Abstract
Streptomyces coelicolor A3(2) ftsI- and ftsW-null mutants produced aerial hyphae with no evidence of septation when grown on a traditional osmotically enhanced medium. This phenotype was partially suppressed when cultures were grown on media prepared without sucrose. We infer that functional FtsZ rings can form in ftsI- and ftsW-null mutants under certain growth conditions.
Rod-shaped bacteria that produce a peptidoglycan cell wall synthesize lateral-wall material during cell elongation and produce septa during cytokinesis. Most rod-shaped bacteria possess separate systems for these processes, each containing a protein of the SEDS (shape, elongation, division, and sporulation) family and a cognate class B penicillin-binding protein (PBP) (7, 9, 11). In Escherichia coli, the protein pairs involved in cell elongation and division are RodA-PBP2 and FtsW-FtsI (PBP3), respectively. However, some bacteria possess three protein pairs, as in Bacillus subtilis, where sporulation-specific division genes exist in addition to those for vegetative functions (15). Streptomyces coelicolor is a gram-positive, filamentous bacterium that requires cell division only for sporulation (13). Its genome possesses four homologous SEDS-PBP pairs (3).
Here we report the characterization of S. coelicolor cell division genes ftsI and ftsW. We show that ftsI and ftsW are dispensable for colony formation but are required for efficient cell division. Similar to the ftsL and divIC mutants (2), the ftsI- and ftsW-null mutants displayed medium-dependent phenotypic defects that are more severe on an osmotically enhanced medium. We suggest that because the ftsI and ftsW mutants are able to divide when grown on certain media, other proteins may compensate for the loss of FtsI and FtsW. Chains of spores are produced under certain growth conditions, implying that ladder-like arrays of Z rings (18) must be stably formed and function in the absence of FtsI and FtsW under certain conditions.
Identification of ftsI and ftsW homologues in S. coelicolor.
The S. coelicolor ftsI and ftsW homologues, ftsISc (StrepDB [http://streptomyces.org.uk/] accession number SCO2090) and ftsWSc (accession number SCO2085), are located in the division and cell wall (dcw) cluster (Fig. 1). We determined the gene sequences prior to the S. coelicolor genome project. ftsISc is predicted to encode a 654-amino-acid, 69.5-kDa bitopic membrane protein with 26% (160/602) of its residues identical to B. subtilis PBP 2B (the FtsI homologue), 31% (188/604) identical to B. subtilis SpoVD (the sporulation-specific FtsI homologue), and 29% (175/586) identical to E. coli FtsI. ftsWSc is predicted to encode a 456-amino-acid, 48-kDa integral membrane protein with 36% (128/351) of its residues identical to B. subtilis SpoVE (the sporulation-specific FtsW homologue), 36% (137/373) identical to B. subtilis FtsW (YlaO), and 31% (114/358) identical to E. coli FtsW. FtsWSc lacks the unique C-terminal extension required for interaction with FtsZ in the related actinomycete Mycobacterium tuberculosis (5), and the three other S. coelicolor SEDS homologues have even shorter C termini. Thus, assignments for ftsWSc and ftsISc are based on sequence similarity and synteny in the dcw cluster, as well as on mutant phenotypes like those of S. coelicolor ftsL and divIC mutants (2). A similar analysis of the same genes was concurrently reported (14).
FIG. 1.
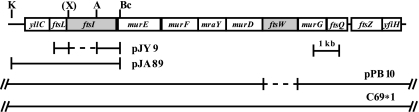
Chromosomal region containing ftsISc and ftsWSc. The S. coelicolor dcw cluster containing ftsISc and ftsWSc is shown. Open boxes indicate gene positions. Key restriction sites used to make mutations and complementation plasmids are shown [A, Acc65I; Bc, BclI; K, KpnI; X, XbaI; (X), introduced by PCR]. The diagram below the map contains horizontal bars representing the inserts of plasmids used in the construction of deletion mutations (pJY9 and pPB10) and those used in genetic complementation experiments, pJA89 and C69*1. Deletion-insertion mutations are indicated by the dashed lines (pJY9 and pPB10).
ftsISc and ftsWSc are dispensable, and mutants have medium-dependent phenotypes.
