Abstract
Knockout mutation of STM4432 resulted in a growth-deficient phenotype of Salmonella enterica serovar Typhimurium in the presence of myo-inositol (MI) as the sole carbon source. STM4432 is part of a 22.6-kb genomic island which spans STM4417 to STM4436 (genomic island 4417/4436) and is responsible for MI degradation. Genome comparison revealed the presence of this island in only six Salmonella strains and a high variability of the iol gene organization in gram-negative bacteria. Upon nonpolar deletion of 11 island loci, the genes involved in six enzymatic steps of the MI pathway were identified. The generation time of S. enterica serovar Typhimurium in minimal medium with MI decreases with higher concentrations of this polyol. Reverse transcriptase PCR showed five separate transcriptional units encompassing the genes iolA-iolB, iolE-iolG1, iolC1-iolC2, iolD1-iolD2-iolG2, and iolI2-iolH. Luciferase reporter assays revealed a strong induction of their promoters in the presence of MI but not glucose. The main regulator, IolR, was identified due to a reduced lag phase of a strain mutated in STM4417 (iolR). Deletion of iolR resulted in stimulation of the iol operons, indicating its negative effect on the iol genes of S. enterica serovar Typhimurium in rich medium at a transcriptional level. Bandshift assays demonstrated the binding of this putative repressor to promoter sequences of iolA, iolC1, and iolD1. Binding of IolR to its own promoter and induced iolR expression in an IolR-negative background demonstrate that its transcription is autoregulated. This is the first characterization of MI degradation in a gram-negative bacterium, revealing a complex transcriptional organization and regulation of the S. enterica serovar Typhimurium iol genes.
More than 60 carbon sources are known to be utilizable by Salmonella enterica serovar Typhimurium (10). Among them is myo-inositol (MI), a substrate that is ubiquitous in soil and plants, where it appears as a free form or as phospholipid derivatives. Old reported that MI utilization by S. enterica serovar Typhimurium strains is temperature dependent and that 95% of all strains investigated fermented MI at 25°C, although they had been designated inositol nonfermenting at 37°C (23).
In addition to S. enterica serovar Typhimurium, growth of gram-negative bacteria on MI has been demonstrated so far for representatives of the genera Serratia and Klebsiella (18) and for Rhizobium leguminosarum (24). The enzymatic steps of the MI degradation were partially analyzed in Aerobacter (reclassified as Klebsiella) aerogenes (3). The genetics and biochemistry of bacterial MI utilization have been described in most detail for Bacillus subtilis (31, 33, 35). In this organism, the iol divergon, comprising the operons iolABCDEFGHIJ and iolRS, and the gene iolT, located elsewhere on the chromosome, were shown to be responsible for MI degradation that finally results in an equimolar mixture of dihydroxyacetone phosphate, acetyl coenzyme A, and CO2 (33). Two transporters belonging to the major facilitator superfamily have been identified in B. subtilis (34). Inactivation of iolT caused an obvious growth defect of B. subtilis, while a mutant with a knockout mutation of iolF, encoding the second MI transporter, showed a significant growth effect only when iolT was mutated simultaneously. IolR is a repressor that regulates the iol divergon of B. subtilis, including iolT (34, 36). It binds to the operator sites within the iol promoters in the absence of MI. If this polyol is present in the medium, it is converted to the intermediate 2-deoxy-5-keto-d-gluconic acid 6-phosphate, which acts as an inducer by antagonizing IolR DNA binding (33). Other bacteria able to grow on MI as the sole carbon source are Corynebacterium glutamicum, Clostridium perfringens, and Lactobacillus casei strain BL23 (14, 17, 30). In C. perfringens, all iol genes except iolR are unidirectionally organized, and a single transcript of 15.6 kb has been identified (14). L. casei BL23 was the first example of a lactic acid bacterium able to utilize MI (30). The iol genes in this organism are located on a 12.8-kb insertion organized in a manner similar to that in C. perfringens. A more complex organization of iol genes was reported for Corynebacterium glutamicum, in which a second gene cluster encodes redundant functions in MI utilization, including oxidation and transport (17). The iol regulon is subjected to carbon catabolite repression mediated by CcpA at least in B. subtilis and L. casei (21, 30).
The molecular genetics of MI degradation by a gram-negative bacterium have not been investigated. Here, we describe that the knockout of several genes in genomic island (GEI) 4417/4436 results in growth-negative phenotypes of S. enterica serovar Typhimurium on MI. The activities of iol gene promoters under various growth conditions were quantified using the luciferase reporter, and the complex transcriptional organization of the iol genes essential for MI degradation was determined by reverse transcriptase PCR (RT-PCR). IolR, encoded by STM4417, was characterized as a negative regulator of Salmonella MI utilization that also regulates its own expression. The binding of this repressor to all but one of the promoters controlling the MI utilization genes was demonstrated by gel mobility shift (GMS) assays.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. S. enterica serovar Typhimurium and Escherichia coli cultures were grown in Luria-Bertani (LB) broth (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl) or in minimal medium (MM) (M9 medium supplemented with 2 mM MgSO4, 0.1 mM CaCl2, and 55.5 mM [1%, wt/vol] MI or 27.8 mM [0.5%, wt/vol] glucose). If necessary, the media were supplemented with the following antibiotics: ampicillin (150 μg/ml), kanamycin (50 μg/ml), chloramphenicol (25 μg/ml), or streptomycin (50 μg/ml). For solid media, 1.5% agar (wt/vol) was added. For all growth and promoter probe experiments, bacterial strains were grown in LB medium overnight at 37°C, washed twice in phosphate-buffered saline, and then adjusted to an optical density at 600 nm (OD600) of 0.005 in the desired liquid growth medium or streaked on agar plates. Growth curves were obtained from bacterial cultures incubated at 37°C under rigorous shaking in 250 ml flasks with 50 ml of MM. The OD600 was measured at appropriate time intervals as indicated.
TABLE 1.
