Abstract
DnaA functions as both a transcription factor and the replication initiator in bacteria. We characterized the DNA binding dynamics of DnaA on a genomic level. Based on cross-linking and chromatin immunoprecipitation data, DnaA binds at least 17 loci, 15 of which are regulated transcriptionally in response to inhibition of replication (replication stress). Six loci, each of which has a cluster of at least nine potential DnaA binding sites, had significant increases in binding by DnaA when replication was inhibited, indicating that the association of DnaA with at least some of its target sites is altered after replication stress. When replication resumed from oriC after inhibition of replication initiation, these high levels of binding decreased rapidly at origin-proximal and origin-distal regions, well before a replication fork could pass through each of the regulated regions. These findings indicate that there is rapid signaling to decrease activation of DnaA during replication and that interaction between DnaA bound at each site and the replication machinery is not required for regulation of DnaA activity in response to replication stress.
Cells use multiple mechanisms to detect and respond to perturbations in replication. In bacteria, the well-characterized RecA-dependent SOS response affects the expression of many genes after disruptions in replication elongation and DNA damage (13). Additionally, there are recA-independent mechanisms that affect gene expression in response to alterations in replication. A significant part of the recA-independent response to perturbations in replication (replication stress) appears to be mediated directly by DnaA (15).
DnaA is widely conserved in bacteria and is best known as the replication initiator (reviewed in references 23, 33, and 40). It binds to a 9-bp site that appears multiple times in the origin of replication in bacterial chromosomes. Binding sites are also found upstream of many genes and throughout the chromosome. At replication origins, DnaA mediates melting of DNA (open complex formation) and helps to recruit proteins required for replication. DnaA is a member of the AAA+ family of ATPases, and the ATP-bound form (DnaA-ATP) is required for replication initiation (36). Newly synthesized DnaA is predominantly in the ATP-bound form, and nucleotide hydrolysis converts DnaA-ATP to the inactive DnaA-ADP form after replication initiation. In Escherichia coli, the levels of DnaA-ATP and DnaA-ADP are linked to replication status, and there can be high levels of DnaA-ATP in cells that are unable to replicate (27).
DnaA is also a transcription factor. DnaA activates or represses transcription of various genes, including its own (1, 3, 26, 34, 39). It is thought that like the case for replication initiation, DnaA-ATP is the active form for transcriptional regulation (reviewed in references 23, 37, and 40), and its ability to function as a transcription factor appears to increase when replication initiation or elongation is inhibited (14, 15). Thus, accumulation of DnaA-ATP during replication stress may result in activation of the transcriptional regulatory activity of DnaA as well as its initiator activity.
In Bacillus subtilis, DnaA appears to control the expression of >50 genes in ∼20 operons in response to perturbations in replication (15). Transcription of these operons changes in response to inhibition of either replication elongation or replication initiation, independently of the well-characterized RecA-mediated SOS response. Furthermore, these operons contain putative DnaA binding sites in their regulatory regions, and DnaA appears to associate with at least six of these in vivo, based on results from chromatin immunoprecipitation (ChIP) and chromatin affinity purification (ChAP-chip) experiments (15, 20). Alterations in DnaA levels during exponential growth also affect the expression of several genes, and a genome-wide analysis of DnaA binding to chromosomal DNA demonstrated strong binding to eight intergenic regions, five of which appear to regulate gene expression (20).
In this study, we used ChIP-chip and ChIP-PCR to monitor binding of B. subtilis DnaA to DNA in vivo during exponential growth and after inhibition of replication. We detected an association of DnaA with at least 17 chromosomal regions, 15 of which correspond to operons whose expression responds to perturbations in DNA replication. Following inhibition of replication elongation or initiation, DnaA binding increased significantly at six chromosomal regions, each of which contains at least nine potential DnaA binding sites. We found that passage of a replication fork through a regulatory region was not required for release of bound DnaA from that region following the resumption of replication. Our results support the notion that DnaA regulates a global response to perturbations in replication and indicate that alterations in DnaA binding to target promoters contribute to the changes in gene expression following inhibition of replication.
MATERIALS AND METHODS
Media and growth conditions.
For all experiments, cells were grown in S750 defined minimal medium (18, 21) with 0.1% glutamate and required amino acids (typically Trp and Phe). Glucose (1%) was the carbon source, except for strains bearing Pxyl-dnaN, for which arabinose (1% [together with xylose at 0.1% as an inducer]) was used. Cells were typically grown at 32°C and shifted to 47°C for temperature-sensitive mutants.
Strains and alleles.
Strains used in this study were the lab wild type, AG174 (trp phe; also called strain JH642); KPL69 [dnaB134(Ts)-zhb83::Tn917 trp phe] (29, 30, 38); KPL73 [dnaD23(Ts) chr::Tn917ΩHU151(mls) trp phe] (29); and AIG200 [Δ(dnaA-oriC-dnaN)::spc amyE::(PxylA-dnaN cat) spoIIIJ::(oriN repN kan) phe trp+] (15).
dnaD23(Ts) and dnaB134(Ts) are temperature-sensitive mutations that prevent initiation of replication at nonpermissive temperatures (7, 8, 10, 24, 30).
dnaA is normally essential, but it can be deleted in strains capable of initiating chromosomal replication from a heterologous origin (2, 19, 22, 32). Therefore, in strain AIG200, the plasmid origin of replication, oriN, along with the plasmid initiator gene repN, was integrated into the chromosome at spoIIIJ, near oriC. This strain also contains a deletion-insertion that removes oriC and the dnaA-dnaN operon and inserts spc. dnaN, the essential gene encoding the β-processivity clamp of DNA polymerase that is normally in an operon with dnaA, is expressed at a heterologous locus from the xylose-inducible promoter Pxyl (Pxyl-dnaN) (2, 15).
ChIP.
