Abstract
Gram-negative bacteria assemble functional amyloid surface fibers called curli. CsgB nucleates the major curli subunit protein, CsgA, into a self-propagating amyloid fiber on the cell surface. The CsgG lipoprotein is sufficient for curlin transport across the outer membrane and is hypothesized to be the central molecule of the curli fiber secretion and assembly complex. We tested the hypothesis that the curli secretion protein, CsgG, was restricted to certain areas of the cell to promote the interaction of CsgA and CsgB during curli assembly. Here, electron microscopic analysis of curli-producing strains showed that relatively few cells in the population contacted curli fibers and that curli emanated from spatially discrete points on the cell surface. Microscopic analysis revealed that CsgG was surface exposed and spatially clustered around curli fibers. CsgG localization to the outer membrane and exposure of the surface domain were not dependent on any other csg-encoded protein, but the clustering of CsgG required the csg-encoded proteins CsgE, CsgF, CsgA, and CsgB. CsgG formed stable oligomers in all the csg mutant strains, but these oligomers were distinct from the CsgG complexes assembled in wild-type cells. Finally, we found that efficient fiber assembly was required for the spatial clustering of CsgG. These results suggest a new model where curli fiber formation is spatially coordinated with the CsgG assembly apparatus.
Escherichia coli and other enteric bacteria assemble highly aggregative fibers on their cell surfaces called curli (1, 9, 14, 24). Curli fibers are critical determinants of attachment during biofilm formation, and they are potent inducers of the host immune response (3, 18, 25, 26). Curli are structurally and biochemically similar to amyloid fibers (8). Amyloid fibers are most readily associated with neurodegenerative ailments such as Alzheimer's disease, yet some cells are able to assemble functional amyloids without eliciting cytotoxicity. Functional amyloids have been identified in organisms ranging from bacteria to mammals, and they fulfill many diverse physiological functions (12).
Curli biogenesis requires the coordinated efforts of proteins encoded by two divergently transcribed operons. csgBAC encode the structural subunits of the fiber, CsgA and CsgB. A third gene in the operon, csgC, has no reported role in E. coli curli biosynthesis, but the csgC homolog in Salmonella enterica serovar Enteritidis may be important for curli ultrastructural properties (13). csgDEFG encode proteins necessary for the production, secretion, and assembly of CsgA and CsgB. CsgD is a transcriptional activator of the csgBAC operon. CsgG is an outer membrane-localized lipoprotein that is required for the secretion of CsgA and CsgB to the cell surface (28). CsgE and CsgF can interact with CsgG at the outer membrane and modulate the stability of CsgA and CsgB, and they are required for efficient curli assembly (8, 28). However, the precise molecular action of CsgE and CsgF remains unclear.
One model of curli assembly is the nucleation-precipitation pathway, which begins with the CsgG-mediated export of preamyloid CsgA and CsgB to the extracellular milieu (2, 21, 28). At the cell surface, CsgB presents an amyloid template to CsgA and “nucleates” soluble CsgA into insoluble amyloid curli fibers (4, 15, 16). Remarkably, CsgA and CsgB do not have to be expressed from the same cell in order for curli to be assembled. Some CsgA expressed by a “donor” cell can polymerize when it contacts a CsgB-expressing “acceptor” cell in a process termed interbacterial complementation (15). In vitro polymerization experiments with purified CsgA and CsgB further suggest that physical contacts between these proteins drive efficient polymerization (15, 16, 31).
We investigated the possibility that the curli secretion protein, CsgG, was restricted to certain areas of the cell to promote the interaction of CsgA and CsgB during curli assembly. Studies with many bacterial systems have revealed a nonuniform distribution of proteins involved in chemotaxis, cell division, and secretion (5, 6, 22). Spatial restriction, rather than random diffusion, may facilitate the protein-protein interactions required to achieve many cellular processes. For example, the enteropathogenic E. coli bundle-forming pilus secretion apparatus is polarly localized (19). Also, the Myxococcus type IV pilus secretin, PilQ, is found exclusively at the poles (23). The ExPortal system in Streptococcus pyogenes, which serves as the major site of protein secretion, is also spatially localized to discrete regions of the cell. Coupled to the ExPortal are chaperone-like proteins that facilitate folding of newly secreted proteins, suggesting spatial and temporal coordination of protein secretion and folding (7, 29). We found that CsgG is organized into foci in curli-producing cells and that this organization requires efficient fiber polymerization. We also found that CsgG contains a domain that is exposed to the cell surface and that it forms a heat- and sodium dodecyl sulfate (SDS)-resistant complex in the outer membrane. Finally, we show that other CsgG-interacting proteins are required for the spatial restriction of CsgG, which provides the first molecular evidence of how CsgG may be modulated by other csg-encoded proteins.
