaks show differences in cap59, while downward peaks show differences in cap67.
GalXM does not colocalize with WGA-binding structures and capsular lipophilic regions.
Recently Rodrigues et al. found that wheat germ lectin (WGA) recognized chitin-like components at opposite poles of C. neoformans cells (30). Additionally, the structures recognized by WGA were associated with the cell wall but also visibly projected into the capsular network. The structures recognized by the WGA lectin appeared as round or hook-like projections that formed an interface between the capsule and bud necks (30). Costaining with GalXM-PA serum and fluorescent WGA revealed that the polyclonal serum and lectin fluorescence did not colocalize (Fig. 6). We also evaluated colocalization of the polyclonal serum and a highly lipophilic carbocyanine dye, DiI, which binds to the capsule in a punctate pattern (Eisenman et al., submitted for publication). The results revealed that some GalXM puncta colocalized with DiI, possibly reflecting GalXM release through vesicles. During these colocalization studies, we noticed accumulation of the polyclonal serum around budding cells. We also noted that for budding C. neoformans cells, the formation of a bridge-like structure over the emergent bud is detected by the GalXM-PA polyclonal serum using indirect immunofluorescence (Fig. 6).
FIG. 6.
GalXM colocalization with WGA-binding molecules and capsular lipophilic regions. C. neoformans strain B3501 was used in all panels. (Top left panel) Fluorescence of GalXM (green) and WGA (red) shows no colocalization. The scale is 10 μm. (Top right panel) Merged bright-field and fluorescence images with GalXM (green) and lipophilic stain DiI (red) show some colocalization. The scale bar is 5 μm. (Lower left panel) Merged bright-field and fluorescence images. GalXM hyperimmune sera (green) detects capsule location overlying the beginning of budding. (Right) Voxx 3D reconstruction of the fluorescent image in the left panel of GalXM (green) and GXM (red). The scale bar is 5 μm for both lower panels.
Association of GalXM with daughter cells.
Given that the GalXM-PA polyclonal sera bound strongly to buds, we evaluated the relative binding of this reagent to mother and daughter cells. To differentiate between them, cryptococcal cell wall was labeled with sulfo-NHS-LC-biotin, whereby the biotin covalently binds to the cell wall and does not segregate to new buds. Hence, the mother cell stains with the Texas red-labeled streptavidin, whereas the nascent budding cells do not. Using this labeling protocol, we consistently found that daughter cells preferentially bound more GalXM-PA serum, implying the presence of GalXM on the nascent polysaccharide capsules (Fig. 7).
FIG. 7.
GalXM association with daughter cells. (Left panel) Indirect immunofluorescence confocal microscopy using GalXM-PA serum detects accumulation of GalXM in budding cells of B3501. This image is the green channel only of the Z-stack shown in the upper right panel of Fig. 6. The scale bar is 5 μm. (Right panel) Immunofluorescence using biotin-streptavidin-Texas red was used to discriminate between mother and bud cells. GalXM (green) is predominantly around the daughter cell. The scale bar is 10 μm.
DISCUSSION
Early attempts to generate a reagent to study GalXM in the capsule partially succeeded with the generation of CN6, a MAb of the IgM isotype. This antibody was generated against GalXM using spheroplast lysate (33). CN6 provided insights that GalXM was near the cell wall, but apart from one publication (33), no additional studies were reported with encapsulated cells. Unfortunately, this antibody is no longer available (33). Given a revival of interest in GalXM, we sought to develop new serological reagents to localize this polysaccharide in the capsule. We were unable to make MAbs due to the toxicity of GalXM (unpublished) but did obtain a hyperimmune serum to a GalXM-PA conjugate that was confirmed to bind GalXM. We also generated a synthetic antibody, scFv D8, to an unknown oxidized carbohydrate epitope that illustrates the antigenic complexity of the capsule. Using these reagents, we have established the location of GalXM in the capsule, associated GalXM secretion with vesicles, and associated GalXM in the nascent capsules of daughter cells, suggesting a possible role in budding.