ChIP of DNA bound to DnaA was done essentially as described previously (15), with minor modifications. Briefly, protein and DNA were cross-linked with formaldehyde, lysed and sheared with 1 mg/ml lysozyme and sonication, and immunoprecipitated with chicken anti-DnaA primary antibodies, donkey anti-chicken secondary antibodies, and protein A-Sepharose beads. Samples were washed and cross-links reversed, and DNA was then eluted, followed by further washing of the beads with 50 mM Tris, pH 8, 10 mM EDTA, 0.8% sodium dodecyl sulfate for 5 min at 37°C. The eluate and wash were combined, treated with proteinase K (0.1 mg/ml, 30 min, 37°C), extracted with phenol-chloroform, precipitated with ethanol, and resuspended in 15 μl double-distilled H2O. Samples of total DNA were taken prior to incubation with primary antibodies, treated with proteinase K (0.1 mg/ml, 30 min, 37°C) in the presence of 0.8% sodium dodecyl sulfate, and then subjected to cross-link reversal, phenol-chloroform extraction, and ethanol precipitation in parallel with the immunoprecipitates. There were at least three biological replicates for each experiment.
(i) ChIP-PCR.
For ChIP-PCR, samples were diluted (typically between 1:10 and 1:150) to give an appropriate level of signal. PCRs were done for 28 cycles, and products were run in 1.5% agarose gels containing ethidium bromide and analyzed and quantified with AlphaEaseFC software v4.0 (Alpha Innotech). The locus yabM was used as a negative control site for background reference. Enrichment at this locus was at background levels in ChIP-chip data (Fig. 1), and it behaved similarly in ChIP-PCR experiments to other loci where DnaA was not expected to bind (not shown). Despite not having specific binding by DnaA, there was a background signal, and this signal was used for normalization. Enrichment was calculated as a ratio of ratios in order to normalize data to the background level, represented by yabM, as follows: (IPi/totali)/(IPyabM/totalyabM), where IPi and totali represent the dilution-adjusted band intensities at the locus of interest.
FIG. 1.
Genome-wide DnaA ChIP-chip profiles for arrested, replicating, and dnaA null cells. The enrichment of each chromosomal region in the DnaA ChIP sample relative to total DNA is plotted versus the chromosomal position relative to oriC at 0°, in the center of the graphs. For each data set, at least three biological replicates were collected, and the median for each locus on the arrays is shown. (A) Wild-type cells (AG174) in exponential growth phase were treated with HPUra for 60 min at 32°C, cross-linked, and harvested for ChIP-chip assay. (B) dnaD(Ts) (KPL73) cells in exponential growth phase at 32°C were shifted to the restrictive temperature (47°C) for 90 min, cross-linked, and harvested for ChIP-chip assay. (C) dnaB(Ts) (KPL69) cells in exponential growth phase at 32°C were shifted to the restrictive temperature (47°C) for 90 min, cross-linked, and harvested for ChIP-chip assay. (D) Wild-type cells (AG174) in exponential growth phase at 32°C were cross-linked and harvested for ChIP-chip assay. (E) dnaA null mutant cells (AIG200) in exponential growth phase at 32°C were cross-linked and harvested for ChIP-chip assay. The signal at 283° (−77° in the figure) is that for yutF; there appears to be a protein bound here that interacts nonspecifically with the anti-DnaA antibody (see text for more details).
(ii) ChIP-chip experiments.
To generate sufficient signals in ChIP-chip experiments, we typically took samples of 100 ml of mid-exponential-phase culture (grown in S750 defined minimal medium). DNA samples were prepared for microarray analysis by labeling samples of the resuspended immunoprecipitated DNA or total DNA with aminoallyl-dUTP, using 13 U Sequenase (USB Corp.) and 5 μg random nonamer as primers. There was no amplification step. The labeled samples were purified and conjugated to Cy3 or Cy5. We used DNA microarrays containing PCR products representing >95% of the open reading frames in the B. subtilis genome and nearly all of the 295 intergenic regions that are >364 bp in length, as described previously (4, 5, 16). Microarray hybridization and data acquisition were performed as described previously (4).
Enrichment of DnaA at each genomic locus represented on the microarrays was calculated as the relative amount of DNA in the immunoprecipitate divided by the amount of total DNA for that locus without immunoprecipitation. This value was normalized to the background signal, calculated as the median value for all chromosomal loci of the amount of DNA in the immunoprecipitate divided by the amount of total DNA. Where presented, enrichment data are plotted according to chromosome position.
RESULTS
Binding of DnaA to several chromosomal regions increases following replication stress.
Previously, using ChIP-PCR, we found that DnaA is associated with several putative target operons whose mRNA levels change in response to perturbations in replication and that contain potential DnaA binding sites (15). Since blocking replication elongation or preventing replication initiation (while allowing ongoing replication to finish) elicits transcriptional responses controlled by DnaA (15), we postulated that binding of DnaA to some targets might increase when replication is inhibited. To test this hypothesis, we monitored DnaA binding at three regulatory targets, dnaA, sda, and ywlC, during replication and after inhibition of replication, using ChIP-PCR. Binding was quantified by measuring the enrichment of a given DNA region in the cross-linked and precipitated material relative to that of other chromosomal regions (see Materials and Methods). We inhibited replication elongation by treating cells with HPUra, a DNA polymerase III inhibitor (6). We inhibited replication initiation by shifting the temperature-sensitive dnaB(Ts) and dnaD(Ts) replication initiation mutants to a nonpermissive temperature and allowing ongoing rounds of replication to finish.
During replication of the wild type and the temperature-sensitive mutants at the permissive temperature, there was low but detectable enrichment for each of the three regions (dnaA, sda, and ywlC) in the DnaA immunoprecipitates relative to other chromosomal regions (Table 1). Sixty minutes after treatment with HPUra to block replication elongation, there was a significant increase in enrichment of each of these three regions, indicating that association of DnaA with these regions was increased (Table 1). There was also a significant increase in enrichment of these regions 90 min after shifting the dnaB(Ts) and dnaD(Ts) mutants to the nonpermissive temperature (47°C) (Table 1), and the dnaD(Ts) mutant typically gave higher enrichment levels than the dnaB(Ts) mutant or treatment with HPUra. There was no increase in enrichment of these regions in wild-type cells incubated at 47°C (Table 1), indicating that the increased binding by DnaA to DNA in the dnaB(Ts) and dnaD(Ts) mutants at the nonpermissive temperature (47°C) was not due to the temperature shift per se.