MATERIALS AND METHODS
Plasmids, strains, and growth conditions.
E. coli strain MC4100 (and its derivations listed in Table 1) was grown on YESCA agar (10 g Casamino Acids, 1 g yeast extract, and 20 g agar per 1 liter) at 26°C for 36 to 48 h to induce curli expression (14). YESCA agar with 10 μg/ml Congo red was used to monitor curli production. CsgG and CsgG-His cloned behind the trc promoter in pTRC99A (plasmids pMC1 and pMC2, respectively, as described in Table 1) were used to overexpress CsgG in cells grown on YESCA plates. Ampicillin was used at 100 μg/ml when appropriate.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source |
|---|---|---|
| Strains | ||
| MC4100 | F−araD139 Δ(argF-lac)U169 rspL150(strR) relA1 flbB5301 deoC1 ptsF25 rbsR | 7a |
| LSR1 | MC4100 csgG::Tn105 | 28 |
| LSR10 | MC4100::ΔcsgA | 8 |
| MHR261 | MC4100::ΔcsgB | 15 |
| MHR592 | MC4100::ΔcsgF | 8 |
| MHR480 | MC4100::ΔcsgE | 8 |
| LSR11 | MC4100::ΔcsgBACcsgDEFG | 28 |
| Plasmids | ||
| pTrc99A | Expression vector | Pharmacia Biotech |
| pMC1 | csgG cloned into pTrc99A | 8 |
| pMC2 | csgG-his6 cloned into pTrc99A | 28 |
| pHL3 | Expression vector | 8 |
| pCsgA | csgA-his6 cloned into pHL3 | 32 |
| pCsgA-ΔR1 | csgA-ΔR1-his6 cloned into pHL3 | 32 |
IFM.
CsgG was visualized by indirect immunofluorescence microscopy (IFM) using a previously described protocol, with some modifications (6). Briefly, cells grown on YESCA plates were suspended in 1× phosphate-buffered saline (PBS) (pH 7.4), and fixed with 2.5% formaldehyde and 0.04% glutaraldehyde, and washed three times in 1× PBS (pH 7.4). Next, fixed cells were incubated with 3% bovine serum albumin (BSA)-1× PBS (pH 7.4) at 37°C for 30 min. Fixed and blocked cells were incubated at 37°C for 1 h with rabbit polyclonal antibodies diluted with 3% BSA-1× PBS (pH 7.4) at the following dilutions: 1/1,000 for anti-CsgG (28), 1/1,000 for anti-CsgA (8), and 1/500 for anti-CsgB (16). Cells were next washed twice with 1× PBS plus 0.1% Tween 20 (pH 7.4), then incubated at 37°C for 30 min with goat anti-rabbit immunoglobulin G (IgG) conjugated to Alexa488 (Molecular Probes, OR) diluted 1/400 in 3% BSA-1× PBS (pH 7.4), and then washed twice in 1× PBS plus 0.1% Tween 20 (pH 7.4). Cells were suspended with 2 mg/ml 4′,6-diamidino-2-phenylindole (DAPI) to stain cellular DNA before being spotted onto a glass slide and viewed with an Olympus BX51 fluorescence microscope. Images of the Alexa488 epifluorescence signal were captured with the following parameters: gain of 80 and offset of 24 for all strains observed and exposures of 500 ms when using anti-CsgG or anti-CsgB antibodies and 250 ms when using anti-CsgA antibodies. The DAPI signal was captured with a 250-ms exposure at the same gain and offset for all strains and conditions. The two images from each field of view were superimposed using Adobe Photoshop software. The number of cells with an Alexa488 signal that colocalized with the DAPI signal was determined. The mean percentage from three independent experiments ± standard error was determined.
Electron microscopy (EM) and immunogold labeling.
