Abstract
BacA is an inner membrane protein associated with maintenance of chronic infections in several diverse host-pathogen interactions. To understand the function of the bacA gene in Mycobacterium tuberculosis (Rv1819c), we insertionally inactivated this gene and analyzed the resulting mutant for a variety of phenotypes. BacA deficiency in M. tuberculosis did not affect sensitivity to detergents, acidic pH, and zinc, indicating that there was no global compromise in membrane integrity, and a comprehensive evaluation of the major lipid constituents of the cell envelope failed to reveal any significant differences. Infection of mice with this mutant revealed no impact on establishment of infection but a profound effect on maintenance of extended chronic infection and ultimate outcome. As in alphaproteobacteria, deletion of BacA in M. tuberculosis led to increased bleomycin resistance, and heterologous expression of the M. tuberculosis BacA homolog in Escherichia coli conferred sensitivity to antimicrobial peptides. These results suggest a striking conservation of function for BacA-related proteins in transport of a critical molecule that determines the outcome of the host-pathogen interaction.
Tuberculosis is a chronic disease of humans that is responsible for more than 2 million deaths annually, and more than one-third of the total human population is latently infected with Mycobacterium tuberculosis (18). The chronic nature of the disease severely complicates attempts to identify better therapeutics or diagnostics for this disease (44). In an attempt to better understand the latency and chronicity of tuberculosis, we have studied what is known about other complex symbiotic and pathogenic relationships.
BacA of Sinorhizobium melliloti, a gram-negative alphaproteobacterial symbiont of legumes, was found to be essential for normal development of alfalfa nodules. BacA was predicted to be a transmembrane protein transporter and showed 64% identity to the SbmA protein of Escherichia coli (17), a protein that sensitizes E. coli to the peptide antibiotics microcins B17, J25, and bleomycin (43). E. coli sbmA-deficient mutants can be complemented by the S. meliloti bacA gene, indicating functional similarity as transporters (20).
In S. meliloti and Brucella abortus, an alphaproteobacterial pathogen that induces spontaneous abortion in chronically infected cows (25), BacA-deficient mutants have been found to show increased resistance to some aminoglycoside antibiotics and bleomycin, as well as enhanced sensitivity to several cell envelope-disrupting agents such as ethanol, detergents, hydrophobic dyes and acidic pH (14, 20, 25). The spectrum of agents to which loss of BacA function conferred hypersensitivity was similar to that previously reported for an S. meliloti lpsB mutant (4), suggesting an abnormality in lipopolysaccharide (LPS). Indeed, LPS isolated from the S. meliloti and B. abortus bacA mutants contained abnormal lipid A moieties, half of which lacked the very-long-chain fatty acid (VLCFA) modification (11). This observation was striking, since BacA shares homology with certain eukaryotic ATP-binding cassette (ABC) transporters, including the human adrenoleukodystrophy protein (11), which is thought to transport activated VLCFA across the peroxisomal membrane (41). Thus, a direct role of BacA in VLCFA transport across the inner membrane has been suggested (11). Subsequently, however, acpXL (encoding an acyl carrier protein) and lpsXL (encoding an acyl transferase), both of which are required for the VLCFA modification of lipid A, were found not to be absolutely required for chronic infection, indicating that is unlikely that the abnormal lipid A of the BacA mutant was solely responsible for the symbiotic defect (12). In addition, an E. coli sbmA mutant did not show alterations in aminoglycoside resistance or sensitivity to detergents or ethanol, suggesting that BacA must have an additional function (20).
The biological substrate for BacA-SbmA is unknown, but increased resistance of deficient mutants against peptide antibiotics strongly suggests that BacA is required for uptake of antimicrobial peptides (20). Recently, it has been reported that E. coli sbmA-null mutants are resistant to the proline-rich antimicrobial peptide Bac7, a cathelicidin-derived peptide from bovine neutrophils. A fluorescently labeled Bac7 variant was internalized in the sbmA mutants at a decreased rate compared with the parental strain, indicating that SbmA protein is required for the import of Bac7 (29).
Among BacA homologs in sequenced microbial genomes, there is a distinct subgroup of proteins named BacA-related proteins. These proteins show a lower similarity (38 to 59%) to the S. meliloti BacA protein and are approximately 200 amino acids longer. At the C termini, they present Walker A and B motifs, as well as an ABC signature sequence (26). They are characterized by the presence of two hydrophobic membrane-spanning domains (MSD) and two cytoplasmic nucleotide-binding domains and can act either as exporters or as importers. In the latter case, there is often an extracytoplasmic region involved, named the substrate-binding domain, that in the case of gram-positive bacteria is anchored to the cell membrane by a lipid tail at the amino end (2, 7, 19, 22, 27, 45).
Curiously, BacA-related proteins are also present in S. meliloti (ExsE) and E. coli (YddA). The exsE gene is part of a cluster involved in succinoglycan biosynthesis in S. meliloti, although ExsE does not play a direct role in the synthesis of this molecule and is not essential for successful symbiosis with alfalfa (28). An ExsE-deficient S. meliloti mutant does not show altered sensitivity to deoxycholic acid (DOC), suggesting that the cell envelope is not compromised in this mutant (16). The YddA protein of E. coli is predicted to be a cytoplasmic membrane protein of unknown function that may facilitate growth under optimal conditions (rich medium at 37°C) (37). So far, the biological function of any BacA-related protein has not been discerned, and it is not known if these proteins play a role similar to those of proteins of the BacA-SbmA group.
The genome of M. tuberculosis encodes 37 apparent ABC transporters, 27 of which appear to be complete (2, 6). The conservation of the essential role of BacA transporter in S. meliloti and B. abortus, two disparate and long-term symbiotic/pathogenic systems, drew our attention to the possibility that the predicted ABC transporter encoded by the Rv1819 gene of M. tuberculosis, which shares 39% similarity with the BacA protein of B. abortus, might play a similar role in the maintenance of chronic tuberculosis infection in the murine model of disease.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The Escherichia coli ElectroMAX DH5α strain (Invitrogen) used for cloning was grown in Luria-Bertani medium with ampicillin (100 μg ml−1) (Sigma), hygromycin (200 μg ml−1) (Invitrogen), kanamycin (50 μg ml−1) (Sigma), tetracycline (10 μg ml−1) (Sigma), or gentamicin (5 μg ml−1) (GibcoBRL) when indicated. E. coli strains RYC1000 (15) and RYC1001 (RYC1000 sbmA [spontaneous], a kind gift from Felipe Moreno) were used for heterologous expression of the mycobacterial BacA in E. coli as described previously (20). M. tuberculosis strains were grown in Middlebrook 7H9 broth (Difco) supplemented with ADC (NaCl, 8.1 g liter−1; bovine serum albumin fraction V [Calbiochem], 50 g liter−1; and d-glucose, 20 g liter−1), 0.02% (vol/vol) glycerol, and 0.05% (vol/vol) Tween 80 (Sigma) or on Middlebrook 7H11 agar (Difco) supplemented with OADC enrichment (same as ADC but also including 0.6 ml liter−1 oleic acid [ICN Biochemicals] and 3.6 mM sodium hydroxide). Where indicated, hygromycin (50 μg ml−1), kanamycin (25 μg ml−1), or 2% (wt/vol) sucrose was added (31). H37Rv (Pasteur) was used as the parental strain of the H37Rv bacA::hyg mutant. In iron uptake experiments glycerol-alanine-salts-Tween (GAST) medium without iron (8) was used either directly (iron-deficient medium) or after treatment overnight with Chelex 100 resin (Bio-Rad) (10 g liter−1) (iron-depleted medium). All iron experiments were performed using sterile polystyrene bottles (Corning).