We constructed a C69-derived cosmid (16) called pBP10 by using in vivo recombination in E. coli containing a deletion-insertion mutation (8), in which an internal 1.3-kb fragment of ftsWSc (codons 5 to 450 of 456) was replaced by an apramycin resistance cassette (Fig. 1). This cosmid was introduced by conjugation into wild-type S. coelicolor strain M145 (Table 1), independently isolated apramycin-resistant, kanamycin-sensitive marker replacement strains were obtained, and one representative was named PFB22 [ΔftsW::acc(3)IV]. Therefore, like ftsZ, ftsQ, ftsL, and divIC (2, 12, 13), ftsWSc was not essential for growth or viability in S. coelicolor. Because murG (located 7 nucleotides downstream of ftsWSc) appears to be an essential gene in S. coelicolor (P. Bidey and J. R. McCormick, unpublished results), strains containing an unmarked, in-frame deletion of ftsWSc (ΔftsW) were isolated by transformation using pPB10Δ (pPB10 treated with FLP recombinase) to avoid the potential effects of transcriptional polarity on downstream genes. One representative PFB22-derived marker replacement strain that was apramycin sensitive and kanamycin sensitive was named PFB26 (ΔftsW). PFB22 and PFB26 were verified by Southern blot analysis and diagnostic PCR, respectively (see the supplemental material). PFB26 (ΔftsW) grew well, producing colonies that were more robust than those of PFB22 [ΔftsW::acc(3)IV] on all media (MM, R2YE, and MS medium [10]) tested, indicating that the aac(3)IV cassette did have a partial polar effect on the expression of murG (and other downstream genes). Colonies of PFB26 produced a slightly less gray pigmented aerial mycelium than the wild type when grown on MS agar (Fig. 2). Consistent with the pale gray aerial mycelium on MS medium, the ftsW mutant produced infrequent chains of spores (data not shown). Unlike ftsZ- and ftsQ-null mutants (12, 13), the ftsWSc-null mutant PFB26 did not overproduce the blue-pigmented antibiotic actinorhodin on MM agar, suggesting that vegetative cross-wall formation is not drastically impaired (data not shown).
TABLE 1.
S. coelicolor strains used in this study
| Straina | Genotype | Reference or source |
|---|---|---|
| DU152 | ΔdivIC::aphI | 2 |
| DU191 | ΔftsL::hyg | 2 |
| HU133 | ΔftsZ::aphI | 13 |
| HU151 | ΔftsQ::addA | 12 |
| J2210 | ΔwhiH::hyg | 17 |
| JBY5 | ΔftsI::aphI | This study |
| M145 | Prototroph SCP1− SCP2− | 10 |
| PFB22 | ΔftsW::acc(3)IV | This study |
| PFB26 | ΔftsW | This study |
All mutant strains are M145 derivatives.
FIG. 2.
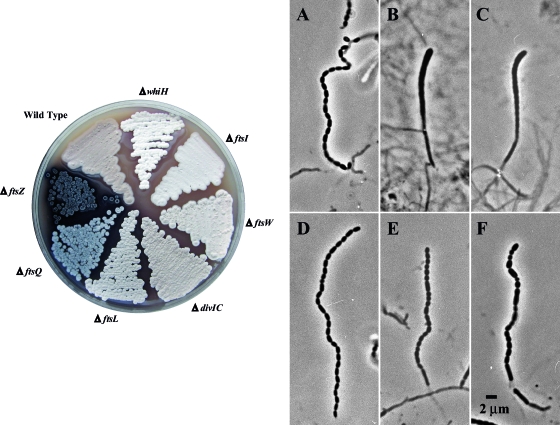
Growth phenotypes of mutants and phase-contrast microscopy of wild-type and mutant aerial hyphae. (Left) MS agar medium was used to visualize the gray pigment associated with spore formation within the aerial mycelium (colony surface). Cultures were incubated for 5 days at 30°C. Strains M145 (wild type), HU133 (ΔftsZ), HU151 (ΔftsQ), DU152 (ΔdivIC), DU191 (ΔftsL), JBY5 (ΔftsI), PFB26 (ΔftsW), and J2210 (ΔwhiH, white mutant control) are shown (see Table 1). (Right) Phase-contrast micrographs of coverslip lifts removed after 6 days of growth on R2YE (A to C) or glucose MM (D to F) are shown. Panels A and D show wild-type spore chains (strain M145). Also shown are aerial hyphae of the ftsWSc-null mutant PFB26 (B, R2YE; E, MM) and ftsISc-null mutant JBY5 (C, R2YE; F, MM). The sporulation phenotype is heterogeneous, and only one category of aerial hyphae produced by the division mutants is shown (see text for details).