Equal numbers of cells grown on YESCA plates at 26°C for 40 h were suspended in 1× PBS (pH 7.4). A 20-μl drop of cell suspension was placed on a piece of Parafilm, and a 200-mesh Formvar-coated grid (Ernest F. Fullam, Inc., NY) was floated on the drop for 2 min. The grid was transferred to a 20-μl drop of 1× PBS and left for an additional 2 min before being stained on a drop of freshly prepared 0.5% uranyl acetate (8). Bacteria were observed using a Phillips CM12 scanning transmission electron microscope. Immunostaining of samples was performed essentially as previously described, with some modifications (30). Briefly, grids with cells bound as described above were subjected to sequential incubation with (20 μl of each solution at room temperature [RT] unless noted otherwise) 1% BSA-1× PBS (5 min); one of the antibody solutions (as indicated in Results) 1/500 anti-CsgG-0.1% BSA-1× PBS, 1/200 anti-CsgA-0.1% BSA-1× PBS, or 1/200 anti-CsgB-0.1% BSA-1× PBS (60 min at 37°C); 0.1% BSA-1× PBS (three times, 2 min each wash); 1/15 anti-rabbit IgG-10-nm gold particles (Sigma, MO)-1% BSA-1× PBS (30 min at 37°C); and 0.1% BSA-1× PBS (three times, 2 min each wash). Preparations were then fixed with 1% glutaraldehyde-1× PBS (5 min) and washed twice with sterile filtered water (5 min each wash). Grids were stained with 0.5% uranyl acetate and viewed with a transmission electron microscope.
Cell fractionation, gel electrophoresis, and immunoblotting.
Cell-free suspensions (CFS) were generated by passing cells twice through a French press at 14,000 lb/in2, followed by centrifugation at 3,000 × g for 15 min to remove unbroken cells. Proteins in the CFS were solubilized with 0.5% Elugent (Calbiochem, Darmstadt, Germany). Cells were also fractionated by detergent extraction into soluble and inner and outer membrane fractions exactly as previously described (28). Elugent-soluble proteins from the Sarkosyl-insoluble outer membrane fraction or the CFS were suspended in loading buffer containing 1.5% SDS and resolved by 5% stacking and 13% or 8% resolving discontinuous SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Blots were probed with anti-CsgG antibodies diluted 1/100,000 in 1% milk-1% BSA plus 1× Tris-buffered saline-Tween 20 (TBST) as described previously (28). Antibodies were removed from blots and reprobed for different antigens by the method described previously (20).
Intact cell dot blotting.
Cells were collected from YESCA plates, suspended in 20 mM Tris-HCl (pH 8), and left unperturbed or sonicated for 30 s before being spotted unto a nitrocellulose membrane. Blots were air dried for 15 min before blocking with 1% milk-1% BSA-1× TBST on a shaker for 2 h at RT or overnight at 4°C. Blots were probed with anti-CsgG antibody diluted 1/100,000 in 1% milk-1% BSA-1× TBST (28), anti-DsbA antibody diluted 1/3,500 in 3% milk-1× TBST (a gift of J. Bardwell), anti-CsgA antibody diluted 1/5,000 in 1% milk-1% BSA-1× TBST (8), anti-CsgB antibody diluted 1/500 in 1% milk-1% BSA-1× TBST (16), or commercially available antihexahistidine antibodies (Covance, NJ) diluted 1/5,000 in 1% milk-1% BSA-1× TBST for 1 h at RT and washed three times (5 min each) in 1× TBST. Next, blots were probed with goat anti-rabbit antibodies conjugated to horseradish peroxidase (Sigma, MO) diluted with 1% milk-1% BSA-1× TBST for 1 h at RT and washed three times (5 min each) in 1× TBST.
RESULTS
CsgG is spatially restricted ino clusters on curli-producing cells.
The major curli fiber subunit, CsgA, is secreted as a soluble protein that is nucleated by outer membrane-associated CsgB into a highly ordered and aggregative amyloid fiber (8, 15, 16, 33). Curli fibers have a distinct morphology that can be readily observed by EM; the fibers are 4- to 6-nm-wide unbranched filaments that form densely tangled masses that interconnect cells (8, 24). EM of MC4100 (wild-type [WT]) cells revealed that curli emanated from discrete regions of the cell surface, although the location varied among individual cells (Fig. 1). Of 660 cells examined, 39% of cells contacted curli fibers, while 61% lacked fibers entirely. The nonuniform distribution of curli fibers within the population of WT cells led us to investigate whether the distribution of CsgG was likewise restricted to curli-producing cells. First, however, we developed methods to detect CsgG spatial distribution in curli-producing cells.