Strains and plasmids used in this study
| Bacterial strain or plasmid | Description and relevant features | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | deoR endA1 gyrA96 hsdR17(rK− mK+) recA1 relA1 supE44 λthi-1 Δ(lacZYA-argFV169) | 11 |
| BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm rne131 | 27 |
| S. enterica serovar Typhimurium strains | ||
| 14028 | Wild-type strain ATCC 14028 | ATCC |
| 14028 STM4417::pIDM1 | Insertion-duplication mutant with STM4417 knockout | This study |
| 14028 STM4432::pIDM1 | Insertion-duplication mutant with STM4432 knockout | This study |
| 14028s | Spontaneous streptomycin-resistant mutant of 14028 | This study |
| 14028s ΔiolR | In-frame iolR (STM4417) deletion mutant | This study |
| 14028s ΔiolB | In-frame iolB (STM4420) deletion mutant | This study |
| 14028s ΔiolA | In-frame iolA (STM4421) deletion mutant | This study |
| 14028s ΔiolE | In-frame iolE (STM4424) deletion mutant | This study |
| 14028s ΔiolG1 | In-frame iolG1 (STM4425) deletion mutant | This study |
| 14028s ΔiolI1 | In-frame iolI1 (STM4427) deletion mutant | This study |
| 14028s ΔiolC | In-frame iolC (STM4429-30) deletion mutant | This study |
| 14028s ΔiolD2 | In-frame iolD2 (STM4432) deletion mutant | This study |
| 14028s ΔiolG2 | In-frame iolG2 (STM4433) deletion mutant | This study |
| 14028s ΔiolI2 | In-frame iolI2 (STM4435) deletion mutant | This study |
| 14028s ΔiolH | In-frame iolH (STM4436) deletion mutant | This study |
| 14028s ΔSTM3253 | In-frame STM3253 deletion mutant | This study |
| Y. enterocolitica W22703 | Wild-type strain, biovar 2, serovar O:9; Nalr Res− Mod+ pYV− | 6 |
| P. luminescens subsp. laumondii TT01 | 8 | |
| Plasmids | ||
| pKD4 | Kanr, pir dependent, FRT sites | CGSC, Yale (7) |
| pKD46 | Lambda Red helper plasmid; Ampr | CGSC, Yale (7) |
| pCP20 | FLP recombinase plasmid; Cmr Ampr | CGSC, Yale (7) |
| pET28b | Expression vector, T7lac promoter; Kanr | Novagen |
| pET28b-iolR | iolR cloned into pET28b for IolR overexpression and purification | This study |
| pIDM1 | Temperature-sensitive plasmid; repA Tetr | 9 |
| pBR322 | Ampr Tetr | Fermentas |
| pBR322-iolR | iolR with putative promoter region cloned into pBR322 for complementation | This study |
| pBR322-iolE | iolE with putative promoter region cloned into pBR322 for complementation | This study |
| pDEW201 | Promoter probe vector; AmprluxCDABE | 28 |
| pDEW201-PiolR | pDEW201 with 299 bp upstream of iolR (STM4417) | This study |
| pDEW201-PiolA | pDEW201 with 288 bp upstream of iolA (STM4421) | This study |
| pDEW201-PiolE | pDEW201 with 321 bp upstream of iolE (STM4424) | This study |
| pDEW201-PiolG1 | pDEW201 with 335 bp upstream of iolG1 (STM4425) | This study |
| pDEW201-PiolC1 | pDEW201 with 301 bp upstream of iolC1 (STM4430) | This study |
| pDEW201-PiolD1 | pDEW201 with 325 bp upstream of iolD1 (STM4431) | This study |
| pDEW201-PiolG2 | pDEW201 with 301 bp upstream of iolG2 (STM4433) | This study |
| pDEW201-PiolI2 | pDEW201 with 301 bp upstream of iolI2 (STM4435) | This study |
| pDEW201-PiolH | pDEW201 with 299 bp upstream of iolH (STM4436) | This study |
| pDEW201-PargS | pDEW201 with 244 bp upstream of argS | This study |
| pDEW201-Pdef | pDEW201 with 350 bp upstream of def | This study |
| pDEW201-′STM0047′ | pDEW201 with 350 bp of STM0047 without promoter homology | This study |
Standard procedures.
DNA manipulations and isolation of chromosomal or plasmid DNA were performed according to standard protocols (25) and following the manufacturers′ instructions. GeneRuler DNA Ladder Mix (Fermentas, St. Leon-Rot, Germany) was used as a marker for DNA analysis. Plasmid DNA was transformed via electroporation by using a Bio-Rad Gene Pulser II as recommended by the manufacturer and as described previously (15). PCRs were carried out with Taq polymerase (Fermentas). As a template for PCR, chromosomal DNA, plasmid DNA, or an aliquot of a single colony resuspended in 100 μl H2O was used. Oligonucleotides used for PCRs are listed in Table S1 in the supplemental material. Strains of an S. enterica serovar Typhimurium mutant library were characterized as described previously (16). S. enterica serovar Typhimurium gene numbers refer to the LT2 annotation (NC 003197). The websites http://globin.cse.psu.edu/enterix and http://www.microbesonline.org/ were used to determinate the distribution of S. enterica serovar Typhimurium open reading frames in the genomes of gram-negative species. Promoter sequences located upstream of the identified genes were predicted with BPROM (Softberry, Inc.).
Phenotypic testing of carbon source utilization.
A set of S. enterica serovar Typhimurium mutants was screened for their ability to utilize a number of 63 different substrates that are possible carbon sources for S. enterica serovar Typhimurium (10). For this purpose, we used a colorimetric assay based on the reduction of tetrazolium violet as final electron acceptor during respiration due to carbon catabolism (4, 22). Salmonella cells were grown overnight in LB medium at 37°C, washed twice with phosphate-buffered saline, resuspended in inoculation solution (M9 medium supplemented with 2 mM MgSO4, 0.1 mM CaCl2, 0.03% pluronic F68, 0.02% gellan gum, and 0.01% tetrazolium violet) and adjusted to an OD600 of 0.3; 90 μl of this cell suspension was then mixed with 10 μl (0.5 M) of each carbon source solution, pipetted in a 96-well microtiter plate, and measured after 24 and 48 h at 37°C in a microtiter plate reader (Tecan, Männedorf, Switzerland) at OD620. Sucrose and lactose, which are not metabolized by S. enterica serovar Typhimurium, served as control substrates.
Construction of deletion mutants and complementing plasmids.
In-frame STM3253, STM4417 (iolR), STM4420 (iolB), STM4421 (iolA), STM4424 (iolE), STM4425 (iolG1), STM4427 (iolI1), STM4430/4429 (iolC1/iolC2), STM4432 (iolD2), STM4433 (iolG2), STM4435 (iolI2), and STM4436 (iolH) deletion mutants were constructed by the one-step method based on the phage λ Red recombinase (7). Briefly, PCR products comprising the kanamycin resistance cassette of plasmid pKD4, including the flanking FRT sites, were generated using pairs of 70-nucleotide-long primers that included 20-nucleotide priming sequences for pKD4 as template DNA. Homology extensions of 50 bp overlapped 18 nucleotides of the 5′ end and 36 nucleotides of the 3′ end of the target gene (19). Five hundred to 1,000 ng of fragment DNA was transferred into S. enterica serovar Typhimurium strain 14028s cells harboring plasmid pKD46. Allelic replacement of the target gene by the kanamycin resistance cassette was controlled by PCR, and nonpolar deletion mutants were obtained upon transformation of pCP20. Gene deletions were verified by PCR analysis and DNA sequencing.
To complement deleted genes, the coding sequences of iolR and iolE plus approximately 300 bp of their upstream region were amplified from chromosomal DNA of strain 14028 with primers listed in Table S1 in the supplemental material. PCR products were digested with EcoRI and SalI (Fermentas) and ligated (T4 DNA ligase; Gibco) into the promoterless vector pBR322 to generate pBR322-iolR and pBR322-iolE, respectively. Their construction was verified by PCR and restriction analysis.