Nucleic acid techniques.
E. coli transformations, cloning, and PCR were based on standard conditions (36). Southern blotting and hybridization procedures were performed as previously described (9). Mycobacterial DNA was isolated using the protocol of Pelicic et al. (31). Transformation of M. tuberculosis was carried out as previously described (39).
Computer analysis.
M. tuberculosis H37Rv, Mycobacterium bovis, Mycobacterium bovis BCG, Mycobacterium ulcerans, Mycobacterium marinum, and Mycobacterium leprae DNA sequences were obtained from the TubercuList, BoviList, BCGList, BuruList, MarinoList, and Leproma servers of the Institut Pasteur (www.pasteur.fr). The M. tuberculosis CDC1551 DNA sequence was from the TIGR Microbial Database (www.tigr.org). Searches for BacA orthologs in other bacterial species were performed using the BLAST program of the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). Alignment of the BacA-homologous protein sequences was carried out using the ClustalW algorithm (40) with 15 as the gap opening penalty and 6.66 as the gap extension penalty. The phylogenetic tree was built using the neighbor-joining algorithm of Saitou and Nei (35) included in the Vector NTI Advance 10 program (Invitrogen).
Construction of the bacA-disrupted mutant and the complemented strain.
Generation of an M. tuberculosis mutant lacking BacA (bacA::hyg) was accomplished by homologous recombination using the system developed by Pelicic et al. (31). Primers bacA-MtF (5′-GGACTAGTCTCCTCGATCCAGCCCTG-3′) and bacA-MtR (5′-GGACTAGTCGAACAGGAGCCCGCCAT-3′), with SpeI sites engineered onto the 5′ end, were used for generating a 3,564-bp PCR fragment containing 979 bp upstream and 1,595 nucleotides of the bacA gene. This fragment was cloned into the vector pBluescript SK (Stratagene). A 1.6-kbp fragment carrying the hyg gene was cloned into the bacA gene at the EcoRV and XbaI sites, replacing a central fragment of 541 bp. Finally the 3.6-kbp fragment harboring the bacA::hyg gene was excised and cloned into the SpeI site of the mycobacterial shuttle vector pPR27 (31). Transformants were plated on 7H11 with 50 μg ml−1 hygromycin at 32°C for 5 weeks. The resulting colonies were grown at 32°C in 10 ml of 7H9 containing 50 μg ml−1 hygromycin and subsequently plated on 7H11 medium with 50 μg ml−1 hygromycin and 2% sucrose at 39°C. DNA from Hygr Sucr temperaturer colonies was digested with SphI, transferred to a Hybond-N nylon membrane (Amersham Pharmacia) by Southern blotting, and hybridized with a 1,994-bp fragment probe of the bacA gene of M. tuberculosis H37Rv Pasteur generated using the primers bacA-3 (5′-TATCGCGATCACGGAAGCGG-3′) and bacA-1 (5′-AAAGACAAACACTCCTC TGCA-3′). For construction of plasmid pKLMt5, a 2,283-bp region of the M. tuberculosis H37Rv chromosome containing the bacA gene flanked by 203 bp upstream and 161 bp downstream was amplified by PCR using primers with EcoRI sites into the 5′ ends. This fragment was cloned into the integrative vector pMV306K (32). After verification of the sequence, the M. tuberculosis bacA::hyg mutant was transformed with plasmid pKLMt5 and plated on 7H11 with 50 μg ml−1 hygromycin and 25 μg ml−1 kanamycin.
Construction of the plasmid for heterologous expression of mycobacterial BacA in E. coli.
The bacA gene was PCR amplified by using primers MtBacA-F (5′-ACTAGTTGGGCCCGAAATTGTTTAAGCCGTCCATCGATTGGT-3′) and MtBacA-R (5′-CTGCAGAGATCGACGGATTCAGCGTATCGCGATCACGGAA-3′), with SpeI and PstI sites, respectively, engineered onto the 5′ ends and pKLMt5 as the template. The resulting 2-kb fragment was cloned in the XbaI-PstI sites of pWSK29, the low-copy-number vector for E. coli (42). The resulting plasmid, pWSK-MtbacA, carries the bacA gene under the control of the lac promoter. After verification of the sequence, pWSK-MtbacA was transformed into RYC1001(pMM100). The plasmid pMM100 is pBR322 carrying lacIq (24).
MIC assays.
The susceptibilities of the M. tuberculosis strains to different compounds were tested by a twofold serial broth dilution method as follows. Cultures of each mutant and the parental strain were grown in 7H9 to an optical density at 650 nm (OD650) of 0.1 (measured in a Novaspec II spectrophotometer [Pharmacia Biotech]). These cultures were used as inocula after a 1:200 dilution. The antituberculosis drugs used were amikacin, cycloserin, ethambutol, ethionamide, isoniazid, kanamycin, ofloxacin, rifampin, streptomycin, tetracycline, and vancomycin, all purchased from Sigma. Other compounds tested were sodium dodecyl sulfate (SDS) (Digene) and zinc sulfate (Aldrich), as well as bleomycin, EDTA, DOC, 2,2′-dipyridyl, and desferoxamine (also acquired from Sigma). The assays were performed in 96-well round-bottom plates (Nunc). The plates were read after 7 and 14 days of incubation, and the MIC was considered the lowest concentration that completely inhibited visible growth. The experiments were performed in duplicate and repeated three times. Susceptibility of the E. coli strains to bleomycin and Bac7(1-16) (1) (synthesized by the Biopolymer Laboratory, Center for Cancer Research, Massachusetts Institute of Technology) was tested in essentially the same manner with minor modifications: Muller-Hinton broth (Sigma) was used as the growth medium, 0.4 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added to the culture to induce the expression of bacA, and the plates were read after 1 day of incubation at 37°C.
Mycolic acid analysis.
Cell wall skeleton-associated mycolic acids were extracted from 200-ml cultures with an OD650 of 0.5 by a procedure described previously (38). For analysis of the mycolic acid associated with glycolipids (trehalose dimycolate [TDM] and trehalose monomycolate), 100-ml cultures of the different M. tuberculosis strains (wild type, bacA::hyg mutant, and bacA::hyg-pKLMt5) were grown to an OD650 of 0.4. Metabolic labeling of fatty acids was achieved by incubating these cultures in the presence of 1 μCi ml−1 of sodium [1-14C]acetate (American Radiolabeled Chemicals; specific activity of ≈57 Ci mol−1, 1 Ci = 3.7 × 1010 Bq) for 24 h prior to lipid extraction. Preparation of mycolic acid-containing glycolipids was carried out by chloroform-methanol (2:1, vol/vol) extraction followed by acetone precipitation according to the procedures described by Slayden and Barry (38). Thin-layer chromatography (TLC) was performed using 250- or 500-μm Silica Gel 60 plates (EM Science). The developing solvent was chloroform-methanol-ammonium hydroxide (80:20:2, vol/vol/vol) for separation of TDM and trehalose monomycolate or petroleum ether-diethyl ether (85:15, vol/vol) (five times) for mycolic acid methyl esters. TLC plates were visualized using a Storm 860 PhosphorImager (Molecular Dynamics).
Mass spectrometry analysis.
Matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) spectra, in the positive mode, were obtained by following the procedure described by Laval et al. (23) using a Voyager RP MALDI-TOF mass spectrometer (Applied Biosystems). Mycolic acid samples were dissolved to a concentration of 1 mM in chloroform, and 1 μl was mixed with the matrix (2,5-dihyroxybenzoic acid [10 mg ml−1] in methanol-chloroform [1:1, vol/vol]) before being analyzed in the Reflectron mode.
Mouse experiments.
Prior to infection, well-dispersed liquid cultures were adjusted to an OD650 nm of 0.5 and stored at −70°C as 20% (vol/vol) glycerol stocks. Inocula were prepared by diluting these stocks to 4 × 106 CFU/ml in phosphate-buffered saline-Tween 80 (0.05%). Eight-week-old C57BL/6 or B6D2/F1 mice (Taconic) were infected using a BioAerosol nebulizing generator (CH Technologies Inc., NJ) for 10 min. Bacteria were enumerated at 1, 14, 49, 63, 126, and 205 days postinfection (four mice per time point) by homogenizing the lungs and spleens of infected mice in 1 ml of 7H9 medium and plating 10-fold serial dilutions on 7H11 medium. An additional 22 mice per group were maintained for survival studies. Survival fractions were calculated using the Kaplan-Meier method (21), and the log rank test was used to determine statistical significance of observed survival differences (GraphPad Prism v3.0; GraphPad Software, CA).
55FeCl3 uptake experiments.
M. tuberculosis strains were grown in 7H9 broth supplemented with ADC to an OD650 of 0.4. These cultures were diluted 1:10 in Fe-depleted GAST medium and incubated for 3 to 4 days until they reach an OD650 of 0.2. At this point half of the cultures were centrifuged (3,000 × g, 10 min), the medium was removed, and pellets were washed twice with Fe-depleted GAST medium. Two sets of cultures, set A (in GAST medium without added Fe) and set B (in GAST medium without added iron and treated overnight with Chelex resin to remove any traces of residual iron) were adjusted to the same OD650. After 4 days of incubation, set A reached an OD650 of around 0.7 and set B reached an OD650 of around 0.45. Both sets of cultures were adjusted again to an OD650 of 0.4, and 0.5 μCi ml−1 of 55FeCl3 (Perkin-Elmer; specific activity of ≈50 mCi ml−1, 1 Ci = 3.7 × 104 MBq) was added. At the indicated times, the OD650 was measured and 500 μl was taken for quantification of 55FeCl3 accumulation. Samples were centrifuged for 5 min at 14,000 × g, and the pellets were washed twice with Hanks balanced salt solution (without magnesium, calcium, and phenol red), Biofluids, and 0.05% Tween 80. Radioactivity was then measured for 5 minutes in the 3H channel of a Beckman LS5801 scintillation counter.
RESULTS
Distribution of BacA homologs in the genus Mycobacterium.
Mycobacterial genes homologous to the bacA sequence from B. abortus are found not only in the two finished M. tuberculosis strains H37Rv (Rv1819c) and CDC1551 (MT1867) but also in other slow-growing members of the genus Mycobacterium, including M. bovis, M. bovis BCG, M. leprae, M. avium-paratuberculosis, M. avium, M. ulcerans, and M. marinum, as well as in fast-growing saprophytic species such as M. smegmatis, M. gilvum, and M. vanbaalenii. The bacA gene of M. tuberculosis encodes a protein of 639 amino acids with six predicted transmembrane helices distributed in two MSD located at the N terminus. Unlike the BacA-SbmA proteins, the mycobacterial BacA protein is predicted to be a complete ABC transporter harboring a nucleotide-binding domain at the carboxy terminus as well as an ABC signature sequence constituting an intact ATPase domain. Among the genus Mycobacterium, the BacA proteins share between 60 and 99% amino acid identity. Likewise, the genomic context of the bacA gene in the different Mycobacterium species is highly conserved (with the exception of Mul3059, which is flanked by a transposase for IS2404). Nevertheless, the M. tuberculosis BacA was slightly more closely related to the BacA-related proteins, such as the ExsE protein of S. meliloti and its homolog in B. abortus, than to SbmA, the E. coli ortholog of BacA.
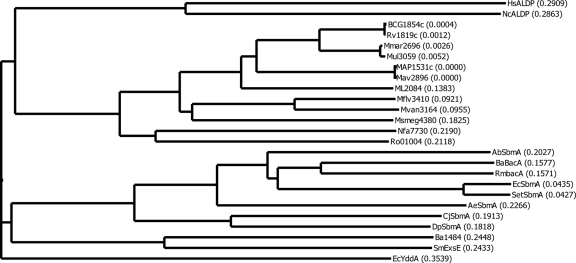
A hierarchical tree with some of the BacA homologs is shown in Fig. 1. Three distinctive groups of the BacA-SbmA proteins are observed. First, the complete mycobacterial BacA-related proteins with sizes of around 639 amino acids form a cluster with different branches for fast- and slow-growing species (the BacA homologs found in other actinobacteria [Nocardia and Rhodococcus] are also included in this group). The second major group contains the BacA proteins from the phylum Proteobacteria, representing five distinct classes. Two discrete arms are apparent: one including the BacA-SbmA proteins ranging from 320 to 420 amino acids containing only an MSD and one including the BacA-related proteins that are complete ABC transporters with sizes of between 551 and 621 amino acids, such as ExsE of S. meliloti, Ba1484 of B. abortus and YddA of E. coli. The two eukaryotic proteins, human adrenoleukodystrophy related protein (ALDP) and its homolog in Neurospora crassa, which are also complete ABC transporters of 740 and 715 amino acids, respectively, represent the third major group of these proteins.
FIG. 1.
Phylogenetic tree of BacA-SbmA proteins. The tree was generated using the neighbor-joining algorithm of Saitou and Nei (35) as indicated in Materials and Methods. Branch distances are indicated in parentheses. HsALDP, ALDP of Homo sapiens (accession no. AJ000327); NcALDP, ALDP of Neurospora crassa (accession no. CAB91246); BCG1854c, BacA of Mycobacterium bovis BCG Pasteur (accession no. YP_977945); Rv1819c, BacA of M. tuberculosis H37Rv (accession no. Q50614); Mmar2696, BacA of M. marinum (accession no. MarinoList MMAR_2696); Mul3059, BacA of M. ulcerans (accession no. YP_906777); MAP1531c, BacA of M. avium subsp. paratuberculosis (accession no. NP_960465); Mav2896, BacA of M. avium (accession no. YP_882082); ML2084, BacA of M. leprae (accession no. CAC31039); Mflv3410, BacA of M. gilvum (accession no. YP_001134674); Mvan3164, BacA of M. vanbaalenii (accession no. YP_953972); Msmeg4380, BacA of M. smegmatis (accession no. YP_888655); Nfa7730, BacA of Nocardia farcinica IFM 10152 (accession no. YP_116982); Ro01004, BacA of Rhodococcus sp. strain RHA1 (accession no. YP_700989); AbSbmA, SbmA of Acinetobacter baumannii (accession no. YP_001085804); BaBacA, BacA of Brucella melitensis biovar Abortus (accession no. AAF76873); RmBacA, BacA of Rhizobium meliloti (accession no. CAA51918); EcSbmA, SbmA of Escherichia coli (accession no. CAA38092); SetSbmA, SbmA of Salmonella enterica subsp. enterica serovar Typhi (accession no. NP_454971); AeSbmA, SbmA of Acidovorax avenae subsp. citrulli (accession no. YP_970563); CjSbmA, SbmA of Campylobacter jejuni subsp. jejuni (accession no. NP_281392); DpSbmA, SbmA of Desulfotalea psychrophila (accession no. YP_066699); Ba1484, Ba1484 of B. abortus (accession no. YP_222173); SmExsE, ExsE of S. meliloti (accession no. CAA12533); EcYddA, YddA of E. coli (accession no. P31826).