FIG. 1.
Curli fibers are nonuniformly distributed on curli-producing cells. Negatively stained electron micrographs of MC4100 (WT) cells show the typical asymmetric display of curli fibers or absence of fibers on some cells. Bars, 500 nm.
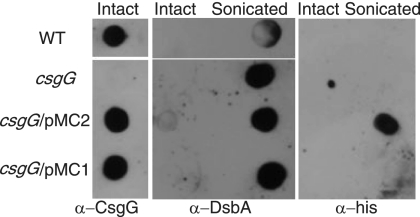
It was proposed that CsgG is localized to the inner leaflet of the outer membrane (21), but the ability of CsgG to support secretion of CsgA into culture supernatants suggested that CsgG might have an extracellular domain (28). Intact-cell immuno-dot blotting was used to test the hypothesis that CsgG was at least partially exposed to the cell surface (Fig. 2). Equal numbers of intact cells were spotted onto a nitrocellulose membrane and probed with anti-CsgG antibodies. Anti-CsgG antibodies reacted with WT intact cells, while no signal was detected in csgG cells. The CsgG signal was restored when the csgG strain was transformed with plasmids encoding CsgG or CsgG-His from pMC1 or pMC2, respectively (Fig. 2, left). Intact cells were also probed with antibodies specific for the periplasmic protein DsbA to assess outer membrane integrity. Anti-DsbA antibodies did not react with intact cells, but sonication of cell samples resulted in detection of anti-DsbA-reactive material after dot blotting (Fig. 2, center). There was no anti-DsbA signal in sonicates of a dsbA mutant strain, JP120 (data not shown). The observation that anti-CsgG antibodies recognized intact cells supported the hypothesis that part of CsgG was exposed to the cell surface. Interestingly, dot blots probed with antihexahistidine antibodies recognized csgG/pMC2 sonicates but not intact cells (Fig. 2, right). Since pMC2 encodes CsgG with a C-terminal hexahistidine epitope, we concluded that the C-terminal region of CsgG was inaccessible to the antibody, possibly due to localization of the C terminus inside the cell.
FIG. 2.
CsgG is exposed to the cell surface. WT cells, csgG cells, and csgG cells containing plasmids encoding CsgG (csgG/pMC1) or CsgG with a C-terminal hexahistidine epitope (csgG/pMC2) were grown on YESCA plates for 48 h at 26°C. Equal numbers of intact or sonicated cells, as indicated, were spotted onto a nitrocellulose membrane before probing with anti-CsgG, anti-DsbA, or antihexahistidine epitope antibodies.
Because CsgG is essential for curlin stability and secretion and since curli assembly requires CsgA and CsgB interaction (21, 28), we predicted that CsgG would be spatially coordinated around the cell so that CsgA and CsgB could efficiently contact each other on the cell surface. Indirect IFM was used to determine if CsgG was localized to distinct regions around the cell. Antibodies directed to CsgG in WT cells revealed distinct immunofluorescent punctate groups (Fig. 3A). We found that approximately 30% of the 1,191 cells examined displayed anti-CsgG-stained foci. WT cells either displayed a punctate signal or lacked a signal altogether, and each cell generally displayed three or fewer foci. Anti-CsgG foci were detected in groups of aggregated bacteria and in isolated bacteria (Fig. 3A). Only about 2.5% of csgG mutant cells displayed anti-CsgG foci (Fig. 3B). In the few csgG cells containing an IFM signal, generally only a single, faint anti-CsgG focus was observed. The punctate anti-CsgG stain was specific for CsgG in WT cells, as omission of the primary antibody dramatically reduced the number of cells with a fluorescent signal (6.2% of 484 cells displayed punctate fluorescent staining [data not shown]).
FIG. 3.