RNA preparation and RT-PCR.
RNA was isolated according to the modified single-step method of Chomczynski and Sacchi (5). Briefly, 15 ml of an S. enterica serovar Typhimurium culture grown in MM supplemented with MI to an OD600 of ∼0.4 was centrifuged, and the cell pellet was resuspended in 1 ml of Trizol (Invitrogen, Karlsruhe, Germany). The cells were disrupted in a Ribolyzer (Hybaid, Heidelberg, Germany) as described recently (12). Following chloroform extraction, nucleic acids were precipitated, washed, and resuspended in 30 μl diethyl pyrocarbonate-treated H2O. DNase treatment was performed with RQ1 DNase I (Promega, Mannheim, Germany) according to the manufacturer's instruction. Annealing of reverse primers (see Table S1 in the supplemental material) was performed in a total volume of 10 μl containing 75 ng of total RNA, 10 pmol reverse primer, and 20 mM deoxynucleoside triphosphate mix using the following protocol: 75°C for 2 min, 70°C for 1 min, 65°C for 1 min, 55°C for 1 min, 50°C for 1 min, 45°C for 1 min, and 42°C for 60 min. Immediately after the mixture reached 42°C, 10 μl RT mix (Promega) with 0.1 M dithiothreitol and 200 U RT (Promega) was added to generate cDNA. Heat inactivation of RT was performed by incubation at 70°C for 15 min, and 2 μl of this sample was then used as the PCR template.
Cloning of promoter fusions.
Putative promoter regions spanning approximately 300 bp upstream of the start codons of the genes iolR (STM4417), iolA (STM4421), iolE (STM4424), iolG1 (STM4425), iolC1 (STM4430), iolD1 (STM4431), iolG2 (STM4433), iolI2 (STM4435), iolH (STM4436), argS (STM1909), and def (STM3406) and an intragenic fragment of STM0047 without promoter homology were amplified from chromosomal DNA of S. enterica serovar Typhimurium 14028 by PCR using the primers listed in Table S1 in the supplemental material. The fragments were then cloned via EcoRI and BamHI or EcoRI and KpnI (Fermentas) upstream of the promoterless luxCDABE genes into the multiple-cloning site of pDEW201. After transformation into E. coli DH5α cells, plasmids containing the correct transcriptional lux fusions were isolated and verified by PCR, restriction analysis, and sequencing.
Quantification of promoter activity.
Bioluminescence measurements were performed in 96-well plates. For growth in MM containing either 27.8 mM glucose or 55.5 mM MI, bacterial cells were grown at 37°C for 11 h (glucose) and 70 h (MI) in 15 ml centrifuge tubes without agitation. At appropriate time points, 200 μl of each sample was transferred to the 96-well plate, and the OD600 and the bioluminescence, measured as relative light units (RLU), were recorded in a Wallac Victor3 1420 multilabel counter (Perkin-Elmer Life Sciences, Turku, Finland).
Overexpression of iolR.
The iolR gene without its stop codon was cloned into plasmid pET28b using the restriction sites XhoI and NhoI, thus introducing a C-terminal fusion of a His6 tag for protein purification purposes. pET28b-iolR was transformed into E. coli BL21, and the expected clone was verified by restriction analysis. An overnight culture of this strain was diluted 1:100 in 100 ml LB medium supplemented with 150 μg/ml ampicillin and incubated for 3 h at 37°C at 180 rpm. Heterologous expression of iolR was then induced by adding 0.1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG). After incubation for 4 h at 37°C and 180 rpm, the cells were harvested by centrifugation at 4°C (30 min, 104 rpm), and the pellet was resuspended in 1 ml buffer A (300 mM NaCl, 50 mM Na3PO4). The cells were subsequently lysed by two passages through a French press (SLM Aminco Instruments, Rochester, NY), and residual cell debris was removed by centrifugation at 4°C (20 min, 1.4 × 104 rpm). After addition of 10 μl of the protease inhibitor phenylmethylsulfonyl fluoride (100 mM), 10 μl of the supernatant containing soluble proteins mixed with 10 μl 2× Laemmli buffer was applied to sodium dodecyl sulfate-polyacrylamide gels to verify IolR-His6 overexpression, and separated proteins were stained with Coomassie blue.
Purification of IolR-His6 and GMS assays.
Protein IolR-His6 was purified using Talon metal affinity resin (Clontech Laboratories, Mountain View, CA). One milliliter of the protein extract was mixed with 1 ml of the resin and incubated for 1 h at room temperature. The probe was then washed 10 times with 0.5 ml buffer A and 5 times with 0.5 ml buffer B (buffer A containing 7.5 mM imidazole). IolR-His6 was eluted 10 times using 0.5 ml buffer C (buffer A with 150 mM imidazole). Fractions containing large amounts of IolR-His6 were pooled, and the buffer was exchanged with GMS buffer (50 mM Tris-HCl [pH 7.5], 50 mM KCl, 10 mM MgCl2, 0.5 mM EDTA, and 10% [vol/vol] glycerol) by gel filtration using PD-10 columns (GE Healthcare, Munich, Germany) (26). The protein concentration was determined in a Nanodrop spectrophotometer (Thermo Fischer Scientific, Langenselbold, Germany), and the purity of eluted fractions was analyzed by separation on a 15% sodium dodecyl sulfate-polyacrylamide gel.
For the GMS assays, putative promoter regions of iolR, iolA, iolE, iolG1, iolC1, iolD1, iolG2, iolI2, and iolH were amplified as described above, and 100 ng of DNA was then mixed with increasing amounts of purified IolR-His6 in GMS buffer. As a control, 100 ng of competitor DNA was added, resulting in a total volume of 20 μl. After incubation for 45 min at room temperature, the samples were loaded with 4 μl of 6× loading dye (Fermentas) on a 9.5% native polyacrylamide gel and separated at 120 V for 3 h in 1× Tris-borate-EDTA buffer precooled at 4°C. DNA was stained in ethidium bromide solution and visualized by UV irradiation.
RESULTS
The MI utilization genes of S. enterica serovar Typhimurium are located on a GEI.