Similar to the ftsL and divIC mutants (2), the strain containing ΔftsW exhibited cell division defects during sporulation, as judged by phase-contrast microscopy. When grown on MM agar containing glucose, the ΔftsW strain displayed a variety of aerial-filament morphologies, including smooth aerial hyphae (10%), aerial hyphae with regularly spaced shallow constrictions (45%), and aerial hyphae containing chains of normal spores, as well as chains of spores with lysed compartments and/or abnormally sized and shaped spores (45%) (Fig. 2E). These morphologies were terminal phenotypes, and hyphae with shallow constrictions were not eventually converted into chains of spores. When PFB26 (ΔftsW) was grown on the osmotically enhanced medium R2YE, the division defect was greatly exacerbated, and the number of aerial filaments observed was reduced. On R2YE, the aerial mycelium contained undifferentiated aerial hyphae (70%) and aerial filaments containing shallow regularly spaced constrictions (30%) (Fig. 2B). The constrictions appeared to be shallower than those produced by ftsL- and divIC-null strains (2).
We constructed a pOJ260-derived plasmid (4) called pJY9 by using in vitro recombination and a restriction site introduced by PCR. pJY9 contains a deletion-insertion mutation, in which an internal 1.2-kb fragment of ftsISc (codons 10 to 406 of 654) was replaced by aphI (Fig. 1). This nonreplicating mutagenic plasmid (pJY9) was introduced by transformation into wild-type S. coelicolor strain M145. Independent deletion-insertion mutants were isolated as neomycin-resistant, apramycin-sensitive transformants, and the representative marker replacement strain JBY5 (ΔftsI::aphI) was used for further study. JBY5 was verified by Southern blot analysis (see the supplemental material). Thus, as with every other division mutant characterized to date, ftsISc is dispensable for growth and viability of S. coelicolor, making this an advantageous system for studying bacterial cytokinesis. We did not anticipate polar effects of the ΔftsI::aphI mutation due to our analysis of a similar ftsL mutant (2). aphI was inserted in the same orientation as ftsISc, and a gap between ftsISc and murE suggests the potential for a promoter in the intercistronic space (208 nucleotides). JBY5 grew well and produced robust colonies on MM, R2YE, and MS media, and its aerial mycelium was paler gray on MS medium than that of the wild type (Fig. 2). Consistent with the pale-gray aerial mycelium on MS medium, the ftsI mutant produced infrequent chains of spores (data not shown) and the vegetative hyphae occasionally contained regions with bulges. Unlike the ftsZ- and ftsQ-null mutants, the ftsISc-null mutant did not overproduce the blue-pigmented antibiotic actinorhodin on MM agar, suggesting that vegetative cross-wall formation is not drastically impaired (data not shown).
When grown on MM, the ftsISc-null strain was essentially indistinguishable from the ftsWSc-null strain, displaying chains of spores (approximately 45% of which was a mixture of normal and aberrant sizes and shapes), as well as aerial filaments with regularly spaced, shallow constrictions (45%) and those completely devoid of constrictions (10%) (Fig. 2F). These morphologies were terminal phenotypes, and hyphae with shallow constrictions were not eventually converted into chains of spores. However, when the strain was grown on the osmotically enhanced medium R2YE, the division defect was greatly exacerbated, and the number of aerial filaments observed was reduced. The phenotype of the ftsISc-null mutant was characterized by short, cane-shaped aerial filaments possessing very shallow, regularly spaced constrictions (30%) (Fig. 2C) and undifferentiated aerial hyphae (70%).
Similar to the ftsL and divIC mutants (2), the ftsISc and ftsWSc mutants were more defective when grown on the osmotically enhanced medium R2YE. To determine if the same parameter affected the division phenotypes of the ftsISc and ftsWSc mutants, we omitted sucrose from R2YE (normal concentration, 0.3 M). We found that the ftsISc- and ftsWSc-null mutants grown on R2YE without sucrose mimicked the phenotypes of the mutants grown on MM (data not shown) (1). Interestingly, the ftsI mutant produced aerial filaments that were shorter than those produced by the ftsW, ftsL, and divIC mutants on the osmotically enhanced medium (data not shown). We speculate that FtsI may also be involved in peptidoglycan synthesis during elongation of the lateral walls of aerial hyphae, resulting in shortened cane-shaped filaments produced by the mutant.