We solved the problem of poor immunogenicity by generating a glycoconjugate of GalXM using a protein carrier, PA from Bacillus anthracis. Protein conjugation is a well-known approach to increase the immunogenicity of polysaccharide antigens. This approach was successfully used to generate antibodies to GXM by creating a conjugate with tetanus toxoid (11). Immunofluorescence studies revealed that GalXM was located on the outer edge of the capsule in encapsulated strains and near the cell wall in acapsular strains. We also observed that the pattern of binding was punctate.
To establish the specificity of the GalXM-PA polyclonal serum for GalXM, we investigated its reactivity with the GalXM-deficient uge1Δ and ugt1Δ mutants that were generated in a serotype A background. The results revealed no binding of GalXM-PA hyperimmune sera to parent strain KN99α or to the mutants (23). Since this result implied the possibility of serotype-related differences in GalXM structure, we generated a uge1Δ strain in the serotype D background of Jec21. The results revealed that the polyclonal antibody bound parent strain Jec21 but not the uge1Δ strain. Based on this result, we concluded that the GalXM-PA hyperimmune serum is specific for serotype D GalXM. This specificity could reflect the fact that the GalXM used in the conjugate was obtained from C. neoformans var. neoformans cap67.
These small discrete pockets observed by immunofluoresence led us hypothesize that GalXM may be potentially delivered in vesicles. Vesicle preparations of cap67 revealed that, except for glucose, which came probably from the culture media, the only monosaccharides detected were galactose, mannose, and xylose, the components that make up GalXM. Although these results do not conclusively establish that GalXM is secreted in vesicles, they are consistent with the notion that some GalXM is vesicle associated.
In a second approach to generate specific antibodies to GalXM by phage display, we inadvertently made scFv antibodies to oxidized polysaccharides. The scFv D8 produced a diffuse punctate pattern around the capsule similar to that observed with GalXM-PA hyperimmune sera. However, the two probes rarely colocalized, implying that they recognized different antigens. We hypothesized that we may have altered the epitopes on GalXM in the screen that isolated scFv D8, which employed periodate-modified biotinylated GalXM for phage display biopanning. Periodate is known to modify the structure of GalXM such that xylose is completely eliminated and two-thirds of galactose and mannose residues are oxidized (7). We did not attempt a phage display technique using nonoxidized GalXM since there are no other reagents that specifically recognize the GalXM molecule.
In an attempt to understand the specificity of D8, we treated C. neoformans cells with proteinase K, DMSO, and sodium metaperiodate and compared the levels of binding of GalXM-PA hyperimmune sera and scFv D8. The results showed that neither proteinase K nor DMSO affected polyclonal or scFv D8 binding to C. neoformans cells. In contrast, periodate significantly decreased the binding of GalXM-PA hyperimmune sera and increased the binding of scFv D8, consistent with destruction of native GalXM epitopes and enrichment for newly oxidized polysaccharide epitopes. Given that scFv D8 was isolated by screening on modified GalXM and taking into account the results of binding to modified C. neoformans cells, we tentatively conclude that D8 recognizes a polysaccharide moiety that is antigenically different from GalXM or GXM. ELISA experiments with D8 showed low affinity to GalXM, as well as some binding to GXM (not shown). Double staining with a MAb to GXM and scFv D8 produced fascinating images of D8-binding material surrounded by GXM consistent with vesicular transport and export. Hence scFv D8 appears to bind to a yet-unidentified oxidized GalXM-like material that is in close association with GXM and may be a new polysaccharide component of the capsule.
We also tested the serum and scFv D8 with serotypes A, B, C, D, and A/D and found that the scFvs, like the serum, bind serotypes A and D, suggesting that they react with polysaccharides from C. neoformans var. grubii and C. neoformans var. neoformans, respectively. We noticed that both reagents reacted more strongly with serotype D. However, neither the GalXM-PA immune serum nor the scFvs bound to C. neoformans gatii serotypes B and C. Another difference was that, unlike the polyclonal serum, scFv D8 does not bind to serotype A/D. Analysis of GalXM from serotype A and D strains by NMR revealed subtle differences consistent with structural differences that could translated into antigenic differences.