TABLE 1.
DnaA binding during exponential growth and replication stress
| Locus | Mean enrichment value ± SE (fold change)a
|
|||||||
|---|---|---|---|---|---|---|---|---|
|
dnaB(Ts)
|
dnaD(Ts)
|
Wild type
|
||||||
| 32°C | 47°C | 32°C | 47°C | No treatment | HPUra treatment | 32°C | 47°C | |
| dnaA | 2.2 ± 2.3 | 42.2 ± 9.5 (18.8) | 8.8 ± 2.4 | 180 ± 130 (20.4) | 4.8 ± 0.9 | 61.4 ± 1.9 (12.7) | 6.4 ± 4.0 | 6.3 ± 3.4 (1.0) |
| sda | 1.7 ± 0.4 | 6.8 ± 1.0 (4.0) | 2.2 ± 0.5 | 15.7 ± 4.2 (7.1) | 2.0 ± 0.5 | 4.4 ± 0.1 (2.2) | 1.3 ± 0.04 | 1.5 ± 0.4 (1.2) |
| ywlC | 2.3 ± 0.3 | 9.7 ± 0.8 (4.3) | 3.4 ± 1.2 | 70.0 ± 3.1 (20.7) | 2.8 ± 0.4 | 14.4 ± 5.9 (5.1) | 1.7 ± 0.38 | 1.6 ± 0.1 (0.9) |
Values presented are the mean (from three independent biological replicates) enrichments ± standard errors for the indicated DNA regions (dnaA, sda, and ywlC), as measured by ChIP-PCR analysis. Data are normalized to data for the yabM locus, which does not bind DnaA specifically (Fig. 1 and data not shown). Strains used were KPL69 [dnaB(Ts)], KPL73 [dnaD(Ts)], and AG174 (wild type [with or without HPUra treatment). Cells were grown at 32°C and treated with HPUra for 60 min. Temperature shifts were to 47°C, and samples were taken after 90 min at that temperature.
Analysis of genome-wide DnaA binding.
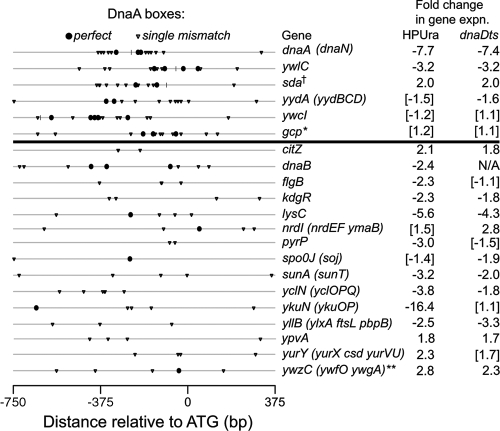
We used ChIP-chip experiments to identify additional chromosomal regions bound by DnaA in vivo. Based on the increase in association of DnaA with dnaA, sda, and ywlC after replication stress, we analyzed DnaA binding by ChIP-chip assay under these conditions as well as during exponential growth. We detected high levels of DnaA binding (∼10- to 70-fold enrichment) at six chromosomal regions following arrest of either replication elongation or initiation (Fig. 1 and 2). The regions include dnaA, sda, ywlC, yydA, and the intergenic regions between the 3′ ends of gcp and ydiF and the 5′ ends of ywcI and vpr. All six of these regions contained at least nine instances of the DnaA box sequence (allowing one mismatch) clustered within a 500-bp region (Fig. 3), and we refer to these as “clustered” sites. Four of the regions with high binding levels (dnaA, sda, ywlC, and yydA) are associated with operons whose transcription changes in response to perturbations in replication (Fig. 3) (15). We postulate that the increased binding of DnaA following inhibition of replication could directly affect transcription of these target operons.
FIG. 2.
DnaA binding at loci with clustered DnaA boxes. Data are shown for specific regions from Fig. 1, plotted for ∼8-kb regions centered around clustered DnaA boxes, where DnaA binding increased substantially upon inhibition of replication. Chromosomal positions are listed as distances (in kilobases) from oriC, and enrichment is shown as described in the legend to Fig. 1. DnaA box locations are indicated by diamonds below the data traces; filled and open diamonds represent perfect and single-mismatch boxes, respectively. Gene locations and orientations are indicated with arrows. Expression of genes with asterisks is altered after inhibition of replication (15). Expression of all genes indicated with asterisks decreases after replication inhibition, except for that of sda, which increases. Data are shown in the vicinities of dnaA (A), ywlC (B), ywcI (C), yydA (D), gcp (E), and sda (F). Filled squares with solid lines represent wild-type cells (AG174) 60 min after treatment with HPUra, filled circles with dotted lines represent untreated wild-type cells (AG174), gray open circles and lines represent dnaA null mutant cells (AIG200), and filled triangles with solid lines represent dnaD(Ts) cells (KPL73) after 90 min at 47°C.
FIG. 3.
Distribution of DnaA boxes at sites bound by DnaA. Shown are locations of perfect (solid circles) and single-mismatch (triangles) DnaA boxes occurring from 750 bp upstream to 375 bp downstream of the start codons of genes where DnaA binding was detected in either ChIP-chip or ChIP-PCR experiments. The first six regions indicated (above the thick line) all have at least nine potential DnaA binding sites with ≤1 mismatch from the consensus and have increased enrichment after inhibition of replication (Table 1; Fig. 1 and 2). Regions indicated below the thick line are those with fewer than nine potential DnaA binding sites. Enrichment at these regions did not consistently increase when replication was inhibited, and DnaA enrichment at flgB, nrdI, sunA, and yllB was greater than that at the background locus yabM but was not statistically significant (Table 2). Listed at right are the transcriptional changes in each of these genes due to inhibition of elongation with HPUra and inhibition of initiation by high-temperature incubation of dnaD(Ts) cells. Listed in parentheses are genes in the same operon that also responded transcriptionally. The data are from reference 15. Bracketed values are not statistically significant. Functions of the genes potentially regulated by DnaA have been summarized previously (15, 20), and they include genes involved in DNA replication (dnaA, dnaN, dnaB, and perhaps ypvA), cell division (ftsL and pbpB), nucleotide metabolism (nrdI, nrdE, and nrdF), pyrimidine biosynthesis (pyrP), lantibiotic biosynthesis (sunA and sunT), sporulation (sda and spo0J), and several unknown functions (“y” genes). †, sda and the yqeGH-aroD-yqeIJK operon are divergently transcribed, and both respond to inhibition of replication (the yqeG operon decreases ∼2 to 3-fold); *, the region shown for gcp is downstream of its 3′ end; **, gene expression values are listed for ywfO (ywzC did not respond to replication inhibition).