CsgG is spatially restricted in the outer membrane. (A and B) Typical images of cells obtained after anti-CsgG indirect immunofluorescence staining of the WT (A) or the isogenic csgG mutant (B). Shown are merged images of cells probed with rabbit anti-CsgG primary antibody and goat anti-rabbit secondary antibody conjugated to Alexa488 (green) and DAPI-stained nucleoid (blue). Bars, 12.5 μm. Quantification of the immunofluorescent foci is presented in Fig. 4B (see text). (C and D) Typical transmission EM images obtained after indirect immunogold labeling with 10-nm gold particles of WT cells probed with anti-CsgG antibodies (C) or without primary antibody (D), as described in Materials and Methods. Cells were grown on YESCA plates at 26°C for 40 h to induce curli expression. Bars, 200 nm.
We next used immuno-EM (IEM) to determine if CsgG colocalized with curli fibers. Cells were grown under curli-inducing conditions, and equal numbers of cells were probed with anti-CsgG antibodies and anti-rabbit secondary antibodies conjugated to 10-nm gold particles before being negatively stained with uranyl acetate. Every curliated cell of the several hundred observed also had gold particles on its surface (Fig. 3C). A majority of these particles were grouped near the cell location from where curli emanated, while very few particles were observed elsewhere on the surface of the cell (Fig. 3C). In addition, we observed gold particles bound to the fibers themselves, which might be due to nonspecific staining or might indicate that CsgG can be dislodged from the cell surface during curli assembly (Fig. 3C). In 17 cells examined at high magnification, 100 anti-CsgG-directed gold beads were observed randomly distributed over the cell surface, while more than 340 gold beads were localized to the region of the cell from where curli fibers emanated. Cell-associated gold particles were rarely observed when the anti-CsgG antibody was omitted from the preparation (Fig. 3D). WT cells in the population that were not observably producing curli fibers either lacked an anti-CsgG signal entirely or contained fewer gold beads which were not spatially restricted in any obvious way (data not shown). Therefore, we concluded that CsgG was clustered into foci in curli-producing cells.
Surface-exposed CsgG is spatially dispersed in csg mutants.
An intriguing aspect of curli biogenesis is the interdependence of protein stabilities between the csg operon-encoded proteins. For example, deletion of csgG or csgE results in decreased stability and secretion of CsgA and CsgB, and conversely, deletion of csgF results in augmented CsgA secretion and stability (8, 16, 21). Furthermore, overexpression studies showed that CsgG interacts with itself and with CsgA, CsgF, and CsgE at the outer membrane (28), although the molecular consequences of these interactions are unclear. Because several csg mutant phenotypes appear to converge at the step of secretion, we examined the spatial organization of CsgG at the outer membrane in different csg mutant backgrounds (Fig. 4). Each of the csg mutant strains examined displayed a dramatic decrease in the number of cells containing CsgG foci (Fig. 4A); 30.0% of WT cells had punctate anti-CsgG staining, compared to 7.2% of csgE, 3.6% of csgF, 2.1% of csgB, and 2.31% of csgA cells (Fig. 4B). We never observed a significant level of diffuse immunofluorescent signal in any of the WT or mutant cells, leading us to further investigate the cause of the loss of IFM signal. Despite the fewer number of CsgG punctate groups in the csg mutant strains compared to the WT, Western blotting indicated that WT and csg mutant strains had similar amounts of CsgG in the outer membrane, inner membrane, cytoplasm/periplasm, and whole-cell fractions (Fig. 4C and data not shown).
FIG. 4.