Additionally we tested whether the polyclonal antibody was binding other previously described structures (30) in the capsule such as chitin-like oligomers and lipids. The results revealed that the polyclonal immune sera did not colocalize with WGA and only partially colocalized with DiI (30). Interestingly, we noted that the GalXM-PA serum stained strongly nascent capsules around budding cells. We distinguished nascent cells from mother cells by labeling with sulfo-NHS-LC-biotin, a reagent that covalently binds to the cell wall and does not segregate to the bud. We confirmed that the polyclonal antibody detects more GalXM around the daughter bud, suggesting that GalXM might be involved in the structural rearrangements of the capsular polysaccharide that accompany budding. An alternate explanation for the preferential binding of the polyclonal antibody to the buds is that the capsule over the buds is less dense, and this may allow for more efficient antibody penetration.
In summary, we have made two new reagents for the study of C. neoformans capsular polysaccharides in the form of a hyperimmune serum to GalXM and an scFv that binds to oxidized carbohydrates. Using these reagents, we established that GalXM was found in the C. neoformans capsule in discrete pockets amid the GXM layers and was abundant in the capsules of nascent buds. Furthermore, isolated vesicle fractions contained galactose, consistent with secretion of GalXM by trans-cell-wall vesicular transport, as has been proposed for GXM (31). Given the association of GalXM with vesicles and the partial colocalization with DiI, this punctate distribution could reflect vesicular transport. The strong staining of GalXM-PA immune sera for the capsules of budding daughter cells also reflects increased vesicular transport at sites of nascent capsule formation or a role in capsular remodeling. Our results are most easily interpreted as indicating that GalXM is primarily an exopolysaccharide, with a possible role in capsule formation during budding, rather than functioning as a structural component of mature capsule. Given the strong immunomodulatory activity of GalXM, one is tempted to speculate that this material is exported for fungal cell defense and could serve an important role for cryptococcal survival in various ecologic niches, including mammalian hosts.
Supplementary Material
Acknowledgments
We thank all participants and the instructors at the 2006 Cold Spring Harbor Phage Display of Proteins and Peptides course for synthesizing the phage display library. Carlos Barbas provided the anti-FITC scFv control. We thank the Complex Carbohydrate Research Center at The University of Georgia for GalXM composition analysis. We also thank Karen Chave at the Wadsworth Center and Johanna Rivera for providing the Bacillus anthracis PA protein, which was prepared with support from the Northeast Biodefense Center (5U54AI057158-05).
The Complex Carbohydrate Research Center is supported by the Department of Energy Center for Plant and Microbial Complex Carbohydrates (DE-FG09-93ER-20097). The instrumentation in the AECOM Structural NMR Resource is supported by the Albert Einstein College of Medicine and in part by grants from the NSF (DBI9601607 and DBI0331934), the NIH (RR017998), and the HHMI Research Resources for Biomedical Sciences. This work was supported by NIH grants AI33774, AI33142, and HL59842-01 to A.C. M.D. was supported by NCI/NIH training grant 2T32CA009173-31(3). This work was supported by a grant from ANR to G.J. (Erapathogenomics programme).
The data in this article are from a thesis to be submitted by Magdia De Jesus in partial fulfillment of the requirements for the degree of doctor of philosophy in the Sue Golding Graduate Division of Medical Science, Albert Einstein College of Medicine, Yeshiva University, Bronx, NY.
Footnotes
Published ahead of print on 24 October 2008.