Insertional inactivation of BacA in M. tuberculosis.
In order to elucidate the role of BacA, we inactivated the M. tuberculosis bacA gene in the virulent strain H37Rv by insertion of a hygromycin resistance cassette within the coding sequence of this gene using homologous recombination (31). Southern blot analysis of chromosomal DNA from the M. tuberculosis bacA::hyg mutant compared with DNA from the H37Rv wild-type parent showed the expected bands for the wild-type (2.4 kbp) and inactivated (3.4 kbp) bacA genes (data not shown). Complementation of this mutant was achieved by cloning a wild-type copy of the bacA gene into an integrative vector. This strain (M. tuberculosis bacA::hyg-PKLMt5) showed two bands in Southern blot analysis, corresponding to the 3.4-kbp disrupted allele as well as the 4.7-kbp wild-type allele inserted into the chromosomal phage insertion site, attB.
BacA inactivation in M. tuberculosis results in bleomycin resistance but does not alter sensitivity to other membrane-disrupting agents.
In order to test the effect of BacA inactivation, we determined the sensitivities of our M. tuberculosis strains to bleomycin, detergents, and zinc. We found that the M. tuberculosis bacA mutant was more resistant to bleomycin than the parental strain, M. tuberculosis H37Rv, and this phenotype was partially reversed in the complemented strain bacA::hyg-pKLMt5 (Table 1). This is consistent with the BacA-related protein of M. tuberculosis having a functional role similar to that of an inward-facing transporter. However, unlike in alphaproteobacteria, but similar to the SbmA protein of E. coli (20), disruption of bacA did not sensitize M. tuberculosis to detergents such as SDS and DOC or zinc sulfate (Table 1). No differences were observed between the M. tuberculosis BacA-deficient and parental strains when they were grown in acidic conditions (pH 5.4) (data not shown), indicating that loss of BacA in M. tuberculosis does not produce gross perturbations in cell envelope integrity.
TABLE 1.
MICs of bleomycin, detergents, and zinc
| Agent | MICa for strain:
|
||
|---|---|---|---|
| H37Rv | H37Rv bacA::hyg | H37Rv bacA::hyg- pKLMt5 | |
| Bleomycin | 9-18 ng ml−1 | 16-32× | 4× |
| SDS | 0.03%, wt/vol | 1× | 1× |
| DOC | 0.01%, wt/vol | 1× | 1× |
| ZnSO4 | 1.5 mg ml−1 | 1× | 1× |
| Bleomycin in Fe-depleted GAST | 9-18 ng ml−1 | 4× | 2× |
Values are expressed as the ratio of the MIC obtained for the mutant to that obtained for the parental strain, H37Rv. Values were determined from three independent experiments, each performed in duplicate.
BacA does not alter resistance to clinically important antituberculosis drugs.
As increased resistance to some aminoglycosides in the BacA mutant of S. meliloti was previously reported (20), we investigated the possible role of the M. tuberculosis BacA in resistance to aminoglycosides used clinically in antituberculosis therapy, including amikacin, kanamycin, and streptomycin along with other commonly used antituberculosis agents. We performed MIC assays with the M. tuberculosis bacA mutant (bacA::hyg), the complemented strain (bacA::hyg-pKLMt5), and the H37Rv parental strain. As in the S. meliloti BacA mutants, a consistent low-level resistance to the three aminoglycosides tested was observed in the M. tuberculosis bacA::hyg strain (Table 2), suggesting a possible role for the BacA-related protein of M. tuberculosis in the transport of these drugs. However, we observed neither resistance nor hypersensitivity to other drugs in the M. tuberculosis bacA mutant strain, suggesting that there is not a substantial role of the BacA-related protein of M. tuberculosis in resistance to these antituberculosis drugs.
TABLE 2.
MICs of various antibiotics used in antituberculosis therapy
| Antibiotic | MICa for strain:
|
||
|---|---|---|---|
| H37Rv | H37Rv bacA::hyg | H37Rv bacA::hyg-pKLMt5 | |
| Amikacin | 0.8 μg ml−1 | 2× | 1-2× |
| Cycloserin | 12.5 μg ml−1 | 1× | 1× |
| Ethambutol | 0.8 μg ml−1 | 1× | 1× |
| Ethionamide | 0.2 μg ml−1 | 1× | 1× |
| Isoniazid | 0.02 μg ml−1 | 1× | 1× |
| Kanamycin | 1.5-3 μg ml−1 | 2-4× | 132×b |
| Ofloxacin | 0.4-0.8 μg ml−1 | 1-2× | 1-2× |
| Rifampin | 0.01-0.02 μg ml−1 | 1× | 1× |
| Streptomycin | 0.4-1.5 μg ml−1 | 2-4× | 2× |
| Tetracycline | 6-12.5 μg ml−1 | 1× | 1× |
| Vancomycin | 25-50 μg ml−1 | 0.5-1× | 1-2× |
Values for the mutants are expressed as the ratio of the MIC obtained for the mutant to that obtained for the parental strain, H37Rv. Values were determined from three independent experiments, each performed in duplicate.
The high level of kanamycin resistance observed in the bacA::hyg-pKLMt5 strain is due to the presence of a kanamycin resistance gene in the vector used for complementing the mutant strain.
Heterologous expression of M. tuberculosis BacA sensitizes E. coli to the antibacterial peptide Bac7.
Recently, it has been shown that the SbmA protein of E. coli is required for the transport of, and susceptibility to, the proline-rich antimicrobial peptide Bac7 and its synthetic derivatives (29). Similarly, the S. meliloti bacA mutant also showed increased resistance against Bac7(1-16), a shorter derivative of Bac7 (H. Kobayashi and G. C. Walker, unpublished data). Because these peptides do not show antibacterial activity against gram-positive bacteria (1), we heterologously expressed the bacA gene of M. tuberculosis in an E. coli sbmA-deficient mutant, RYC1001, to assess the ability of M. tuberculosis BacA to mediate the influx of Bac7. Expression of M. tuberculosis bacA significantly sensitized RYC1001 to both bleomycin and Bac7(1-16) (Table 3), further suggesting the functional similarity of M. tuberculosis BacA to SbmA/BacA of gram-negative bacteria.
TABLE 3.
Sensitivity of E. coli strains to bleomycin and Bac7(1-16)
| Strain | Relevant characteristics | MIC (μM)a
|
|
|---|---|---|---|
| Bleomycin | Bac7 (1-16) | ||
| RYC1000 | sbmA+ | 9 | 0.8 |
| RYC1001 | sbmA | 18 | 2 |
| RYC1001(pWSK-29) (pMM100) | sbmA(empty vector) (lacIq plasmid) | 18 | 2 |
| RYC1001(pWSK-MtBacA) (pMM100) | sbmA(Plac-bacA+Mt) (lacIq plasmid)b | 4 | 0.1 |
Values were determined from three independent experiments, each performed in duplicate.
bacA+Mt, bacA+ gene from M. tuberculosis.
Lack of BacA alters the virulence properties of M. tuberculosis.