Finally, because a significant fraction of the aerial hyphae of the ftsISc and ftsWSc mutants develop into chains of separated spores on MM, we infer that functional FtsZ rings form, producing evenly spaced sporulation septa under certain conditions. This is in contrast to Mycobacterium smegmatis, in which it has been shown that FtsZ localized to the midcell when FtsW was depleted but cell separation was not completed (6).
Genetic complementation experiments.
Complementation experiments were performed with the most restrictive medium (osmotically enhanced R2YE) to ensure that the division defects observed were imparted by the introduced mutations. pJA89 (2), a low-copy-number plasmid containing Pdcw-yllC-ftsL-ftsISc (Fig. 1), partially restored sporulation to the ftsISc-null mutant, indicating that the phenotype is due in part to the absence of ftsISc (data not shown) (1). C69*1 (a matable version of cosmid C69) (Fig. 1), which contains the dcw cluster and flanking regions, was able to complement the division defect imparted by the ftsWSc-null mutation when integrated by homologous recombination (data not shown) (1). We conclude that the division phenotype was linked to the ΔftsW mutation.
Septal morphologies of the ftsISc- and ftsWSc-null mutants.
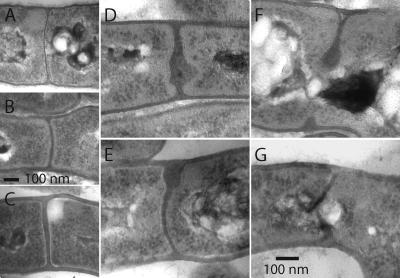
We examined the morphology of the septa in hyphae by transmission electron microscopy (TEM) as previously described (2), using strains grown on R2YE, where the division defect was most severe. Approximately half of all vegetative cross-walls examined in the ftsISc and ftsWSc mutants appeared to be normal (Fig. 3, compare panels A to C). Otherwise, the cross-walls in the mutants were aberrant, with cell wall material on both sides of the division site that had not met in the middle (Fig. 3F), or completed cross-walls were wavy and/or thickened (Fig. 3D, E, and G). Complete cross-walls were reported in a concurrent study of ftsI and ftsW mutants (14). However, those observations were done with cells grown on a very unusual choice of medium for Streptomyces (2× yeast-tryptone [YT] medium), and perhaps the salt concentration may have affected the observed phenotypes. Unexpectedly, no sporulation septa could be definitively identified for either mutant. The surfaces of ftsWSc and ftsISc mutant colonies were difficult to embed for TEM, and the sporadic population of aerial filaments observed by phase-contrast microscopy may have been too infrequent in the TEM sections. The mutants produced mostly aerial hyphae that lacked constrictions, and constrictions are used to unambiguously identify aerial hyphae.
FIG. 3.
TEMs showing vegetative cross-walls in wild-type and division mutant filaments. Strains were grown for 6 days on R2YE, the medium showing the largest defect in division, and then prepared for TEM examination. (A) A wild-type vegetative cross-wall (strain M145), which can vary in thickness, is shown. (B and C) Normal vegetative cross-walls (50%) are shown for ΔftsISc (JBY5) and ΔftsWSc (PFB26) mutants, respectively. (D to G) The other half of the cross-walls are formed aberrantly, as shown for the ftsISc-null mutant (D and F) and the for ftsWSc-null mutant (E and G).
Summary and conclusions.
Of the six division genes of S. coelicolor genetically analyzed to date (2, 12-14; this study), only two are more strictly required for division under most circumstances (ftsQ and ftsZ) and only one is absolutely required (ftsZ). By analogy to the divisome described for other systems, it appears that proteins presumably recruited late in the division pathway of S. coelicolor are not absolutely required for cell division under certain conditions. Because S. coelicolor is predicted to encode four SEDS/PBP pairs, one other pair must be able to substitute for FtsW/FtsI to support cell division under some growth conditions. One of the SEDS proteins presumably moves lipid II-linked precursors across the membrane (19), and a PBP must synthesize septal peptidoglycan.
Nucleotide sequence accession numbers.
The sequences of ftsISc and ftsWSc were deposited in GenBank under accession numbers AF123319 and U10879, respectively. Subsequently, the sequences determined by the genome project agreed (3).