DnaA was also bound at the six clustered sites in exponentially growing wild-type cells (Fig. 1C and 2) and in dnaB(Ts) and dnaD(Ts) cells at the permissive temperature (not shown). However, the enrichment values of ∼2- to 8-fold over background indicated that DnaA binding was significantly lower than that following replication inhibition (∼10- to 70-fold). As described above, binding did not increase in wild-type cells incubated at 47°C (not shown).
The increase in DnaA binding at clustered sites after replication was blocked was not due to an increase in the intracellular concentration of DnaA. The increase in binding occurred despite transcriptional autorepression of dnaA (15, 34), which should decrease DnaA levels in response to replication inhibition. As expected, we found that levels of DnaA as a fraction of total protein had decreased approximately 35 to 50% 60 min after inhibition of replication elongation with HPUra (data not shown). Thus, the increase in DNA binding by DnaA was not due to increased levels of DnaA but rather was likely due to increased activity of DnaA in response to replication inhibition.
To verify that the signal in the ChIP-chip experiments was specific to DnaA, we performed similar experiments in the presence and absence of HPUra in a dnaA null mutant in which DnaA was rendered nonessential by the replacement of oriC with the plasmid replication origin oriN (2, 15, 19, 22, 32). There was no detectable enrichment of the six clustered loci in the absence of DnaA (Fig. 1E and 2). There was enrichment detected from yutF in both dnaA+ and dnaA null cells (Fig. 1E), indicating that this signal is not due to specific immunoprecipitation of DnaA and is perhaps due to a cross-reacting protein. The enrichment value appeared to be higher for dnaA null cells, probably due to the reduced background signal throughout the chromosome caused by the absence of DnaA and the subsequent lack of nonspecific binding throughout the genome. We excluded yutF from further analyses and discussion.
In addition to the six strong DnaA binding regions identified above, the ChIP-chip data indicated that there were other regions that were weakly and/or inconsistently enriched. Comparison of ChIP-chip data for three conditions, namely, exponentially growing cells, cells after inhibition of replication initiation, and cells after inhibition of replication elongation, indicated that only two additional regions had enrichment levels of >2-fold (relative to both the genomic background and the ChIP-chip data from the dnaA null mutant) under at least two of the three conditions. One region, near yrhC, is in the middle of an operon, and its transcription did not appear to change in response to replication stress; this region was not analyzed further. The other, near spo0J, did respond transcriptionally to replication stress (15), and it is discussed further below. An independent genome-wide analysis of DnaA binding in untreated cells also identified the six clustered sites plus dnaN and thdF (20). We included dnaN in the analysis of dnaA because the two genes constitute an operon and the sites are also part of oriC. thdF did not qualify as significantly enriched in our analysis due to variability, although it was enriched 2.05-fold in arrested dnaB(Ts) cells. It is possible that our microarray probe for thdF was poorly placed to detect bound DnaA.
Analysis by ChIP-PCR of DnaA binding to regions affected by inhibition of replication.
Several chromosomal regions previously found to be enriched in DnaA immunoprecipitates in ChIP-PCR experiments (15) were not identified in our ChIP-chip experiments or in ChAP-chip experiments (20). This discrepancy indicates that ChIP-PCR experiments are probably more sensitive than ChIP-chip or ChAP-chip experiments. Previously, we observed a difference in sensitivity between ChIP-PCR and ChIP-chip assays for the DNA binding protein Spo0J (4). If the difference in sensitivity between ChIP-chip and ChIP-PCR assays exists for DnaA, as we expect, then there are likely to be chromosomal regions bound by DnaA that were not detected by the ChIP-chip and ChAP-chip approaches.
Using ChIP-PCR, we tested for DnaA binding at almost all of the operons previously postulated to be regulated by DnaA in response to inhibition of replication (15) for which ChIP-chip results were negative. mRNA levels from these operons change in response to inhibition of replication, in a RecA-independent manner, and the regulatory regions have at least two matches to the DnaA box consensus (allowing for one mismatch per box) within 500 bp of the start codon of the first gene (15). We found that 9 of the 14 loci tested had statistically significant enrichment in the DnaA immunoprecipitates during replication and/or after inhibition of replication elongation (Table 2). These regions include citZ, lysC, yurY, yclN, dnaB, ypvA, kdgR, and ykuN (Table 2). Additionally, spo0J, which responds to perturbation of replication stress but has only one DnaA box within 500 bp of the beginning of its operon (15), was found to bind DnaA in the ChIP-chip experiments (Table 2; see above). The coding sequence of the spo0J operon contains three DnaA boxes.
TABLE 2.