The role of BacA in the virulence of M. tuberculosis was studied using a low-dose aerogenic murine model of infection. B6D2/F1 mice were infected with approximately 140 CFU of either the bacA::hyg mutant or the parental H37Rv strain. The average initial numbers of bacteria implanted were found to be 123 ± 17 and 153 ± 27 (average ± standard error of the mean), respectively. The growth kinetics of the two strains were identical throughout the infection, indicating that loss of BacA did not affect initial bacterial replication or containment of replication in the lungs and spleens of these animals (Fig. 2A and B). The 22 mice remaining in each group were observed until they died in survival experiments that extended over a period of 340 days. This experiment demonstrated that although the bacA mutant was not impaired for growth in B6D2/F1 mice, the mice infected with this strain survived longer (mean, 221 days) than those infected with the parental H37Rv strain (mean, 156 days) (P < 0.0001) (Fig. 2C). This result confirmed the results obtained in a previous experiment in which mice infected with 117 ± 23 CFU/lung of the wild-type strain showed a mean survival time of 173 days, while mice infected with the bacA::hyg mutant (58 ± 10 CFU/lung) displayed a mean survival time of 263 days (P < 0.0001) (data not shown). In a third experiment, B6D2/F1 mice were infected with the parental strain H37Rv and the complemented bacA::hyg-pKLMt5 strain (43 ± 11 and 108 ± 17 CFU/lung, respectively); the mean survival times for the infected mice were 184 and 166 days, respectively (P = 0.2475) (data not shown).
FIG. 2.
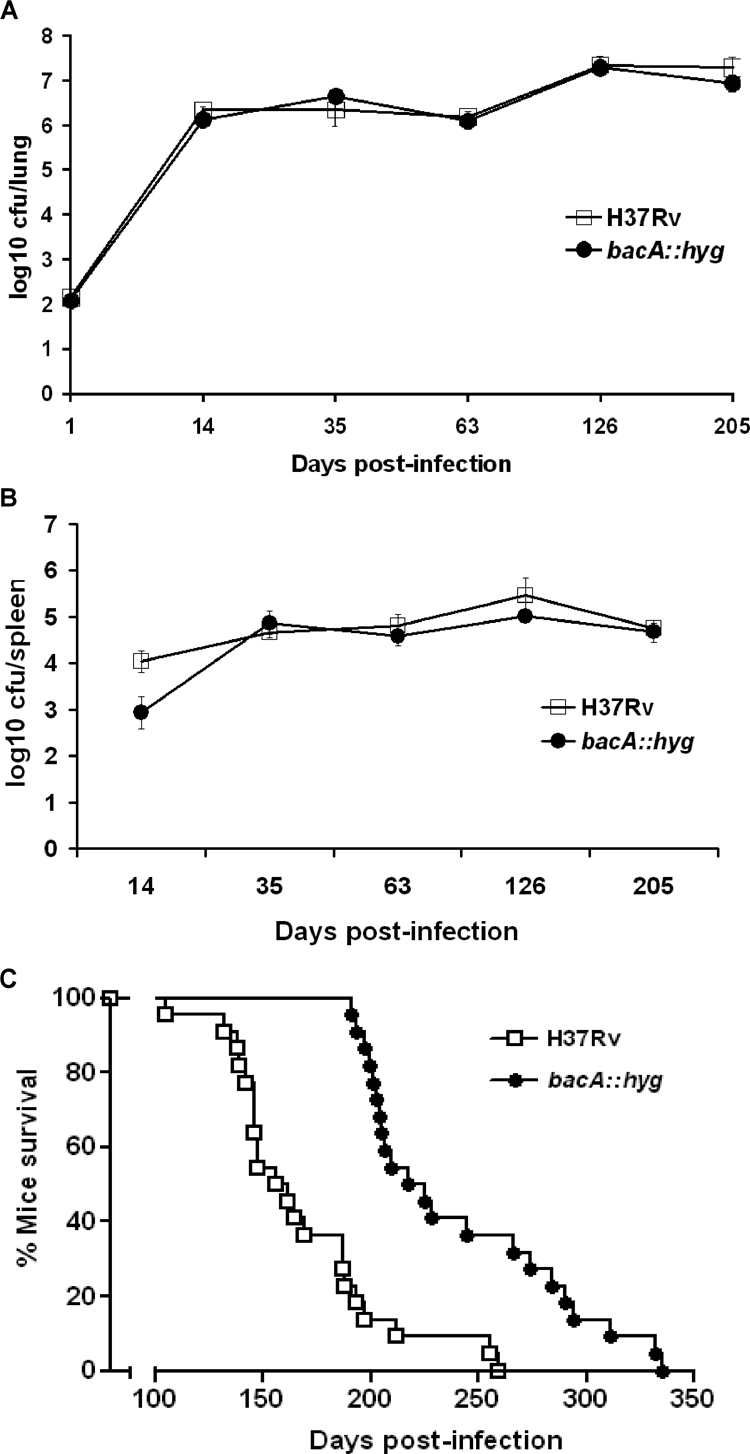
BacA contributes to virulence of M. tuberculosis in mice. B6D2/F1 mice were infected aerogenically with either the bacA::hyg mutant or the parental strain H37Rv. Bacterial numbers were monitored at the indicated times postinfection by harvesting lungs (A) and spleens (B) of infected mice. Results are expressed as averages of log10 CFU/lung along with the standard deviations obtained from replicate platings. (C) Mice were evaluated in a time-to-death experiment with the bacA::hyg mutant and the parental strain H37Rv. Analysis of these data was done using the Kaplan-Meier method, and a log rank test was used to determine statistical significance of observed survival differences (GraphPad Prism v3.0; GraphPad Software, CA).
Inactivation of the bacA gene does not modify mycolic acid composition and does not produce dramatic changes in the lipid content of the M. tuberculosis cell envelope.
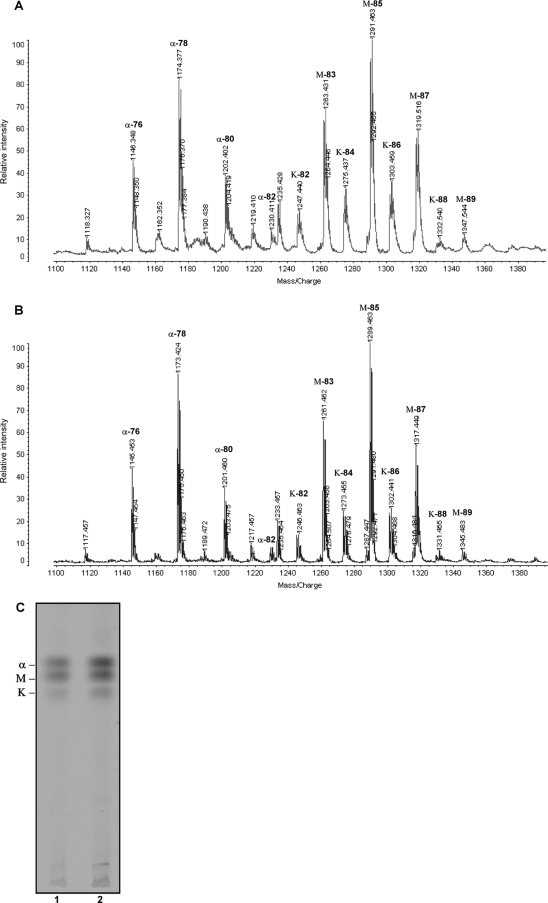
Species of the genus Mycobacterium do not posses LPS in the cell envelope, but very-long-chain fatty acids are present in many of the abundant lipids of the mycobacterial cell wall, with mycolic acids being the longest fatty acid present, so we first examined the mycolic acids for possible modifications. Covalent cell wall skeleton-associated mycolic acids were extracted from the cells of the M. tuberculosis bacA::hyg and H37Rv strains and analyzed by MALDI-TOF mass spectrometry. Figure 3A and B show the spectra obtained from this analysis. In both strains the three expected polarity classes, i.e., methoxy-, keto-, and α-mycolic acids, were seen as peaks corresponding to homologous series of acids differing by two methylene units. Both the relative abundance of each major class and the overall length of the mycolates within each class appeared to be identical between the wild-type and mutant strains.