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Andris-Widhopf, J., C. Rader, P. Steinberger, R. Fuller, and C. F. Barbas III. 2000. Methods for the generation of chicken monoclonal antibody fragments by phage display. J. Immunol. Methods 242159-181. [DOI] [PubMed] [Google Scholar]
- 2.Barbas, C. F., D. R. Burton, J. K. Scott, and G. J. Silvermann. 2001. Phage display: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 3.Bose, I., A. J. Reese, J. J. Ory, G. Janbon, and T. L. Doering. 2003. A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryot. Cell 2655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryan, R. A., O. Zaragoza, T. Zhang, G. Ortiz, A. Casadevall, and E. Dadachova. 2005. Radiological studies reveal radial differences in the architecture of the polysaccharide capsule of Cryptococcus neoformans. Eukaryot. Cell 4465-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casadevall, A., J. Mukherjee, and M. D. Scharff. 1992. Monoclonal antibody based ELISAs for cryptococcal polysaccharide. J. Immunol. Methods 15427-35. [DOI] [PubMed] [Google Scholar]
- 6.Cherniak, R., L. C. Morris, B. C. Anderson, and S. A. Meyer. 1991. Facilitated isolation, purification, and analysis of glucuronoxylomannan of Cryptococcus neoformans. Infect. Immun. 5959-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherniak, R., E. Reiss, and S. Turner. 1982. A galactoxylomannan antigen of Cryptococcus neoformans serotype A. Carbohydr. Res. 103239-250. [Google Scholar]
- 8.Cherniak, R., and J. B. Sundstrom. 1994. Polysaccharide antigens of the capsule of Cryptococcus neoformans. Infect. Immun. 621507-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Jesus, M., E. Hackett, M. Durkin, P. Connolly, A. Casadevall, R. Petraitiene, T. J. Walsh, and L. J. Wheat. 2007. Galactoxylomannan does not exhibit cross-reactivity in the Platelia Aspergillus enzyme immunoassay. Clin. Vaccine Immunol. 14624-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Jesus, M., C. G. Park, Y. Su, D. L. Goldman, R. M. Steinman, and A. Casadevall. 2008. Spleen deposition of Cryptococcus neoformans capsular glucuronoxylomannan in rodents occurs in red pulp macrophages and not marginal zone macrophages expressing the C-type lectin SIGN-R1. Med. Mycol. 46153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devi, S. J. N., R. Schneerson, W. Egan, T. J. Ulrich, D. Bryla, J. B. Robbins, and J. E. Bennett. 1991. Cryptococcus neoformans serotype A glucuronoxylomannan-protein conjugate vaccines: synthesis, characterization, and immunogenicity. Infect. Immun. 593700-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28350-356. [Google Scholar]
- 13.Eckert, T. F., and T. R. Kozel. 1987. Production and characterization of monoclonal antibodies specific for Cryptococcus neoformans capsular polysaccharide. Infect. Immun. 551895-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goren, M. B. 1967. Experimental murine cryptococcosis: effect of hyperimmunization to capsular polysaccharide. J. Immunol. 98914-922. [PubMed] [Google Scholar]
- 15.Ikeda, R., T. Shinoda, Y. Fukazawa, and L. Kaufman. 1982. Antigenic characterization of Cryptococcus neoformans serotypes and its application to serotyping of clinical isolates. J. Clin. Microbiol. 1622-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.James, P. G., and R. Cherniak. 1992. Galactoxylomannans of Cryptococcus neoformans. Infect. Immun. 601084-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levitz, S. M., and C. A. Specht. 2006. The molecular basis for the immunogenicity of Cryptococcus neoformans mannoproteins. FEMS Yeast Res. 6513-524. [DOI] [PubMed] [Google Scholar]
- 18.Litvintseva, A. P., R. Thakur, L. B. Reller, and T. G. Mitchell. 2005. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in Sub-Saharan Africa. J. Infect. Dis. 192888-892. [DOI] [PubMed] [Google Scholar]