ChIP-PCR analysis of DnaA binding at putative regulatory targetsa
| Locusb | No HPUra treatment
|
HPUra treatment
|
Overall P value | ||
|---|---|---|---|---|---|
| Mean enrichment ± SE | P value | Mean enrichment ± SE | P value | ||
| citZ | 1.3 ± 0.8 | 0.72 | 2.5 ± 0.4 | 0.03 | 0.40 |
| dnaB | 26.4 ± 9.2 | 0.01 | 0.9 ± 0.4 | 0.72 | 0.002 |
| flgB | 2.0 ± 1.1 | 0.32 | 1.0 ± 0.3 | 0.97 | 0.31 |
| kdgR | 1.8 ± 0.1 | 0.01 | 3.3 ± 1.6 | 0.17 | 0.34 |
| lysC | 1.7 ± 0.4 | 0.16 | 4.7 ± 1.1 | 0.02 | 0.05 |
| nrdI (nrdEF ymaB) | 1.8 ± 0.3 | 0.07 | 2.2 ± 1.1 | 0.18 | 0.69 |
| pyrP | 2.6 ± 0.6 | 0.04 | 3.5 ± 2.4 | 0.13 | 0.63 |
| spo0Jc (soj) | 3.9 ± 0.4 | 0.001 | 6.3 ± 1.7 | 0.02 | 0.62 |
| sunA (sunT) | 4.4 ± 1.7 | 0.06 | 8.6 ± 4.8 | 0.08 | 0.42 |
| yclN (yclOPQ) | 5.4 ± 1.6 | 0.03 | 6.8 ± 2.1 | 0.03 | 0.62 |
| ykuN (ykuOP) | 5.4 ± 1.2 | 0.02 | 4.8 ± 3.3 | 0.10 | 0.85 |
| yllB (ylxA ftsL pbpB) | 1.8 ± 0.5 | 0.18 | 1.7 ± 0.9 | 0.35 | 0.91 |
| ypvA | 3.1 ± 0.7 | 0.05 | 2.7 ± 0.3 | 0.02 | 0.65 |
| yurY (yurX csd yurVU) | 21.7 ± 5.2 | 0.01 | 5.2 ± 1.1 | 0.02 | 0.01 |
| ywzC (ywfO ywgA) | 4.2 ± 1.7 | 0.05 | 3.2 ± 1.6 | 0.12 | 0.62 |
Wild-type cells (AG174) left untreated or treated with HPUra for 60 min were harvested and analyzed by ChIP-PCR for DnaA binding at the indicated loci. Values presented are the mean enrichments ± standard errors for at least three independent biological replicates, normalized to the yabM locus, which is not specifically bound by DnaA. P values were determined by t test. The overall P value compares results with and without HPUra treatment.
Operons that respond transcriptionally to replication stress and have at least two potential DnaA binding sites were chosen for analysis. Loci are sorted alphabetically by the first gene. Additional genes in a given operon that are also affected by replication stress are indicated in parentheses.
Data for spo0J are taken from the ChIP-chip results (Fig. 1).
DnaA binding changed notably at only three of these nine regions following inhibition of replication elongation. The lysC region was reproducibly increased in enrichment after inhibition of replication elongation (Table 2). In contrast, the enrichment in the dnaB and yurY regions significantly decreased after inhibition of replication elongation and was quite high during ongoing replication. This decrease might indicate modulation of DnaA by factors that do not affect binding at other regions. Alternatively, it might indicate different binding specificities for the two different nucleotide-bound forms of DnaA, DnaA-ATP and DnaA-ADP.
In contrast to the chromosomal regions with clustered DnaA boxes that had significant increases in DnaA binding after inhibition of replication, most of these regions (Table 2) did not consistently show significant changes in levels of DnaA binding following treatment with HPUra. However, given the uncertainty in the data, we cannot distinguish whether DnaA binding is relatively constant regardless of replication status or whether increases occur that are not detectable with this approach.
The ChIP-PCR results for the dnaB and yurY regions differed from the ChIP-chip results. Enrichment in these regions was approximately 20-fold in exponentially growing cells in the ChIP-PCR experiments, but neither was found to be significantly enriched in the ChIP-chip experiments: DnaA enrichment at dnaB was generally ∼2-fold, but it was not statistically significant due to variability, and no enrichment was detected at yurY. It is possible that these loci had unusually large differences in sensitivity between the two techniques for reasons such as the locations of the probes relative to DnaA binding sites, as the ChIP-PCR primers targeted the upstream region containing binding sites and the array probes corresponded to open reading frames. The hybridization intensity for the dnaB spot on the microarrays was also below average and not of high quality, perhaps contributing to the unreliability of the data.
Five of the 14 loci tested gave inconclusive results. They had enrichment levels at least 1.6-fold above background, but the levels were not statistically significant (Table 2).
Rapid changes in DnaA binding upon resumption of replication.
In E. coli, Hda is required for the regulatory inactivation of DnaA during replication elongation (see references 23 and 27 and references therein). It is not known if regulatory inactivation of DnaA is completely distributive or requires passage of a replication fork through chromosomal regions bound by DnaA-ATP. We suspected that the increases in DnaA binding and activity that occurred when replication was blocked in B. subtilis were likely to be reversible. We directly tested binding of DnaA after resumption of replication and found that DnaA was released from DNA and that release occurred in a distributive manner at all regions tested before passage of a replication fork through each region.
We monitored binding of DnaA to five regions with clusters of binding sites—dnaA, yydA, ywlC, ywcI, and sda—using ChIP and quantitative PCR while replication was blocked and when replication resumed. Samples were taken (i) during exponential growth, (ii) after inhibition of replication initiation, and (iii) at various times following release of the replication block during a synchronized round of replication. If replication forks must interact with DnaA bound at the various regions to facilitate release of DnaA, then DnaA binding should decrease at regions closer to the replication origin before decreasing at distal regions. Alternatively, if DnaA bound at these regions is able to exchange rapidly in response to the presence of active replication forks, then DnaA binding should decline similarly at all regions, before the replication fork reaches each region.