FIG. 3.
Inactivation of bacA does not affect mycolic acid composition. (A and B) Arbinogalactan-linked mycolic acids were extracted from M. tuberculosis H37Rv (A) and bacA::hyg (B) strains, and MALDI-TOF analysis was performed. (C) TLC of trehalose-linked mycolic acids (TDM). After TLC separation of [1-14C]acetate-labeled glycolipids containing mycolic acids, TDM was scraped from the plate and extracted. Following hydrolysis and subsequent methylation, mycolic acid methyl esters were separated five times in petroleum ether-diethyl ether (85:15, vol/vol) as the solvent system. Lane 1, M. tuberculosis H37Rv; lane2, M. tuberculosis bacA::hyg mutant. α, α-mycolic acids; K, ketomycolic acids; M, methoxymycolic acids.
Because TDM is abundant on the cell surface, we also analyzed the mycolic acid composition of TDM specifically. For this, cultures were labeled with [1-14C]acetate at an OD650 of 0.4, and glycolipids containing noncovalently cell wall-associated lipids were extracted and the TDM purified by preparative TLC. After hydrolysis, the TDM-derived mycolic acids were then analyzed by TLC (Fig. 3C). No differences were observed in the ratio of the three class of mycolic acid present in TDM in the bacA::hyg strain compared to the wild type.
Further comparisons of apolar and polar lipid extracts from both the cells and the media using a wide variety of TLC systems and detection strategies failed to reveal any significant differences. Gas chromatography-mass spectrometry analysis of the shorter-chain fatty acids contained in both polar and apolar total lipid extracts likewise did not reveal any substantial changes (data not shown). However, the limitations for detecting the relative abundance of each fatty acid in the different molecules present in complex mixed samples do not permit us to exclude the possibility that small changes have occurred.
BacA deficiency does not have an effect on iron acquisition.
ABC transporters involved in metal uptake are present in both gram-negative and gram-positive bacteria. As bleomycin requires, among other cofactors, a reduced transition metal (either Fe2+ or Cu1+) for activity (5), we hypothesized that the bleomycin resistance of the BacA mutant might be the consequence of reduced transport of a metal or metal chelate such as the mycobactin siderophore. To address this question, we determined MICs of the cation-chelating agent EDTA and the iron-chelating agents 2,2′-dipyridyl and desferoxamine for all three strains. Neither EDTA nor CuSO4 MICs varied between the three strains tested, while two- to fourfold decreases in resistance to 2,2′-dipyridyl and desferoxamine were observed in the M. tuberculosis bacA mutant (Table 4). This result combined with the fact that the resistance to bleomycin observed in the M. tuberculosis bacA::hyg strain in medium without iron was significantly reduced (only up to fourfold higher) compared to that in the wild type also grown in iron-depleted medium (Table 1) suggested that BacA might in fact be involved in iron import. We therefore performed 55FeCl3 accumulation assays using the parental and BacA-deficient strains either in iron-deficient medium (that is, standard medium without any added iron) or in iron-depleted medium in which all trace amounts of iron were removed by treatment with an iron-scavenging resin (Chelex). As expected, iron uptake was faster and more efficient in cultures that were totally depleted of iron than in cultures that were simply iron deficient (Fig. 4). Both strains under both conditions showed the same rate of 55FeCl3 accumulation, indicating that iron uptake was independent of BacA.
TABLE 4.
MICs of chelating agents
| Agent | MICa for strain:
|
||
|---|---|---|---|
| H37Rv | H37Rv bacA::hyg | H37Rv bacA::hyg- pKLMt5 | |
| EDTA | 100 μM | 1× | 1× |
| Dypiridyl | 250 mM | 0.5× | 1× |
| Deferoxamine | 2-4 mM | 0.25-0.5× | 0.5× |
| CuSO4 | 10 mg ml−1 | 1× | 1× |
Values for the mutants are expressed as the ratio of the MIC obtained for the mutant to that obtained for the parental strain, H37Rv. Values were determined from three independent experiments, each performed in duplicate.
FIG. 4.
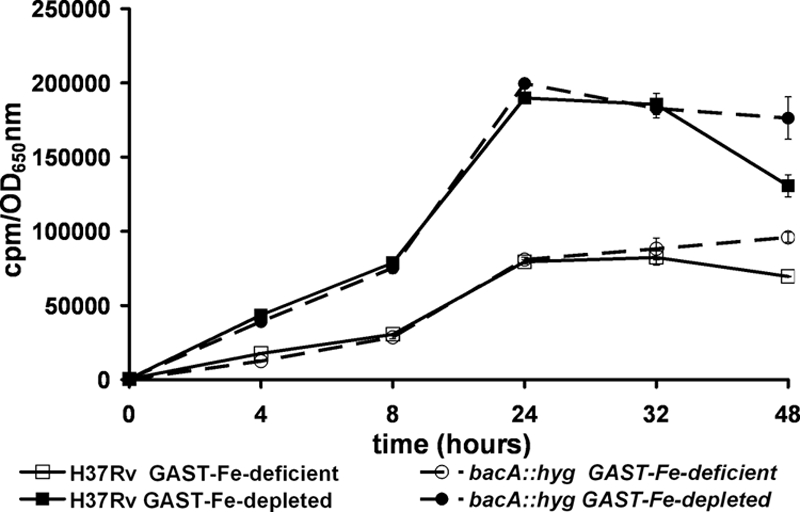
BacA deficiency does not have an effect on iron accumulation. 55FeCl3 uptake experiments were performed with the bacA::hyg mutant or the parental strain H37Rv. After 4 days of growth in iron-deficient medium, cultures were adjusted to the same bacterial concentration and 55FeCl3 was added to a final concentration of 0.5 μCi ml−1. At the indicated times, the bacterial concentration was assessed by OD650 measurements and samples were collected. The iron uptake was quantified by measurements of the radioactivity accumulated in the bacterial pellets. The results are expressed as the ratio of the radioactivity measured divided by the OD650 value obtained at the corresponding time. Two different iron conditions were tested, i.e., iron-depleted medium and iron-deficient medium (see Materials and Methods for details).
DISCUSSION
In this study, we have attempted to elucidate the role of BacA in the genus Mycobacterium by inactivating this gene in M. tuberculosis. The M. tuberculosis BacA-deficient strain shows an intriguing phenotype. It shares some attributes with the BacA mutants of the alphaproteobacteria, such as increased resistance to bleomycin and aminoglycosides, and alters the outcome of a chronic infection within the host. However, unlike in alphaproteobacteria and similar to the case in E. coli, the integrity of the cell envelope is unaffected by the absence of BacA. However, the M. tuberculosis BacA does seem to be capable of mediating resistance to certain antimicrobial peptides such as bleomycin and Bac7. Our results show that, like the BacA proteins of the alphaproteobacteria, the M. tuberculosis BacA is involved in maintaining a long-term interaction with the host. Our results therefore suggest more functional similarity between the M. tuberculosis BacA and the BacA-SbmA proteins than would be expected based upon the closer homology to the ExsE protein of S. meliloti. To our knowledge, this is the first report that describes a comparable role for a BacA-related protein and the BacA-SbmA proteins.