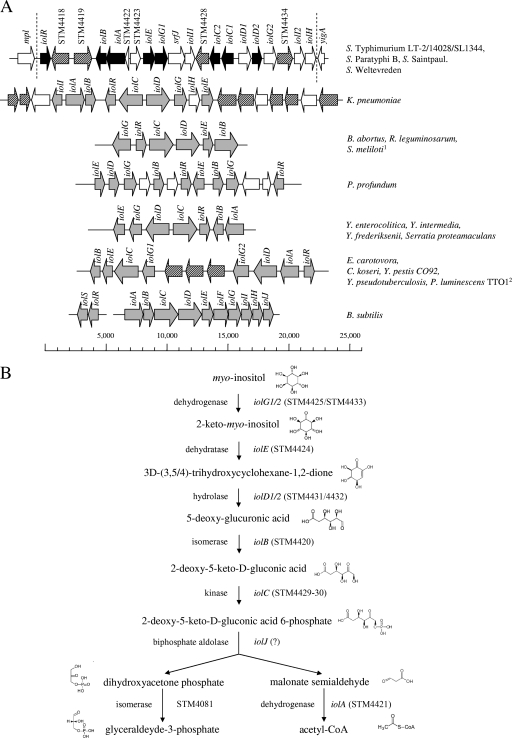
A total of 177 clones of a mutant library of strain 14028 had been characterized with respect to the site of an insertional knockout (15). Fifty mutated genes were predicted to be involved in carbohydrate metabolism and transport, and each mutant was therefore tested for its ability to respire in the presence of one of 66 carbon sources, including MI. One mutant was not able to use this substrate as the sole carbon source, and a second mutant appeared to metabolize MI within 24 h in comparison to the wild-type strain, which showed a significant respiration signal only after 48 h. The strains are mutated in the genes STM4432 and STM4417, which belong to a GEI of 22.6 kb that starts with STM4417 and ends with STM4436 (20). Homology searches using BLAST programs (1) revealed 12 genes to be possibly involved in MI metabolism by S. enterica serovar Typhimurium (Fig. 1A). The island also carries four genes coding for putative sugar transporters, two hypothetical regulatory genes, one gene of unknown function, and srfJ. The last one is regulated by the two-component system SsrAB, that also controls the expression of Salmonella pathogenicity island 2 genes (29). The annotation of the MI utilization genes of S. enterica serovar Typhimurium shown in Fig. 1A is essentially based on homologies to B. subtilis and is also in line with a comparative annotation performed recently (30). Frameshifts have split the S. enterica serovar Typhimurium iolC and iolD homologues into two open reading frames annotated iolC1/2 and iolD1/2. The frameshift in iolC might result in two functional proteins, because IolC2 represents an intact enzyme domain with a putative kinase function. The enzymes IolG2, IolD1, and IolD2 are predicted to require thiamine pyrophosphate as a cofactor. The GC content of GEI 4417/4436 (50.7%) does not significantly differ from that of the whole genome (52.2%), a finding that argues against a recent acquisition of the Salmonella MI utilization island by horizontal gene transfer.
FIG. 1.
(A) Examples of iol divergons. GEI 4417/4436 (22.6 kb) of S. enterica serovar Typhimurium is presented in comparison to the structural organization of iol genes from B. subtilis and several gram-negative bacteria. Salmonella genes experimentally demonstrated in this study to belong to the inositol divergon are depicted in black; their homologues in other pathogens are shown in gray. Genes encoding putative permeases are hatched. 1, in this organism, iolG is transcribed in the same orientation as the other genes; 2, the iol cluster of P. luminescens is similar to that of E. carotovora but lacks two of three putative permease genes. (B) Reconstruction of the pathway for MI degradation in S. enterica serovar Typhimurium. Seven stepwise reactions are involved in MI degradation to glyceraldehyde-3-phosphate and acetyl coenzyme A (acteyl-CoA). None of the genes from GEI 4417/4436 encodes a homologue of a biphosphate aldolase. Chemical structures were taken from the Kyoto encyclopedia of genes and genomes (13).
iol genes of gram-negative bacteria.
GEI 4417/4436 is present in the genomes of the S. enterica serovar Typhimurium strains LT2, 14028, SL1344; S. enterica serovar Saintpaul strain SARA23; S. enterica serovar Weltevreden strain HI_N05-537; and S. enterica serovar Paratyphi B strain SPB7. Most genes of this island are absent in the genomes of the E. coli strains K-12, O157:H7, CFT073, 042, and E2348/69; of Shigella flexneri, Shigella dysenteriae M131649, and Shigella sonnei 53G; of eight S. enterica serovars (serovars Typhi CT18/Ty2, Paratyphi A/C, Enteritidis, Dublin, Gallinarum, and Diarizonae); of Salmonella bongori, and of Vibrio cholerae. A chromosomal fragment of Klebsiella pneumoniae carries nine genes involved in MI degradation and seven genes encoding putative transporters. Permeases that might play a role in MI uptake were also found in an iol gene cluster present in Erwinia carotovora, Citrobacter koseri, Yersinia pestis, Yersinia pseudotuberculosis, and Photorhabdus luminescens. Yersinia enterocolitica and two nonpathogenic Yersinia species share the same organization of iol genes, as do Brucella abortus, Rhizobium leguminosarum, and Sinorhizobium meliloti.
We therefore tested Y. enterocolitica and P. luminescens for their ability to utilize MI as the only carbon source. Indeed, we observed growth of psychrotrophic Y. enterocolitica at 15°C and 22°C, but not at 37°C, and of P. luminescens at 30°C (data not shown). This result shows that six genes, as exemplified by Yersinia spp., are sufficient for MI degradation by gram-negative species.
The pathway of MI degradation in S. enterica serovar Typhimurium is depicted in Fig. 1B. The annotation and the functional assessment of the genes essentially follow enzymological studies of B. subtilis and K. aerogenes (2, 3).
Growth properties of S. enterica serovar Typhimurium in the presence of MI.
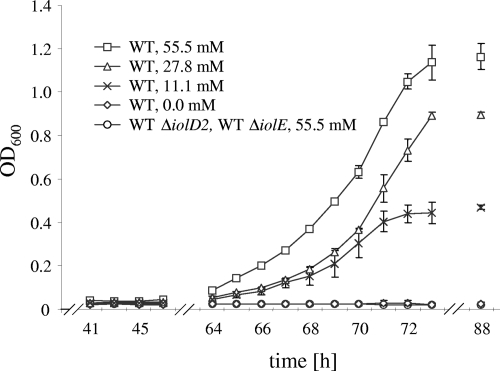
We investigated the growth behavior of strain 14028 in MM supplemented with various concentrations of MI. Three features appeared to be characteristic of S. enterica serovar Typhimurium growth under these conditions: (i) the maximum OD600 in the presence of MI parallels that of the strain grown in glucose (data not shown), (ii) the growth is dose dependent, and (iii) the lag phase is prolonged to approximately 60 h (Fig. 2). As already reported by Old, the wild-type strain produced abundant amounts of a brown pigment (23). This pigmentation appeared only in stationary phase. It was absent when the strain was cultivated anaerobically and might therefore result from metabolite oxidation.
FIG. 2.
Growth curves of the S. enterica serovar Typhimurium wild-type strain 14028 in MM without or with increasing concentrations of MI. The iolD2 and iolE deletion mutants were cultivated in the presence of 55.5 mM MI. Zero growth of the wild-type strain and the two mutants in the absence of this carbon source was monitored for at least 100 h after inoculation. Standard deviations from at least three independent experiments are shown. The molarity of MM with respect to MI is indicated. WT, wild type.
Phenotypes of deletion mutants.