Supplementary Material
Acknowledgments
We thank Joseph Suhan (Carnegie Mellon University) for providing assistance with electron microscopy and Berthold Gust for advice concerning lambda Red recombination.
This work was supported by grant GM56915 from the National Institutes of Health (to J.R.M.) and an award from Beta Beta Beta National Biology Honor Society (to A.B.C.).
Footnotes
Published ahead of print on 31 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bennett, J. A. 2006. Molecular genetic analysis of division and development in Streptomyces coelicolor. Ph.D. dissertation. Duquesne University, Pittsburgh, PA.
- 2.Bennett, J. A., R. M. Aimino, and J. R. McCormick. 2007. Streptomyces coelicolor genes ftsL and divIC play a role in cell division but are dispensable for colony formation. J. Bacteriol. 1898982-8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417141-147. [DOI] [PubMed] [Google Scholar]
- 4.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 11643-49. [DOI] [PubMed] [Google Scholar]
- 5.Datta, P., A. Dasgupta, S. Bhakta, and J. Basu. 2002. Interaction between FtsZ and FtsW of Mycobacterium tuberculosis. J. Biol. Chem. 27724983-24987. [DOI] [PubMed] [Google Scholar]
- 6.Datta, P., A. Dasgupta, A. K. Singh, P. Mukherjee, M. Kundu, and J. Basu. 2006. Interaction between FtsW and penicillin-binding protein 3 (PBP3) directs PBP3 to mid-cell, controls cell septation and mediates the formation of a trimeric complex involving FtsZ, FtsW and PBP3 in mycobacteria. Mol. Microbiol. 621655-1673. [DOI] [PubMed] [Google Scholar]
- 7.den Blaauwen, T., M. A. de Pedro, M. Nguyen-Disteche, and J. A. Ayala. 2008. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 32321-344. [DOI] [PubMed] [Google Scholar]
- 8.Gust, B., G. Chandra, D. Jakimowicz, T. Yuqing, C. J. Bruton, and K. F. Chater. 2004. Lambda Red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv. Appl. Microbiol. 54107-128. [DOI] [PubMed] [Google Scholar]
- 9.Ikeda, M., T. Sato, M. Wachi, H. K. Jung, F. Ishino, Y. Kobayashi, and M. Matsuhashi. 1989. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J. Bacteriol. 1716375-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, England.
- 11.Matsuhashi, M., M. Wachi, and F. Ishino. 1990. Machinery for cell growth and division: penicillin-binding proteins and other proteins. Res. Microbiol. 14189-103. [DOI] [PubMed] [Google Scholar]
- 12.McCormick, J. R., and R. Losick. 1996. Cell division gene ftsQ is required for efficient sporulation but not growth and viability in Streptomyces coelicolor A3(2). J. Bacteriol. 1785295-5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCormick, J. R., E. P. Su, A. Driks, and R. Losick. 1994. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol. Microbiol. 14243-254. [DOI] [PubMed] [Google Scholar]
- 14.Mistry, B. V., R. Del Sol, C. Wright, K. Findlay, and P. Dyson. 2008. FtsW is a dispensable cell division protein required for Z-ring stabilization during sporulation septation in Streptomyces coelicolor. J. Bacteriol. 1905555-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Real, G., A. Fay, A. Eldar, S. M. Pinto, A. O. Henriques, and J. Dworkin. 2008. Determinants for the subcellular localization and function of a nonessential SEDS protein. J. Bacteriol. 190363-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Redenbach, M., H. M. Kieser, D. Denapaite, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 2177-96. [DOI] [PubMed] [Google Scholar]
- 17.Ryding, N. J., G. H. Kelemen, C. A. Whatling, K. Flardh, M. J. Buttner, and K. F. Chater. 1998. A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 29343-357. [DOI] [PubMed] [Google Scholar]
- 18.Schwedock, J., J. R. McCormick, E. R. Angert, J. R. Nodwell, and R. Losick. 1997. Assembly of the cell division protein FtsZ into ladder-like structures in the aerial hyphae of Streptomyces coelicolor. Mol. Microbiol. 25847-858. [DOI] [PubMed] [Google Scholar]
- 19.van Dam, V., R. Sijbrandi, M. Kol, E. Swiezewska, B. de Kruijff, and E. Breukink. 2007. Transmembrane transport of peptidoglycan precursors across model and bacterial membranes. Mol. Microbiol. 641105-1114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.