In alphaproteobacteria, BacA deficiency has a pleiotropic phenotypic effect, including increased resistance to bleomycin, increased sensitivity to cell envelope-disrupting agents (detergents, hydrophobic dyes, and ethanol), and impairment in establishing a chronic intracellular infection in the corresponding host (17, 20, 25). The augmented sensitivity to some chemical compounds has been explained by the fact that the BacA mutants show an altered lipid A composition in which approximately half of the lipid A molecules lack VLCFA (27-OH-C28:0 and 29-OH-C30) (11). This lipid A alteration severely compromises the outer membranes of alphaproteobacteria, rendering theses mutants more sensitive to these compounds. In fact, the same phenotype has been observed in S. meliloti AcpXL and LpxXL mutants, which completely lack VLCFA modification of the lipid A (12). However, this change in lipid A composition does not account for the impairment of the S. meliloti BacA-deficient strain in establishing chronic infection in the host plant, since the S. meliloti AcpXL and LpxXL mutants are still able to establish a functional symbiosis with alfalfa plants, although not as efficiently as the wild-type strain (12).
We hypothesized that the mycobacterial BacA could also be involved in transport of one of the long-chain fatty acids characteristic of the genus. The most abundant and characteristic fatty acids present the genus Mycobacterium are the mycolic acids. These are very long (C70 to C90) α-alkyl β-hydroxy fatty acids that can exist in the wall in two basic forms: covalently bound to the arabinogalactan skeleton and esterified to a variety of carbohydrate-containing molecules, mainly trehalose, with TDM being the most abundant form in M. tuberculosis (3). Our studies showed that none of the major lipid components of the mycobacterial cell envelope were altered in abundance or structure in a strain lacking the BacA protein. Minor components of the cell envelope can, of course, have significant effects on permeability and function, and we cannot rule out subtle alterations that could not be detected in our assays.
However, as was the case with bacA mutation in alphaproteobacteria, the M. tuberculosis bacA mutant was compromised for maintenance of a persistent infection state in mice. The mechanism by which BacA deficiency cause attenuation is unknown, but recently it has been reported that the B. abortus bacA mutant induce larger amounts of the proinflammatory cytokines tumor necrosis factor alpha and interleukin-12 than the parental strain, in either in vivo or in vitro experiments using peritoneal macrophages of C57BL/6 mice (30). These results suggest that the BacA protein could transport a molecule that can directly or indirectly modulate the proinflammatory host response. This phenotype has been described as characteristic of a more general class of “immunopathology” mutants, including, for example, those with mutations of the pks1-15 gene, which encodes a polyketide synthase involved in the synthesis of the phenol glycolipid that inhibits the innate immune response. Mice infected with this mutant show a longer survival time than those infected with the parental strain in spite of it not being impaired for in vivo growth (33).
Similar to the SbmA/BacA mutants of gram-negative bacteria, the M. tuberculosis BacA-deficient strain also shows increased resistance to bleomycin. In S. meliloti, BacA-mediated sensitivity to bleomycin is independent of the VLCFA modification of lipid A, since introduction of a bacA-null mutation in the S. meliloti lpxXL mutant strain confers resistance to bleomycin without changing the lipid A composition (13). Since bleomycins are glycopeptide antibiotics that require activation by binding transition metals (Fe2+ or Cu1+), we hypothesized that BacA may be involved in the transport of any of these metals or in the transport of molecules that bind these metals. M. tuberculosis produces two forms of salicylic acid-derived siderophores, the lipophilic mycobactin T and the hydrophilic mycobactin T or carboxymycobactin. The ferri-siderophore complex therefore seemed a plausible candidate as a substrate for BacA. Nevertheless, in the M. tuberculosis BacA-deficient strain, iron accumulation, as measured by uptake of 55FeCl3, remained unaltered. The Rv1348 and Rv1349 genes of M. tuberculosis were found to be required for iron acquisition and virulence (34). These two genes have been identified as the irtAB system, and it has been postulated that IrtAB act as an Fe-carboxymycobactin transporter, a suggestion that has recently been supported experimentally (10).
In conclusion, in this study we have shown that BacA, the M. tuberculosis homolog of an ABC transporter that contributes to the maintenance of chronic infections in systems as diverse as B. abortus infection of cattle and S. meliloti infection of alfalfa, also plays an important role in the outcome of chronic infections by M. tuberculosis in mice. This striking conservation of function across three disparate symbiotic/pathogenic relationships suggests an important underlying mechanism. While this protein does not appear to directly contribute to drug efflux or efflux of components of the cell wall of M. tuberculosis, we provide compelling evidence that the protein may be involved in the transport of certain antimicrobial peptides. While the precise biological substrate transported by BacA has remained elusive, our studies suggest that this substrate is likely to be a host-derived antimicrobial peptide that plays an important role in determining whether an infection is progressive or latent in a given individual.
Acknowledgments
We thank C. Guilhot for providing us with the mycobacterial allelic exchange system and J. Gonzales for technical assistance.
This research was supported by the Intramural Research Programs of the NIH, NIAID. This work was also supported by National Institutes of Health grant GM31010 to G.C.W. G.C.W. is an American Cancer Society Research Professor.
Footnotes
Published ahead of print on 7 November 2008.