To experimentally demonstrate that genes of GEI 4417/4436 are responsible for MI degradation by S. enterica serovar Typhimurium, nonpolar deletions of iolB, iolA, iolE, iolG1, iolI1, iolC1/2, iolD2, iolG2, iolI2, iolH, and STM3253 were constructed as described above. Growth of these 11 mutants was monitored at least for 5 days, or until the cultures reached stationary phase. No growth deficiencies of the mutants were observed in LB medium or in MM supplemented with glucose (data not shown). The mutants 14028s ΔiolIB, 14028s ΔiolA, 14028s ΔiolE, 14028s ΔiolG1, 14028s ΔiolC, and 14028s ΔiolD2 did not grow in liquid MM containing MI as the sole carbon source, clearly demonstrating the role of the deleted genes in MI utilization. In contrast, growth deficiencies of the mutants 14028s ΔSTM3253, 14028s ΔiolI1, 14028s ΔiolI2, 14028s ΔiolH, and 14028s ΔiolG2 in comparison to the wild type were not observed under these conditions. IolI1 and IolI2 might be functionally redundant, and the role of IolH remains to be disclosed. Two genes coding for proteins with homology to IolG from B. subtilis (IolG1) and to a putative MI dehydrogenase of Lactobacillus plantarum WCFS1 (IolG2) are present in GEI 4417/4436, a redundancy that might explain the wild-type-like growth of the iolG2 mutant. STM3253 encodes a protein with a significant homology of 38% to B. subtilis IolJ. IolJ is responsible for the formation of dihydroxyacetone phosphate and malonate semialdehyde from 2-deoxy-5-keto-d-gluconic acid 6-phosphate (Fig. 1B), but GEI 4417/4436 does not encode such a biphosphate aldolase. The wild-type-like phenotype of the deletion mutant (data not shown), however, excludes a role of STM3253 in MI degradation. All mutants were also streaked on MM agar plates containing 55.5 mM MI and incubated for 64 h. The phenotype of the iolE deletion mutant could be complemented with pBR322-iolE as shown by growth on MM agar plates and in liquid medium (data not shown). These results indicate that iolB, iolA, iolD2, iolE, iolG1, and iolC1/2 are required for MI degradation as indicated in Fig. 1A, thus confirming the pathway reconstruction in Fig. 1B.
Differential expression of genes involved in MI degradation.
In order to investigate the regulation of the genes in GEI4417/4436, fragments of approximately 300 bp located upstream of the start codons of STM4417, iolA, iolE, iolG1, iolC1, iolD1, iolG2, iolI2, and iolH were cloned into the promoter probe vector pDEW201 carrying the luxCDABE cassette. Promoter fragments of def and argS, encoding peptide deformylase and arginyl-tRNA synthetase, respectively, were cloned as positive controls, and a 350-bp intragenic fragment of STM0047 without promoter homology served as a negative control. No promoter sequences could be found upstream of iolB, iolC2, and iolD2. Recombinant plasmids were transformed into strain 14028s and a mutant with a deletion of STM4417, and bioluminescence was measured during growth experiments in LB broth or in MM containing either MI or glucose until the cells reached stationary phase. Regardless of carbon source and growth phase, the reporter did not respond to the fragments upstream of iolG1, iolG2, and iolH in a wild-type-like background in comparison to negative controls with a nonsense sequence cloned into pDEW201 or the empty vector pDEW201 (Table 2). In the presence of glucose and in rich medium, the promoters of STM4417 and iolC1 were transcriptionally active, emitting approximately 2 × 105 to 3 × 105 RLU/OD600 unit. The putative iolD1 promoter region resulted in light emission slightly above the threshold level set by the control construct with the nonsense fragment STM0047, whose activity ranges from 1.25 × 104 to 1.87 × 104 RLU/OD600 unit. The strong induction of the iolA promoter under the same growth conditions is probably due the role of IolA in alanine, aspartate, and propanoate metabolism (13). In MM with MI, the promoters of STM4417, iolE, iolC1, iolD1, and iolI2 were at least 10-fold induced and the iolA promoter 3-fold induced during the exponential growth phase. A similar promoter induction pattern was observed when a mutant lacking STM4417 was grown in MM with glucose, indicating a STM4417-mediated negative regulation of the genes required for MI degradation. Due to its obvious negative regulatory function in MI degradation, we annotated STM4417 as iolR in accordance with the MI repressor protein IolR of gram-positive bacteria. The iolE and iolI2 promoters are also strongly induced in the iolR-negative background in comparison to their transcriptional activity in the presence of glucose. However, their absolute RLU/OD600 unit values are at least 2 orders of magnitude lower than those of the IolR-regulated promoters of iolA, iolC1, and iolD1. The high induction rates of PiolE and PiolI2 in the presence of MI compared to glucose, however, point to a role of both promoters in MI utilization, and they might indirectly have been repressed by IolR. The iolR promoter itself is induced in the absence of IolR in the presence of glucose as well as MI, indicating an autoregulatory activity of this repressor. Taken together, these findings indicate that the promoters of iolR, iolE, iolC1, iolD1, and iolI2 are strongly induced in MM with MI during the exponential growth phase, while being repressed in the presence of glucose or in rich medium, and the iolA promoter is active under each condition tested here.
TABLE 2.
Quantification of iol promoter activitiesa
| Fragment cloned into pDEW201 | 14028s grown in MM + glucose
|
14028s grown in MM + MI
|
14028s ΔiolR grown in MM + glucose
|
|||||
|---|---|---|---|---|---|---|---|---|
| RLU/OD600 unitb | SD (%) | RLU/OD600 unit | SD (%) | Fold inductionc | RLU/OD600 unit | SD (%) | Fold induction | |
| PiolR | 1.92 × 105 | 3.1 | 2.01 × 106 | 2.8 | 10.5 | 2.46 × 106 | 5.3 | 12.8 |
| PiolA | 4.16 × 106 | 2.4 | 1.27 × 107 | 7.8 | 3.1 | 1.78 × 107 | 3.7 | 4.3 |
| PiolE | 9.27 × 102 | 67.9 | 2.97 × 106 | 0.8 | 3,202.8 | 2.16 × 104 | 17.8 | 23.3 |
| PiolG1 | 4.93 × 103 | 7.4 | 1.55 × 104 | 7.4 | 3.2 | 2.65 × 104 | 10.4 | 5.4 |
| PiolC1 | 2.91 × 105 | 2.8 | 9.88 × 106 | 6.7 | 33.9 | 1.90 × 107 | 2.2 | 65.3 |
| PiolD1 | 2.95 × 104 | 7.3 | 5.72 × 106 | 2.6 | 194.0 | 7.42 × 106 | 5.4 | 251.7 |
| PiolG2 | 5.61 × 102 | 14.0 | 6.29 × 103 | 27.0 | 11.2 | 1.69 × 104 | 26.3 | 30.1 |
| PiolI2 | 8.32 × 102 | 24.2 | 1.40 × 107 | 0.7 | 16,864.2 | 3.25 × 104 | 11.9 | 39.0 |
| PiolH | 1.66 × 103 | 58.9 | 1.17 × 103 | 60.2 | 0.7 | 1.83 × 103 | 32.5 | 1.1 |
| Controls | ||||||||
| PargS | 2.96 × 106 | 4.0 | 1.85 × 106 | 18.3 | 0.6 | 3.41 × 106 | 10.1 | 1.2 |
| Pdef | 1.13 × 106 | 2.9 | 1.11 × 106 | 5.5 | 1.0 | 1.35 × 106 | 2.8 | 1.2 |
| ′STM0047′ | 1.62 × 104 | 6.6 | 1.25 × 104 | 3.9 | 0.8 | 1.87 × 104 | 4.0 | 1.2 |
| None | 1.58 × 103 | 9.0 | 1.36 × 103 | 11.3 | 0.9 | 1.00 × 103 | 50.4 | 0.6 |
Samples were taken from the late exponential phase.