Spatial restriction of CsgG requires other csg-encoded proteins. (A) Typical images obtained after anti-CsgG IFM staining of csgE, csgF, csgB, or csgA cells grown on YESCA plates for 40 h at 26°C. Shown are merged images of cells stained with DAPI (blue) and Alexa488 anti-CsgG signal (green), although no cell-associated Alexa488 signal is evident. Bars, 12.5 μm. (B) Quantification of cell-associated foci after IFM staining. The percentage of cells with anti-CsgG immunofluorescence stain was determined for WT (1,191 cells examined), csgG (G−; 528 cells examined), csgE (E−; 553 cells examined), csgF (F−; 567 cells examined), csgB (B−; 549 cells examined), or csgA (A−; 329 cells examined) cells by counting the percentage of DAPI-stained cells colocalized with Alexa488 punctate dots. The standard error of the mean from three independent experiments is indicated above each bar in the graph. (C) WT or csgE cells were grown on YESCA plates for 48 h at 26°C, and cell fractions were prepared as described in Materials and Methods. Equal proportions of fractions containing whole cells (lane 1), high-speed supernatants containing cytosol and periplasm (lane 2), inner and outer membranes (lane 3), Sarkosyl-soluble inner membranes (lane 4), or Sarkosyl-insoluble outer membranes (lane 5) were resolved by SDS-PAGE before Western blotting with anti-CsgG antibodies. Similar results were seen for all csg mutant strains examined (data not shown). (D) WT, csgG, csgE, csgF, csgB, csgA, or csgBACcsgDEFG/pMC1 cells were grown on YESCA plates for 48 h at 26°C before equal an equal number of cells was spotted onto a nitrocellulose membrane and probed with anti-CsgG antibodies, as indicated. (E) Typical transmission EM image obtained after indirect immunogold labeling with 10-nm gold particles of csgA cells probed with anti-CsgG antibodies as described in Materials and Methods. Cells were grown on YESCA plates at 26°C for 40 h to induce expression of csgB and csgDEFG operons. Similar results were obtained with other csg mutant strains tested (see Results). Bar, 200 nm.
Since protein instability or mislocalization did not account for the decrease in CsgG IFM stain that was observed in the csg mutant strains, we tested if CsgG surface accessibility was altered in the csg mutant strains. The polyclonal anti-CsgG antibody reacted with intact cells of each strain tested (Fig. 4D), indicating that CsgG was still surface exposed in csg mutant strains. A slight decrease in anti-CsgG signal was evident in csgE and csgF samples. To test the possibility that the outer membrane prevented the anti-CsgG antibody from reaching CsgG in any of the csg mutant strains, we performed anti-CsgG IFM on permeabilized cells. However, no change in the relative levels of anti-CsgG IFM signal was evident in any strain when membranes were permeabilized with Triton X-100 before probing with antibodies (data not shown). Therefore, the loss of CsgG foci was not due to gross changes in CsgG steady-state levels, mislocalization, or exposure of CsgG to the cell surface. To further test the hypothesis that exposure of CsgG to the cell surface was independent of any csg-encoded protein, we also probed intact csgBACcsgDEFG cells expressing CsgG from the plasmid pMC1 with anti-CsgG antibodies. The anti-CsgG antibody reacted readily with csgBACcsgDEFG/pMC1 cells (Fig. 4D).
Taken together, these results indicated that the CsgG produced in csgA, csgB, csgE, and csgF cells was spatially dispersed in the outer membrane and not localized into punctate clusters. We used anti-CsgG IEM to confirm this notion. We saw two distinct populations of csg mutant cells by IEM: some cells showed many anti-CsgG beads, while many cells had very few anti-CsgG beads or lacked gold beads altogether. A similar pattern was observed for WT cells (Fig. 3). As shown in Fig. 4E, anti-CsgG-directed gold beads, when present, were randomly distributed on the cell surface of csg mutant strains. Therefore, we conclude that csg-encoded proteins were required for spatial restriction of CsgG and that the loss of anti-CsgG IFM signal in the csg mutant strains was due to a more uniform spatial distribution of CsgG on the surface of the cells, which lowered the IFM signal intensity to below background levels.
CsgG multimers are resistant to denaturation with heat and detergent.
Resistance to denaturation is a property of many outer membrane-localized multimeric complexes, and two distinct forms of assembly-dependent electrophoretic mobility have been described for outer membrane proteins (17, 27). We have previously shown that differently tagged CsgG constructs can copurify and coimmunoprecipitate when overexpressed in the same cell, suggesting that CsgG can interact with itself at the outer membrane (28). To examine the possibility that an assembly defect contributed to the loss of spatial clustering of CsgG expressed in the csg mutant strains, we examined the assembly status of CsgG in the various CsgG-expressing strains. WT and isogenic csgE, csgF, csgB, and csgA strains were grown on YESCA plates at 26°C for 48 h, and then CFS were prepared as described in Materials and Methods. Freshly prepared CFS was suspended in SDS sample buffer and incubated for 10 min at various temperatures between RT and 95°C before loading onto a discontinuous 5% to 8% SDS-polyacrylamide gel (Fig. 5). CsgG solubilized from WT cells migrated as a single high-molecular-mass species above the 170-kDa standard in the 8% resolving gel at temperatures of below 55°C (Fig. 5A and B, lanes 1 and 2; see Fig. S1A in the supplemental material). An intermediate-molecular-mass CsgG species that migrated between the 170- and 130-kDa standards was detected when WT samples were heated at temperatures greater than 55°C (Fig. 5A and B, lanes 3; see Fig. S1A in the supplemental material). The 30-kDa CsgG monomer was detected only when the samples were heated to 95°C for 10 min (Fig. 5A and B, lanes 4).