- 19.Maxson, M. E., E. Dadachova, A. Casadevall, and O. Zaragoza. 2007. Radial mass density, charge, and epitope distribution in the Cryptococcus neoformans capsule. Eukaryot. Cell 695-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McFadden, D. C., M. De Jesus, and A. Casadevall. 2006. The physical properties of the capsular polysaccharides from Cryptococcus neoformans suggest features for capsule construction. J. Biol. Chem. 2811868-1875. [DOI] [PubMed] [Google Scholar]
- 21.Merkle, R. K., and I. Poppe. 1994. Carbohydrate composition analysis of glycoconjugates by gas-liquid chromatography/mass spectrometry. Methods Enzymol. 2301-15. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell, T. G., and J. R. Perfect. 1995. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin. Microbiol. Rev. 8515-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moyrand, F., T. Fontaine, and G. Janbon. 2007. Systematic capsule gene disruption reveals the central role of galactose metabolism on Cryptococcus neoformans virulence. Mol. Microbiol. 64771-781. [DOI] [PubMed] [Google Scholar]
- 24.Moyrand, F., and G. Janbon. 2004. UGD1 encoding the Cryptococcus neoformans UDP-glucose dehydrogenase is essential for growth at 37°C and for capsule biosynthesis. Eukaryot. Cell 31601-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukherjee, J., A. Casadevall, and M. D. Scharff. 1993. Molecular characterization of the humoral responses to Cryptococcus neoformans infection and glucuronoxylomannan-tetanus toxoid conjugate immunization. J. Exp. Med. 1771105-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukherjee, J., M. D. Scharff, and A. Casadevall. 1992. Protective murine monoclonal antibodies to Cryptococcus neoformans. Infect. Immun. 604534-4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perfect, J. R. 2005. Cryptococcus neoformans: a sugar-coated killer with designer genes. FEMS Immunol. Med. Microbiol. 45395-404. [DOI] [PubMed] [Google Scholar]
- 28.Pericolini, E., E. Cenci, C. Monari, M. De Jesus, F. Bistoni, A. Casadevall, and A. Vecchiarelli. 2006. Cryptococcus neoformans capsular polysaccharide component galactoxylomannan induces apoptosis of human T-cells through activation of caspase-8. Cell. Microbiol. 8267-275. [DOI] [PubMed] [Google Scholar]
- 29.Petter, R., B. S. Kang, T. Boekhout, B. J. Davis, and K. J. Kwon-Chung. 2001. A survey of heterobasidiomycetous yeasts for the presence of the genes homologous to virulence factors of Filobasidiella neoformans, CNLAC1 and CAP59. Microbiology 1472029-2036. [DOI] [PubMed] [Google Scholar]
- 30.Rodrigues, M. L., M. Alvarez, F. L. Fonseca, and A. Casadevall. 2008. Binding of the wheat germ lectin to Cryptococcus neoformans suggests an association of chitinlike structures with yeast budding and capsular glucuronoxylomannan. Eukaryot. Cell 7602-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigues, M. L., L. Nimrichter, D. L. Oliveira, S. Frases, K. Miranda, O. Zaragoza, M. Alvarez, A. Nakouzi, M. Feldmesser, and A. Casadevall. 2007. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot. Cell 648-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaishnav, V. V., B. E. Bacon, M. O'Neill, and R. Cherniak. 1998. Structural characterization of the galactoxylomannan of Cryptococcus neoformans Cap67. Carbohydr. Res. 306315-330. [DOI] [PubMed] [Google Scholar]
- 33.van de Moer, A., S. L. Salhi, R. Cherniak, B. Pau, M. L. Garrigues, and J. M. Bastide. 1990. An anti-Cryptococcus neoformans monoclonal antibody directed against galactoxylomannan. Res. Immunol. 14133-42. [DOI] [PubMed] [Google Scholar]
- 34.Villena, S. N., R. O. Pinheiro, C. S. Pinheiro, M. P. Nunes, C. M. Takiya, G. A. DosReis, J. O. Previato, L. Mendonca-Previato, and C. G. Freire-de-Lima. 2008. Capsular polysaccharides galactoxylomannan and glucuronoxylomannan from Cryptococcus neoformans induce macrophage apoptosis mediated by Fas ligand. Cell. Microbiol. 101274-1285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.