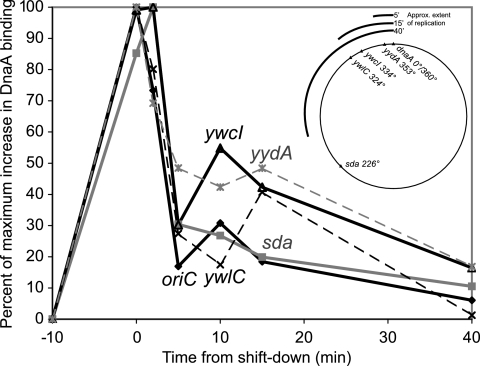
To block replication, we shifted exponentially growing dnaD(Ts) cells to the restrictive temperature and waited for 60 min to allow most ongoing rounds of replication to finish. As expected, enrichment of the five tested regions in the DnaA immunoprecipitates increased 60 min after inhibition of replication initiation (Fig. 4). Replication was allowed to reinitiate from oriC by shifting the dnaD(Ts) cells back to the permissive temperature (32°C). By 5 minutes after the temperature shift, binding of DnaA to the chromosomal regions had already decreased about 50% or more. After 15 min at the permissive temperature, binding of DnaA at each region was near its original level (Fig. 4). The positions of replication forks were estimated based on previous work with synchronized B. subtilis cultures (38). The rate of replication elongation is ∼0.5 kb/s or ∼2.6°/min. A fully synchronous round of bidirectional replication initiating at oriC (0°/360°) would result in fork positions at approximately 13° and 347° at 5 min, 39° and 321° at 15 min, and 104° and 256° at 40 min post-replication initiation (Fig. 4). However, cells do not actually initiate in a fully synchronous manner. Rather, initiation occurs over a period of 5 to 10 min after release. Thus, in the cell population, replication forks are spread over the approximately 15° to 20° trailing the positions given above.
FIG. 4.
Release of DnaA binding after resumption of replication. dnaD(Ts) cells (KPL73) in exponential growth phase were shifted to the restrictive temperature for 60 min and then shifted back to the permissive temperature in order to generate a synchronous round of replication initiation. Samples were taken for ChIP-PCR analysis immediately before and at the end of the incubation at the restrictive temperature and 2, 5, 10, 15, and 40 min after the shift back to the permissive temperature. DnaA binding was analyzed at oriC/dnaA (solid diamonds and black lines), yydA (gray asterisks and gray dashed lines), ywcI (solid triangles and black lines), ywlC (black X characters and black dashed lines), and sda (filled gray squares and solid gray lines). Data were normalized according to the basal and maximal levels of signal detected to facilitate comparison of binding kinetics at sites with different levels of DnaA binding (see, for example, levels at dnaA/oriC and sda in Fig. 2). Data are shown in the vicinities of mntH (A), yloB (B), pksN (C), nrdE (D), sunA (E), yrhC (F), ywpH (G), spo0J (H), and ysmB (I). The map in the upper right shows the position of each analyzed site on the chromosome and the approximate extent of replication 5, 15, and 40 min after release.
The similar behaviors of these five sites irrespective of chromosomal location indicate that direct interaction between the replication machinery and the DnaA-DNA nucleoprotein complex at each site along the chromosome is not required to control the level of DnaA binding to DNA at each region. Rather, it seems likely that DnaA can exchange rapidly in vivo and respond globally to the presence of an active replication fork.
DISCUSSION
We found that DnaA associates with multiple regions of chromosomal DNA in vivo and that association at the regions that have clustered DnaA boxes increases in response to inhibition of replication. We readily detected binding at the clustered regions, and most were detected previously using ChIP-PCR (15) and ChAP-chip assays (20). DnaA association with other regions was more difficult to detect with the genomic approaches (ChIP-chip and ChAP-chip assays) but was detected using ChIP-PCR. We did not detect significant changes in the level of DnaA association with most of these regions in response to perturbations in replication. Clearly, current techniques are limited in the ability to detect many of the DnaA binding sites in vivo. Nonetheless, useful information was obtained from analysis of the available data from in vivo experiments.
DnaA binding sites.
A combination of in vivo and in vitro analyses demonstrated that the DnaA binding site is a somewhat degenerate 9-bp sequence, TT(A/T)TNCACA (23, 31), found in multiple copies in bacterial origins of chromosomal replication. Allowing for one mismatch at any of the nine positions, there are ∼12,000 potential binding sites throughout the B. subtilis genome. Comparative DNA sequence analyses have been used to estimate the conservation, and hence significance, of some of these sequences (9, 15, 20). In addition, studies of the physical association of DnaA with chromosomal DNA revealed binding to several regions that contain multiple potential binding sites (15, 20). The set of DnaA-bound sites that we observed by ChIP-chip assay and ChIP-PCR very clearly fell into two groups, distinguished on both the functional and sequence levels (Fig. 3).
Clustered DnaA binding sites.
Regions with clustered DnaA binding sites, which had more than nine potential DnaA binding sites with at most one mismatch, behaved differently with respect to DnaA binding than did regions with only a few potential DnaA binding sites. Enrichment of the regions with clustered DnaA binding sites increased dramatically in response to inhibition of replication. Increased enrichment of these regions with clusters of sites indicates that either the number of bound DnaA molecules increased or the cross-linking efficiency of individual molecules increased, perhaps due to a conformational change. These possibilities are not mutually exclusive. DnaA appears to form a polymeric nucleoprotein filament (12), and it is quite possible that this type of structure allows for more efficient binding and/or cross-linking to DNA. Such filamentation was originally proposed for DnaA at oriC, but it is reasonable to extend this notion to other chromosomal regions with clusters of DnaA binding sites analogous to the number of sites in oriC. (Other than oriC, the regions with clustered sites do not function as replication origins.) Both increased binding and a conformational change could be affected by the nucleotide-bound state of DnaA (see below), and changes in binding and conformation are not mutually exclusive; we suspect that both occur.
Nonclustered DnaA binding sites.
The other targets of DnaA that we analyzed have fewer potential binding sites, with fewer than nine in a 500-bp region (Fig. 3). We focused on those regions upstream of genes whose expression changes in response to replication stress (15). In most of these regions, there was little or no detectable change in DnaA binding following inhibition of replication. Because the levels of DnaA binding at these regions were generally low relative to the detection limit of the technique, we cannot conclude that the level of DnaA binding did not change. A change of around 50% or less would be very difficult to detect by ChIP because of the variability between replicates but potentially could cause a significant change in gene expression. If the amount of binding does not actually change at these regions, then another mechanism must be involved in controlling the genes downstream of these regulatory regions. For example, the nucleotide-bound state of DnaA could change and affect gene expression, perhaps via interactions with the transcriptional machinery or alterations in the DNA. Alternatively, there could be other regulatory proteins involved in expression of some of the target operons.