FIG. 5.
CsgG forms a detergent-stable high-molecular-weight multimer. MC4100 (WT) or isogenic csgG, csgE, csgF, csgA, and csgB mutant strains were grown on YESCA plates for 40 h at 26°C, and CFS were generated as described in Materials and Methods. Proteins were solubilized in 0.5% Elugent prior to being resuspended in 1× SDS loading buffer with 2-mercaptoethanol. Samples were incubated for 10 min at RT (lanes 1, 5, 6, and 10), 37°C (lanes 2, 7, and 11), 55°C (lanes 3, 8, and 12), or 95°C (lanes 4, 9, and 13) and electrophoresed by 5% to 8% discontinuous SDS-PAGE. The gel was transferred to a polyvinylidene difluoride membrane and probed with anti-CsgG antibodies.
Surprisingly, CsgG in unheated samples obtained from csgE, csgF, csgB, or csgA strains resolved by SDS-PAGE did not migrate exclusively as a single, high-molecular-mass band, as was detected in WT samples. Rather, various amounts of the intermediate-molecular-mass CsgG species migrating between the 170- and 130-kDa standards were detected in unheated samples from csg mutant cells (Fig. 5A, lanes 6 and 10, and B, lanes 6 and 13). Notably, detection of monomeric CsgG in any strain required that that the samples be heated to 95°C prior to electrophoresis (Fig. 5A, lanes 3, 9, and 13, and B, lanes 4, 9, and 13), suggesting that the intermediate species was an SDS-stable CsgG oligomer. A 45-kDa CsgG species was detected in heated and unheated csgE, csgF, and csgB samples (Fig. 5A, lanes 6 to 13, and B, lanes 10 to 13). We did not observe this band in the WT, csgG, or csgA samples. The 45-kDa band did not react with antibodies raised against CsgA (data not shown), and its identity remains unclear. The loss of the single high-molecular-mass CsgG-containing complex in the csg mutant strains suggests that all of these proteins are either directly or indirectly required for the assembly of the high-molecular-mass CsgG species. Importantly, antibodies raised against CsgA, CsgE, or CsgF did not react with the high- or intermediate-molecular-mass CsgG bands (data not shown).
CsgG expressed from plasmid pMC1 in the csgBACcsgDEFG strain migrated predominately as the intermediate-molecular-mass species between the positions of the 170- and 130-kDa standards when incubated at temperatures of below 95°C, which suggested that an SDS-resistant CsgG species can assemble in the absence of all of the other csg-encoded proteins (see Fig. S1A, lanes 6 to 9, in the supplemental material). We next overexpressed CsgG in each of csg mutant strains to test whether variations in CsgG protein concentration could account for the shift in CsgG electrophoretic mobility in these strains. Overexpression of CsgG in any strain did not increase the amount of the high-molecular-mass CsgG species, although the intermediate-molecular-mass species accumulated (see Fig. S1B in the supplemental material and data not shown), raising the possibility that a stoichiometric relationship between CsgG and other proteins may be required for the stable production of the high-molecular-mass species. The presence of the intermediate-molecular-mass CsgG species in the absence of any or all csg proteins suggests that a CsgG oligomer can assemble independently of other curli biogenesis proteins but that the properties of this oligomer are distinct from those of the CsgG complexes formed in WT cells. These data also suggest that since CsgG expressed in the various csg mutant strains is spatially dispersed, the CsgG complex represented by the intermediate-molecular-mass species observed in the csg mutant strains is likewise dispersed in the outer membrane.
Fiber assembly is required for CsgG clustering.