Data are averages from three independent experiments.
Fold induction was calculated with respect to the RLU/OD600 unit values for the wild-type strain grown in glucose.
Transcriptional organization of iol genes.
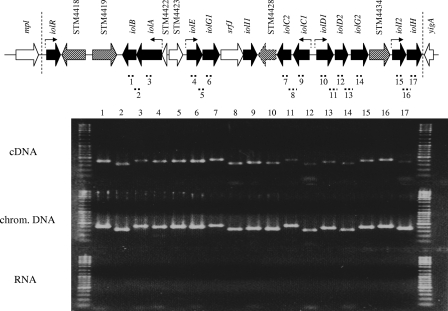
In contrast to the unidirectional organization of iol genes of B. subtilis and C. perfringens, the genes involved in MI degradation by gram-negative bacteria are not transcribed polycistronically (Fig. 1A). As demonstrated above, iolB, iolG1 iolC2, and iolD2 are obligate for MI degradation but do not possess a separate promoter (Table 2), suggesting a cotranscription of iolB with iolA, of iolG1 with iolE, of iolC2 with iolC1, and of iolD2/iolG2 with iolD1. To reveal the transcriptional organization of the iol genes within GEI4417/4436, we performed RT-PCR with RNA isolated from strain 14028 grown in MM with MI. The RNA was demonstrated to be DNA free, and cDNA of 17 regions spanning approximately 300 to 500 bp was amplified. Two oligonucleotides hybridizing to STM4434 and iolI2 did not result in a PCR product from cDNA, thus validating the approach. All PCRs with cDNA as the template revealed a DNA fragment whose length corresponds to the PCR fragments amplified from genomic DNA (Fig. 3). In line with the data for the luciferase reporter fusions, iolG2 and iolH are under the control of the promoters located upstream of iolE and iolI2. In summary, iolA-iolB, iolE-iolG1, iolC1-iolC2, iolD1-iolD2-iolG2, and iolI2-iolH are transcriptionally coupled. Thus, these five operons of S. enterica serovar Typhimurium comprise all genes required for MI utilization as well as three functionally dispensable or redundant genes, iolG2, iolI2, and iolH.
FIG. 3.
Transcriptional organization of S. enterica serovar Typhimurium MI utilization genes. Strain 14028s was grown in MM with 55.5 mM MI at 37°C, and mRNA was extracted at an OD600 of 0.4. cDNA was amplified with reverse primers listed in Table S1 in the supplemental material. RT-PCR was performed with primer pairs specific for the indicated regions 1 to 17. All PCR products were separated by 2% agarose gel electrophoresis. As controls, PCR amplification products with genomic DNA and DNase-treated RNA samples as template are shown. Line numbers correspond to PCR product numbers depicted above the gel lanes. Arrows indicate promoters identified in this study.
IolR acts as transcriptional repressor.
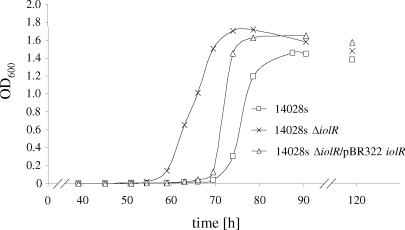
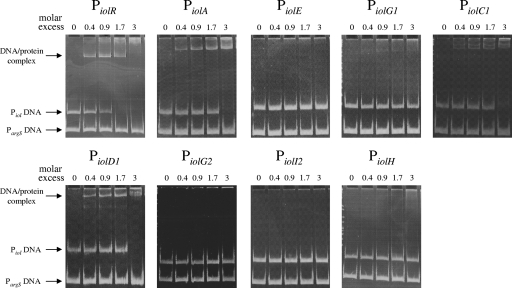
Homology searches with the protein sequence of IolR (STM4417) revealed a putative transcriptional regulator with an HTH-6 motif belonging to the RpiR family. This domain is N terminal to a sugar isomerase domain that is predicted to bind phosphosugars. No homologies to the characterized IolR proteins of B. subtilis or C. glutamicum were observed, but there were homologies to putative regulators which might play a similar role in regulation of MI degradation by K. pneumoniae, Y. enterocolitica, Y. pestis, and P. luminescens (Fig. 1). In MM supplemented with MI, a nonpolar deletion of iolR resulted in a lag phase of 10 h shorter than the one measured for the wild-type strain (Fig. 4). The 14028s ΔiolR phenotype could be complemented by gene expression of iolR from pBR322 (Fig. 4). Induction of the promoters of iolR, iolA, iolC1, and iolD1 in the iolR deletion mutant in the presence of glucose, and the identification of an HTH motif in the IolR sequence, prompted us to perform promoter binding studies. For that purpose, IolR was overexpressed in E. coli BL21(DE3) and purified as described above. The putative promoter fragments of iolR, iolA, iolE, iolG1, iolC1, iolD1, iolG2, iolI2, and iolH fragments were incubated without or with increasing amounts of the purified IolR proteins, and the protein-DNA complexes were separated on a 9.5% native polyacrylamide gel (Fig. 5). A retarded DNA band with decreased motility representing the IolR-DNA complex was observed with the iolR, iolA, iolC, and iolD1 fragments. Thus, the binding of IolR to the respective promoters results in repression of these genes during growth of S. enterica serovar Typhimurium in glucose-rich medium. Binding of IolR to its own promoter demonstrates its autoregulatory function. In contrast, complex formation was observed neither with a control fragment of the argS promoter indicating the IolR binding specificity, nor with fragments upstream of iolE, iolI2, iolH, iolG1, and iolG2. IolI2 is not required for MI degradation (see above), and its expression might therefore not be regulated by IolR. The bandshift experiments with the iolE and iolI2 promoters are in line with the data in Table 2.
FIG. 4.