REFERENCES
- 1.Benincasa, M., M. Scocchi, E. Podda, B. Skerlavaj, L. Dolzani, and R. Gennaro. 2004. Antimicrobial activity of Bac7 fragments against drug-resistant clinical isolates. Peptides 252055-2061. [DOI] [PubMed] [Google Scholar]
- 2.Braibant, M., P. Gilot, and J. Content. 2000. The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 24449-467. [DOI] [PubMed] [Google Scholar]
- 3.Brennan, P. J., and H. Nikaido. 1995. The envelope of mycobacteria. Annu. Rev. Biochem. 6429-63. [DOI] [PubMed] [Google Scholar]
- 4.Campbell, G. R., B. L. Reuhs, and G. C. Walker. 2002. Chronic intracellular infection of alfalfa nodules by Sinorhizobium meliloti requires correct lipopolysaccharide core. Proc. Natl. Acad. Sci. USA 993938-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J., and J. Stubbe. 2005. Bleomycins: towards better therapeutics. Nat. Rev. Cancer. 5102-112. [DOI] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry, 3rd, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulson, K. Taylor, S. Whithead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393537-544. [DOI] [PubMed] [Google Scholar]
- 7.Dassa, E., and P. Bouige. 2001. The ABC of ABCS: a phylogenetic and functional classification of ABC systems in living organisms. Res. Microbiol. 152211-229. [DOI] [PubMed] [Google Scholar]
- 8.De Voss, J. J., K. Rutter, B. G. Schroeder, H. Su, Y. Zhu, and C. E. Barry III. 2000. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. USA 971252-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domenech, P., M. C. Menendez, and M. J. Garcia. 1994. Restriction fragment length polymorphisms of 16S rRNA genes in the differentiation of fast-growing mycobacterial species. FEMS Microbiol. Lett. 11619-24. [DOI] [PubMed] [Google Scholar]
- 10.Farhana, A., S. Kumar, S. S. Rathore, P. C. Ghosh, N. Z. Ehtesham, A. K. Tyagi, and S. E. Hasnain. 2008. Mechanistic insights into a novel exporter-importer system of Mycobacterium tuberculosis unravel its role in trafficking of iron. PLoS One 3e2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferguson, G. P., A. Datta, J. Baumgartner, R. M. Roop II, R. W. Carlson, and G. C. Walker. 2004. Similarity to peroxisomal-membrane protein family reveals that Sinorhizobium and Brucella BacA affect lipid-A fatty acids. Proc. Natl. Acad. Sci. USA 1015012-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson, G. P., A. Datta, R. W. Carlson, and G. C. Walker. 2005. Importance of unusually modified lipid A in Sinorhizobium stress resistance and legume symbiosis. Mol. Microbiol. 5668-80. [DOI] [PubMed] [Google Scholar]
- 13.Ferguson, G. P., A. Jansen, V. L. Marlow, and G. C. Walker. 2006. BacA-mediated bleomycin sensitivity in Sinorhizobium meliloti is independent of the unusual lipid A modification. J. Bacteriol. 1883143-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson, G. P., R. M. Roop, 2nd, and G. C. Walker. 2002. Deficiency of a Sinorhizobium meliloti BacA mutant in alfalfa symbiosis correlates with alteration of the cell envelope. J. Bacteriol. 1845625-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genilloud, O., M. C. Garrido, and F. Moreno. 1984. The transposon Tn5 carries a bleomycin resistance determinant. Gene 32225-233. [DOI] [PubMed] [Google Scholar]
- 16.Gibson, K. E., G. R. Campbell, J. Lloret, and G. C. Walker. 2006. CbrA is a stationary-phase regulator of cell surface physiology and legume symbiosis in Sinorhizobium meliloti. J. Bacteriol. 1884508-4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glazebrook, J., A. Ichige, and G. C. Walker. 1993. A Rhizobium meliloti homolog of the Escherichia coli peptide-antibiotic transport protein SbmA is essential for bacteroid development. Genes Dev. 71485-1497. [DOI] [PubMed] [Google Scholar]
- 18.Harries, A. D., and C. Dye. 2006. Tuberculosis. Ann. Trop. Med. Parasitol. 100415-431. [DOI] [PubMed] [Google Scholar]
- 19.Higgins, C. F. 2001. ABC transporters: physiology, structure and mechanism—an overview. Res. Microbiol. 152205-210. [DOI] [PubMed] [Google Scholar]
- 20.Ichige, A., and G. C. Walker. 1997. Genetic analysis of the Rhizobium meliloti bacA gene: functional interchangeability with the Escherichia coli sbmA gene and phenotypes of mutants. J. Bacteriol. 179209-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan, E. L., and P. Meier. 1958. Nonparametric estimation for incomplete observations. J. Am. Stat Assoc. 53457-481. [Google Scholar]
- 22.Lage, H. 2003. ABC-transporters: implications on drug resistance from microorganisms to human cancers. Int. J. Antimicrob. Agents 22188-199. [DOI] [PubMed] [Google Scholar]
- 23.Laval, F., M. A. Laneelle, C. Deon, B. Monsarrat, and M. Daffe. 2001. Accurate molecular mass determination of mycolic acids by MALDI-TOF mass spectrometry. Anal. Chem. 734537-4544. [DOI] [PubMed] [Google Scholar]
- 24.Lavina, M., A. P. Pugsley, and F. Moreno. 1986. Identification, mapping, cloning and characterization of a gene (sbmA) required for microcin B17 action on Escherichia coli K12. J. Gen. Microbiol. 1321685-1693. [DOI] [PubMed] [Google Scholar]
- 25.LeVier, K., R. W. Phillips, V. K. Grippe, R. M. Roop II, and G. C. Walker. 2000. Similar requirements of a plant symbiont and a mammalian pathogen for prolonged intracellular survival. Science 2872492-2493. [DOI] [PubMed] [Google Scholar]
- 26.LeVier, K., and G. C. Walker. 2001. Genetic analysis of the Sinorhizobium meliloti BacA protein: differential effects of mutations on phenotypes. J. Bacteriol. 1836444-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linton, K. J., and C. F. Higgins. 1998. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 285-13. [DOI] [PubMed] [Google Scholar]
- 28.Long, S., S. McCune, and G. C. Walker. 1988. Symbiotic loci of Rhizobium meliloti identified by random TnphoA mutagenesis. J. Bacteriol. 1704257-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mattiuzzo, M., A. Bandiera, R. Gennaro, M. Benincasa, S. Pacor, N. Antcheva, and M. Scocchi. 2007. Role of the Escherichia coli SbmA in the antimicrobial activity of proline-rich peptides. Mol. Microbiol. 66151-163. [DOI] [PubMed] [Google Scholar]
- 30.Parent, M. A., R. Goenka, E. Murphy, K. Levier, N. Carreiro, B. Golding, G. Ferguson, R. M. Roop II, G. C. Walker, and C. L. Baldwin. 2007. Brucella abortus bacA mutant induces greater pro-inflammatory cytokines than the wild-type parent strain. Microbes Infect. 955-62. [DOI] [PubMed] [Google Scholar]
- 31.Pelicic, V., M. Jackson, J. M. Reyrat, W. R. Jacobs, Jr., B. Gicquel, and C. Guilhot. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 9410955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Primm, T. P., S. J. Andersen, V. Mizrahi, D. Avarbock, H. Rubin, and C. E. Barry III. 2000. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J. Bacteriol. 1824889-4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reed, M. B., P. Domenech, C. Manca, H. Su, A. K. Barczak, B. N. Kreiswirth, G. Kaplan, and C. E. Barry III. 2004. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 43184-87. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez, G. M., and I. Smith. 2006. Identification of an ABC transporter required for iron acquisition and virulence in Mycobacterium tuberculosis. J. Bacteriol. 188424-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4406-425. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Serina, S., F. Nozza, G. Nicastro, F. Faggioni, H. Mottl, G. Deho, and A. Polissi. 2004. Scanning the Escherichia coli chromosome by random transposon mutagenesis and multiple phenotypic screening. Res. Microbiol. 155692-701. [DOI] [PubMed] [Google Scholar]
- 38.Slayden, R. A., and C. E. Barry III. 2001. Analysis of the lipids of Mycobacterium tuberculosis, p. 229-245. In T. Parish and N. G. Stoker (ed.), Mycobacterium tuberculosis protocols, vol. 54. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 39.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 41911-1919. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wanders, R. J., W. F. Visser, C. W. van Roermund, S. Kemp, and H. R. Waterham. 2007. The peroxisomal ABC transporter family. Pflugers Arch. 453719-734. [DOI] [PubMed] [Google Scholar]
- 42.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100195-199. [PubMed] [Google Scholar]
- 43.Yorgey, P., J. Lee, J. Kordel, E. Vivas, P. Warner, D. Jebaratnam, and R. Kolter. 1994. Posttranslational modifications in microcin B17 define an additional class of DNA gyrase inhibitor. Proc. Natl. Acad. Sci. USA 914519-4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Young, D. B., M. D. Perkins, K. Duncan, and C. E. Barry, 3rd. 2008. Confronting the scientific obstacles to global control of tuberculosis. J. Clin. Investig. 1181255-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young, J., and I. B. Holland. 1999. ABC transporters: bacterial exporters—revisited five years on. Biochim. Biophys. Acta 1461177-200. [DOI] [PubMed] [Google Scholar]