The 9-bp DnaA binding site consensus sequence is quite common when one mismatch is allowed (11,983 occurrences in the genome), and thus there are many potential binding sites throughout the B. subtilis genome. A 500-bp region has a 5.5% chance (Poisson statistics) of containing at least four matches. In contrast, the chance of a cluster of sites with nine or more boxes in a 500-bp region is 0.0018%. Thus, throughout the genome, there are many collections of potential binding sites that are similar in sequence and number to those in the lower part of Fig. 3. Given the limited sensitivity of the ChAP-chip (20) and ChIP-chip data, it is likely that some (perhaps many) of these sites are bound by DnaA in vivo. For many transcription factors, degenerate binding sequences are more common than demonstrable regulatory activity (e.g., see references 11, 17, and 28), and DnaA is no different. If these sites are in fact bound by DnaA and serve any function, then it is possible that regulation of expression of nearby genes occurs under specific conditions that have not been tested. Alternatively, these sites may play a more general role in some aspect of DnaA function, such as titrating the levels of DnaA available to bind sites in regulatory regions that directly influence gene expression or replication.
Nucleotide binding and regulation of DnaA.
DnaA is a member of the AAA+ family of ATPases (25), and based on work with E. coli DnaA, changes in the nucleotide binding state of DnaA serve as a regulatory switch; the ATP-bound form is fully active, and the ADP-bound form is not, although it binds DNA (23, 37, 40). Applying this model to transcriptional regulatory sites leads to the notion that inhibition of replication causes the accumulation of DnaA in its active, ATP-bound form, thereby causing an increase in binding and/or a conformational change resulting in changes in transcription of target operons.
When replication resumes, the association of DnaA with chromosomal regions with clusters of sites rapidly returns to normal levels, well before a replication fork has had time to traverse each of the clustered binding regions. These findings indicate that there is a diffusible signal that controls the activity of DnaA in response to ongoing replication and/or that DnaA rapidly associates with and dissociates from DNA in vivo. The off rate of DnaA dissociation from its binding site in vitro is a few minutes (35), consistent with the possibility of this type of regulation. For example, if the nucleotide-bound state of DnaA is affected by passage of a replication fork through sites to which DnaA is bound, then there would be a local conversion of DnaA-ATP to DnaA-ADP. Rapid association and dissociation in vivo could allow for such local changes in DnaA to quickly equilibrate with the pool of DnaA throughout the cell and affect sites far from the replication fork. We are not aware of any measurements of the in vivo exchange of DnaA on and off DNA.
Whereas much is known for E. coli about regulation of the activity of DnaA at oriC and the regulatory factors involved (see references 23 and 40 and references therein), many of these factors are not present in B. subtilis and other gram-positive organisms or in many gram-negative organisms (40). Our findings indicate that the association of DnaA with some of its target sites significantly changes in response to replication stress in B. subtilis. One of the present challenges is to determine how the activity of DnaA is modulated in response to replication stress.
Acknowledgments
This work was supported by NIH grant GM41934 to A.D.G. and NIH Kirschstein NRSA fellowship 5 F32 G-076950 to A.M.B.
We thank Stephen P. Bell, Catherine A. Lee, Wiep Klaas Smits, Richard B. Weart, and Lyle A. Simmons for critical readings of the manuscript, Alexi I. Goranov and Melanie M. Berkmen for helpful conversations, and Hannah Blitzblau for technical advice.
Footnotes
Published ahead of print on 14 November 2008.
REFERENCES
- 1.Atlung, T., E. S. Clausen, and F. G. Hansen. 1985. Autoregulation of the dnaA gene of Escherichia coli K12. Mol. Gen. Genet. 200442-450. [DOI] [PubMed] [Google Scholar]
- 2.Berkmen, M. B., and A. D. Grossman. 2007. Subcellular positioning of the origin region of the Bacillus subtilis chromosome is independent of sequences within oriC, the site of replication initiation, and the replication initiator DnaA. Mol. Microbiol. 63150-165. [DOI] [PubMed] [Google Scholar]
- 3.Braun, R. E., K. O'Day, and A. Wright. 1985. Autoregulation of the DNA replication gene dnaA in E. coli K-12. Cell 40159-169. [DOI] [PubMed] [Google Scholar]
- 4.Breier, A. M., and A. D. Grossman. 2007. Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol. Microbiol. 64703-718. [DOI] [PubMed] [Google Scholar]
- 5.Britton, R. A., P. Eichenberger, J. E. Gonzalez-Pastor, P. Fawcett, R. Monson, R. Losick, and A. D. Grossman. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 1844881-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, N. C. 1970. 6-(p-Hydroxyphenylazo)-uracil: a selective inhibitor of host DNA replication in phage-infected Bacillus subtilis. Proc. Natl. Acad. Sci. USA 671454-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruand, C., A. Sorokin, P. Serror, and S. D. Ehrlich. 1995. Nucleotide sequence of the Bacillus subtilis dnaD gene. Microbiology 141321-322. [DOI] [PubMed] [Google Scholar]
- 8.Bruand, C., M. Velten, S. McGovern, S. Marsin, C. Serena, S. D. Ehrlich, and P. Polard. 2005. Functional interplay between the Bacillus subtilis DnaD and DnaB proteins essential for initiation and re-initiation of DNA replication. Mol. Microbiol. 551138-1150. [DOI] [PubMed] [Google Scholar]
- 9.Burkholder, W. F., I. Kurtser, and A. D. Grossman. 2001. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell 104269-279. [DOI] [PubMed] [Google Scholar]
- 10.Callister, H., S. Le Mesurier, and R. G. Wake. 1977. Initiation of deoxyribonucleic acid replication in germinating spores of Bacillus subtilis 168 carrying the dnaB(Ts)134 mutation. J. Bacteriol. 1301030-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Comella, N., and A. D. Grossman. 2005. Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis. Mol. Microbiol. 571159-1174. [DOI] [PubMed] [Google Scholar]
- 12.Erzberger, J. P., M. L. Mott, and J. M. Berger. 2006. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 13676-683. [DOI] [PubMed] [Google Scholar]
- 13.Friedberg, E., G. Walker, W. Siede, R. Wood, R. Schultz, and T. Ellenberger. 2005. DNA repair and mutagenesis, 2nd ed. American Society for Microbiology, Washington, DC.