Growth curve of strain 14028s ΔiolR. Exponential growth of the strain lacking the repressor IolR starts approximately 10 h earlier than that of the wild-type strain. The phenotype of 14028s ΔiolR could partially be complemented upon in trans expression of iolR via pBR322. Average values from three independent experiments are shown. Standard deviations are not given due to a variable lag phase, and all growth curves were normalized to a lag phase ending 60 h after inoculation.
FIG. 5.
Promoter binding activity of IolR. The interaction of IolR with the regulatory region of nine genes of GEI 4417/4436 is shown. One hundred nanograms of DNA was used in each experiment. The promoter fragments were incubated without or with increasing amounts (7 ng, 14 ng, 28 ng, and 49 ng [221 fM, 442 fM, 884 fM, and 1547 fM, respectively]) of the purified IolR protein. No bandshift was observed when a maximum of 210 to 280 ng IolR, corresponding to a maximal 17-fold molar excess, was incubated with promoter DNAs of iolE, iolG2, iolI, and iolH (data not shown). Protein-DNA complexes were separated on a 9.5% native polyacrylamide gel. A 200-bp sequence of the argS promoter served as a negative control.
DISCUSSION
Although a large number of putative iol genes are present in gram-negative genomes, little is known about their functionality and their organization in comparison to their gram-positive counterparts. The systematic knockout of GEI 4417/4436 genes revealed that this S. enterica serovar Typhimurium island encodes all the enzymatic activities required for MI catabolism, leading to the production of acetyl coenzyme A and dihydroxyacetone phosphate. The genes iolR, iolB, iolA, iolE, iolG1, iolC1/2, and iolD2 provide an MI-negative phenotype upon deletion and encode the key functions to utilize MI. These genes are also present in the genomes of K. pneumoniae, Yersinia spp., and P. luminescens, while homologues of other GEI 4417/4436 iol genes could not be identified in these strains. As experimentally demonstrated in this study, these genes enable Y. enterocolitica and P. luminescens to catabolize MI. iolI and iolH were found only in the genome of K. pneumoniae. The putative inosose isomerase IolI has been demonstrated to convert 2-keto-MI to 1-keto-d-chiro-inositol (32). Together with its very strong induction in the presence of MI, these results suggest a role of IolI in providing IolE substrates. The function of IolH remains to be elucidated.
S. enterica serovar Typhimurium exhibits similar growth phenotypes in MM containing glucose or MI with respect to the generation time during exponential phase and the OD600 in stationary phase. However, a remarkable difference is the extended lag phase of approximately 60 h in the presence of MI as the sole carbon source. Such a retarded metabolic switch has not been reported for gram-positive bacteria able to grow on MI. For example, growth of C. glutamicum in MI-containing medium starts within a few hours after inoculation (17). 2-Deoxy-5-keto-d-gluconic acid has long been considered a key step in MI degradation (2). Only recently, 2-deoxy-5-keto-d-gluconic acid 6-phosphate, another intermediate of MI degradation (Fig. 1B) was identified to antagonize IolR binding to the promoter of the B. subtilis iol operon (33). Thus, the long lag phase of S. enterica serovar Typhimurium in the presence of MI might be the result of a tighter repression of its iol genes, but other mechanisms of IolR antagonizing or an additional regulatory factor cannot be excluded. This assumption is supported by the obviously IolR-independent iolE and iolI2 regulation as shown by the bandshift experiments (Fig. 5). Addition of glucose during exponential phase had no effect on the promoter activity of iolE and iolI2 in the ΔiolR background (data not shown), excluding the possibility that they are under catabolite repression. iolE, encoding the dehydratase that catalyzes the second step in MI degradation, might be positively induced by MI or a related substance rather than by an antagonistically acting intermediate such as 2-deoxy-5-keto-d-gluconic acid 6-phosphate.
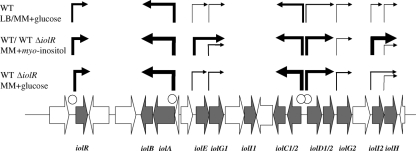
RT-PCR, reporter fusions, and GMS revealed a consistent picture of the transcriptional organization, the regulation, and the promoter activities of the Salmonella iol genes. The data obtained are summarized in Fig. 6. IolR binds to the iolR promoter and regulates its own expression (Fig. 4 and 5). This is in line with the finding that in B. subtilis, inactivation of iolR results in a constitutive transcription of the iol divergon including iolJ (33). IolR was demonstrated in this study to negatively regulate the transcription of three gene clusters required for MI utilization by S. enterica serovar Typhimurium, namely, iolA-iolB, iolC1-iolC2, and iolD1-iolD2-iolG2. A lack of IolR binding to the putative promoter of the iolE-iolG1 operon, which encodes the first two steps in MI degradation, hints at an additional, IolR-independent regulatory mechanism. The autoregulatory activity of the MI repressor and substrate antagonism might explain that iolR of S. enterica serovar Typhimurium is induced under conditions in which the genes involved in MI degradation are transcribed, an observation already described for C. perfringens (14). However, iolR of C. glutamicum is not upregulated in the presence of MI (17).
FIG. 6.
Regulation of MI utilization in S. enterica serovar Typhimurium. The wild-type strain and an iolR deletion mutant carrying recombinant pDEW201-constructs were grown in LB or in MM with MI or glucose. Promoter induction is depicted by arrows of different sizes (from smallest to largest): <104 RLU/OD600 unit, <105 RLU/OD600 unit, <106 RLU/OD600 unit, <107 RLU/OD600 unit, and <108 RLU/OD600 unit. Induction of iol genes was similar in both strains in the presence of MI. Binding sites of IolR are indicated by open circles. WT, wild type.
In gram-positive bacteria, the genes encoding enzymes for MI degradation are mostly unidirectionally organized, resulting in polycistronically transcribed operons (14, 30). Two iol clusters putatively encoding redundant functions were identified in C. glutamicum and L. plantarum (17, 30). Genome comparison revealed a more complex transcriptional organization of iol genes in S. enterica serovar Typhimurium and other gram-negative bacteria belonging to the genera Yersinia, Photorhabdus, Citrobacter, Erwinia, Brucella, Photobacterium, and Rhizobium (Fig. 1A). Besides Salmonella, genes encoding putative redundant enzymatic functions have been found only in the Photobacterium profundum genome (iolE and iolG), which also carries an iolR duplication, and in the island exemplified by E. carotovora. Remarkably, Y. enterocolitica, Y. intermedia, and Y. frederiksenii on the one hand and Y. pestis and Y. pseudotuberculosis on the other hand carry distinct iol gene clusters, suggesting their independent acquisition by Yersinia ancestor strains. Taken together, these data support a repeated acquisition and chromosomal rearrangement of iol genes in gram-negative bacteria.
Open questions that are currently being addressed for S. enterica serovar Typhimurium are the transport mechanisms for MI or derivatives, further regulatory mechanisms contributing to MI utilization, and the identification of MI-related substrates metabolized by the Iol enzymes.
Supplementary Material
Acknowledgments
We thank Siegfried Scherer for financial support.
We thank Siegfried Scherer for helpful discussions and Sarah Schaaf for support with GMS experiments. Patrick Schiwek and Theresa Käuferle are acknowledged for technical assistance.
Footnotes
Published ahead of print on 14 November 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, W. A., and B. Magasanik. 1971. The pathway of myo-inositol degradation in Aerobacter aerogenes. Conversion of 2-deoxy-5-keto-d-gluconic acid to glycolytic intermediates. J. Biol. Chem. 2465662-5675. [PubMed] [Google Scholar]
- 3.Berman, T., and B. Magasanik. 1966. The pathway of myo-inositol degradation in Aerobacter aerogenes. J. Biol. Chem. 241807-813. [PubMed] [Google Scholar]
- 4.Bochner, B. R., P. Gadzinski, and E. Panomitros. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 111246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162156-159. [DOI] [PubMed] [Google Scholar]
- 6.Cornelis, G., and C. Colson. 1975. Restriction of DNA in Yersinia enterocolitica detected by recipient ability for a derepressed R factor from Escherichia coli. J. Gen. Microbiol. 87285-291. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer-Le Saux, M., V. Viallard, B. Brunel, P. Normand, and N. E. Boemare. 1999. Polyphasic classification of the genus Photorhabdus and proposal of new taxa: P. luminescens subsp. luminescens subsp. nov., P. luminescens subsp. akhurstii subsp. nov., P. luminescens subsp. laumondii subsp. nov., P. temperata sp. nov., P. temperata subsp. temperata subsp. nov. and P. asymbiotica sp. nov. Int. J. Syst. Bacteriol. 491645-1656. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs, T. M., J. Klumpp, and K. Przybilla. 2006. Insertion-duplication mutagenesis of Salmonella enterica and related species using a novel thermosensitive vector. Plasmid 5539-49. [DOI] [PubMed] [Google Scholar]
- 10.Gutnick, D., J. M. Calvo, T. Klopotowski, and B. N. Ames. 1969. Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhimurium LT-2. J. Bacteriol. 100215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 12.Jakob, K., P. Satorhelyi, C. Lange, V. F. Wendisch, B. Silakowski, S. Scherer, and K. Neuhaus. 2007. Gene expression analysis of Corynebacterium glutamicum subjected to long-term lactic acid adaptation. J. Bacteriol. 1895582-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa, M., and S. Goto. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2827-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawsar, H. I., K. Ohtani, K. Okumura, H. Hayashi, and T. Shimizu. 2004. Organization and transcriptional regulation of myo-inositol operon in Clostridium perfringens. FEMS Microbiol. Lett. 235289-295. [DOI] [PubMed] [Google Scholar]
- 15.Klumpp, J., and T. M. Fuchs. 2007. Identification of novel genes in genomic islands that contribute to Salmonella typhimurium replication in macrophages. Microbiology 1531207-1220. [DOI] [PubMed] [Google Scholar]
- 16.Knuth, K., H. Niesalla, C. J. Hueck, and T. M. Fuchs. 2004. Large-scale identification of essential Salmonella genes by trapping lethal insertions. Mol. Microbiol. 511729-1744. [DOI] [PubMed] [Google Scholar]
- 17.Krings, E., K. Krumbach, B. Bathe, R. Kelle, V. F. Wendisch, H. Sahm, and L. Eggeling. 2006. Characterization of myo-inositol utilization by Corynebacterium glutamicum: the stimulon, identification of transporters, and influence on l-lysine formation. J. Bacteriol. 1888054-8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Legakis, N. J., J. T. Papavassiliou, and M. E. Xilinas. 1976. Inositol as a selective substrate for the growth of Klebsiellae and Serratiae. Zentralbl. Bakteriol. 235453-458. [PubMed] [Google Scholar]
- 19.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 1796228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413852-856. [DOI] [PubMed] [Google Scholar]
- 21.Miwa, Y., and Y. Fujita. 2001. Involvement of two distinct catabolite-responsive elements in catabolite repression of the Bacillus subtilis myo-inositol (iol) operon. J. Bacteriol. 1835877-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosmann, T. 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 6555-63. [DOI] [PubMed] [Google Scholar]
- 23.Old, D. C. 1972. Temperature-dependent utilization of meso-inositol: a useful biotyping marker in the genealogy of Salmonella typhimurium. J. Bacteriol. 112779-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Primrose, S. B., and C. W. Ronson. 1980. Polyol metabolism by Rhizobium trifolii. J. Bacteriol. 1411109-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 26.Schaaf, S., and M. Bott. 2007. Target genes and DNA-binding sites of the response regulator PhoR from Corynebacterium glutamicum. J. Bacteriol. 1895002-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 18560-89. [DOI] [PubMed] [Google Scholar]
- 28.Van Dyk, T. K., and R. A. Rosson. 1998. Photorhabdus luminescens luxCDABE promoter probe vectors. Methods Mol. Biol. 10285-95. [DOI] [PubMed] [Google Scholar]
- 29.Worley, M. J., K. H. Ching, and F. Heffron. 2000. Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol. Microbiol. 36749-761. [DOI] [PubMed] [Google Scholar]
- 30.Yebra, M. J., M. Zuniga, S. Beaufils, G. Perez-Martinez, J. Deutscher, and V. Monedero. 2007. Identification of a gene cluster enabling Lactobacillus casei BL23 to utilize myo-inositol. Appl. Environ. Microbiol. 733850-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida, K., M. Yamaguchi, H. Ikeda, K. Omae, K. Tsurusaki, and Y. Fujita. 2004. The fifth gene of the iol operon of Bacillus subtilis, iolE, encodes 2-keto-myo-inositol dehydratase. Microbiology. 150571-580. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida, K., M. Yamaguchi, T. Morinaga, M. Ikeuchi, M. Kinehara, and H. Ashida. 2006. Genetic modification of Bacillus subtilis for production of d-chiro-inositol, an investigational drug candidate for treatment of type 2 diabetes and polycystic ovary syndrome. Appl. Environ. Microbiol. 721310-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshida, K., M. Yamaguchi, T. Morinaga, M. Kinehara, M. Ikeuchi, H. Ashida, and Y. Fujita. 2008. myo-Inositol catabolism in Bacillus subtilis. J. Biol. Chem. 28310415-10424. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida, K., Y. Yamamoto, K. Omae, M. Yamamoto, and Y. Fujita. 2002. Identification of two myo-inositol transporter genes of Bacillus subtilis. J. Bacteriol. 184983-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida, K. I., D. Aoyama, I. Ishio, T. Shibayama, and Y. Fujita. 1997. Organization and transcription of the myo-inositol operon, iol, of Bacillus subtilis. J. Bacteriol. 1794591-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshida, K. I., T. Shibayama, D. Aoyama, and Y. Fujita. 1999. Interaction of a repressor and its binding sites for regulation of the Bacillus subtilis iol divergon. J. Mol. Biol. 285917-929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.