- 14.Gon, S., J. E. Camara, H. K. Klungsoyr, E. Crooke, K. Skarstad, and J. Beckwith. 2006. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 251137-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goranov, A. I., L. Katz, A. M. Breier, C. B. Burge, and A. D. Grossman. 2005. A transcriptional response to replication status mediated by the conserved bacterial replication protein DnaA. Proc. Natl. Acad. Sci. USA 10212932-12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goranov, A. I., E. Kuester-Schoeck, J. D. Wang, and A. D. Grossman. 2006. Characterization of the global transcriptional responses to different types of DNA damage and disruption of replication in Bacillus subtilis. J. Bacteriol. 1885595-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamoen, L. W., W. K. Smits, A. de Jong, S. Holsappel, and O. P. Kuipers. 2002. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 305517-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harwood, C. R., and S. M. Cutting. 1990. Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, England.
- 19.Hassan, A. K., S. Moriya, M. Ogura, T. Tanaka, F. Kawamura, and N. Ogasawara. 1997. Suppression of initiation defects of chromosome replication in Bacillus subtilis dnaA and oriC-deleted mutants by integration of a plasmid replicon into the chromosomes. J. Bacteriol. 1792494-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishikawa, S., Y. Ogura, M. Yoshimura, H. Okumura, E. Cho, Y. Kawai, K. Kurokawa, T. Oshima, and N. Ogasawara. 2007. Distribution of stable DnaA-binding sites on the Bacillus subtilis genome detected using a modified ChIP-chip method. DNA Res. 14155-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaacks, K. J., J. Healy, R. Losick, and A. D. Grossman. 1989. Identification and characterization of genes controlled by the sporulation-regulatory gene spo0H in Bacillus subtilis. J. Bacteriol. 1714121-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadoya, R., A. K. Hassan, Y. Kasahara, N. Ogasawara, and S. Moriya. 2002. Two separate DNA sequences within oriC participate in accurate chromosome segregation in Bacillus subtilis. Mol. Microbiol. 4573-87. [DOI] [PubMed] [Google Scholar]
- 23.Kaguni, J. M. 2006. DnaA: controlling the initiation of bacterial DNA replication and more. Annu. Rev. Microbiol. 60351-375. [DOI] [PubMed] [Google Scholar]
- 24.Karamata, D., and J. D. Gross. 1970. Isolation and genetic analysis of temperature-sensitive mutants of B. subtilis defective in DNA synthesis. Mol. Gen. Genet. 108277-287. [DOI] [PubMed] [Google Scholar]
- 25.Katayama, T. 2008. Roles for the AAA+ motifs of DnaA in the initiation of DNA replication. Biochem. Soc. Trans. 3678-82. [DOI] [PubMed] [Google Scholar]
- 26.Kucherer, C., H. Lother, R. Kolling, M. A. Schauzu, and W. Messer. 1986. Regulation of transcription of the chromosomal dnaA gene of Escherichia coli. Mol. Gen. Genet. 205115-121. [DOI] [PubMed] [Google Scholar]
- 27.Kurokawa, K., S. Nishida, A. Emoto, K. Sekimizu, and T. Katayama. 1999. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 186642-6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazazzera, B., T. Palmer, J. Quisel, and A. D. Grossman. 1999. Cell density control of gene expression and development in Bacillus subtilis, p. 27-46. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, DC.
- 29.Lemon, K. P., I. Kurtser, J. Wu, and A. D. Grossman. 2000. Control of initiation of sporulation by replication initiation genes in Bacillus subtilis. J. Bacteriol. 1822989-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mendelson, N. H., and J. D. Gross. 1967. Characterization of a temperature-sensitive mutant of Bacillus subtilis defective in deoxyribonucleic acid replication. J. Bacteriol. 941603-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Messer, W. 2002. The bacterial replication initiator DnaA. DnaA and oriC, the bacterial mode to initiate DNA replication. FEMS Microbiol. Rev. 26355-374. [DOI] [PubMed] [Google Scholar]
- 32.Moriya, S., A. K. Hassan, R. Kadoya, and N. Ogasawara. 1997. Mechanism of anucleate cell production in the oriC-deleted mutants of Bacillus subtilis. DNA Res. 4115-126. [DOI] [PubMed] [Google Scholar]
- 33.Mott, M. L., and J. M. Berger. 2007. DNA replication initiation: mechanisms and regulation in bacteria. Nat. Rev. Microbiol. 5343-354. [DOI] [PubMed] [Google Scholar]
- 34.Ogura, Y., Y. Imai, N. Ogasawara, and S. Moriya. 2001. Autoregulation of the dnaA-dnaN operon and effects of DnaA protein levels on replication initiation in Bacillus subtilis. J. Bacteriol. 1833833-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaper, S., and W. Messer. 1995. Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem. 27017622-17626. [DOI] [PubMed] [Google Scholar]
- 36.Sekimizu, K., D. Bramhill, and A. Kornberg. 1987. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell 50259-265. [DOI] [PubMed] [Google Scholar]
- 37.Speck, C., C. Weigel, and W. Messer. 1999. ATP- and ADP-dnaA protein, a molecular switch in gene regulation. EMBO J. 186169-6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, J. D., G. M. Sanders, and A. D. Grossman. 2007. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128865-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, Q. P., and J. M. Kaguni. 1987. Transcriptional repression of the dnaA gene of Escherichia coli by dnaA protein. Mol. Gen. Genet. 209518-525. [DOI] [PubMed] [Google Scholar]
- 40.Zakrzewska-Czerwinska, J., D. Jakimowicz, A. Zawilak-Pawlik, and W. Messer. 2007. Regulation of the initiation of chromosomal replication in bacteria. FEMS Microbiol. Rev. 31378-387. [DOI] [PubMed] [Google Scholar]