Since we detected spatially restricted CsgG foci in curli-producing WT cells only, we investigated the relationship between fiber assembly and CsgG spatial restriction. Because CsgG clusters were absent from CsgG-expressing strains deficient in curlin secretion (csgE) or fiber assembly (csgE, csgB, csgA, and csgF), we reasoned that CsgA polymerization into curli fibers was essential for CsgG spatial localization. We examined CsgG clustering by IEM and IFM in csgA cells expressing CsgA or a CsgA mutant that was unable to polymerize into fibers. CsgA-ΔR1 is missing 23 residues that comprise the first of five CsgA amyloidogenic domains and is unable to polymerize into fibers after it is secreted from the cell (32) (Fig. 6B). When csgA cells were transformed with plasmid pCsgA-His expressing WT CsgA, 23% of 579 cells contained anti-CsgG foci by IFM (Fig. 6C, inset), and anti-CsgG-directed gold beads were clustered with curli fibers in csgA/pCsgA cells as observed with IEM (Fig. 6C). In contrast, only 10% of 567 csgA cells expressing CsgA-ΔR1-His displayed anti-CsgG foci and clusters by IFM and IEM (Fig. 6D). Therefore, curli subunit secretion alone is not sufficient to mediate CsgG clustering, and secretion must be accompanied by CsgA polymerization into a fiber for CsgG clusters to form.
FIG. 6.
Fiber polymerization is required for CsgG clustering. (A) Diagram of WT CsgA functional domains showing the N-terminal CsgG secretion signal (blue) and the five amyloid domains, R1 to R5. (B) Diagram of CsgA-ΔR1, which lacks the first of five amyloid domains. (C and D) Typical transmission EM images obtained after indirect immunogold labeling with 10-nm gold particles of csgA cells expressing CsgA (C) or CsgA-ΔR1 (D) probed with anti-CsgG antibodies as described in Materials and Methods. Cells were grown on YESCA plates at 26°C for 40 h to induce expression of csgB and csgDEFG operons. The insets show images obtained after anti-CsgG IFM staining of csgA/pCsgA cells (579 cells examined) or csgA/pCsgA-ΔR1 cells (567 cells examined), as indicated, grown on YESCA plates for 46 h at 26°C. Shown are merged images of cells stained with DAPI (blue) and Alexa488 anti-CsgG signal (green). The percentage of cells examined which contained anti-CsgG foci in each strain is indicated.
DISCUSSION
Curli fiber assembly occurs by a process termed nucleation-precipitation, where soluble CsgA and CsgB interact at the cell surface to form insoluble amyloid fiber aggregates. A key event preceding nucleation-precipitation is the transportation of CsgA and CsgB to, and across, the outer membrane. CsgA and CsgB stability and secretion depend on the outer membrane-localized lipoprotein CsgG (21, 28). Here, we report that CsgG is spatially restricted on the cell surface and that other csg-encoded proteins are required for organization of CsgG around the cell.
We found that CsgG formed SDS-resistant multimers and was clustered into spatially discrete foci that were exposed to the extracellular surface. Furthermore, we observed that curli fibers emanated from spatially discrete regions of WT cells (Fig. 1). Immunogold labeling revealed that CsgG was most abundant at the point(s) of the cell where curli emanated from the surface (Fig. 3). Additionally, we observed some binding of the gold-labeled anti-CsgG antibodies to the fibers themselves, suggesting that CsgG may become dislodged from the membrane during curli assembly. However, fiber-associated CsgG is likely only a minor fraction of the population, since CsgG-His is not recognized by an anti-His antibody added to the outside of cells (Fig. 2). The spatial restriction of CsgG into foci around curli fibers suggests a model where CsgA and CsgB fiber assembly is coordinated with CsgG spatial organization.
What mechanisms may be responsible for the clustering of CsgG around curli fibers? One possibility is that CsgG is spatially restricted prior to CsgA or CsgB secretion. In this scenario, CsgA and CsgB are secreted to the same location of the cell surface, perhaps facilitating the high efficiency of curli assembly. Another possibility is that CsgG is not restricted into foci until after secretion of CsgA and CsgB begins. The postsecretion aggregation of CsgA and CsgB into fibers may itself result in the clustering of CsgG around the fibers. Our data favor the later model. First, we did not observe spatially clustered CsgG foci in CsgG-expressing strains deficient in curlin secretion or curli assembly (Fig. 4). Further, CsgG foci were not evident when we transformed csgA cells with a plasmid encoding CsgA-ΔR1, a CsgA mutant protein that is stable and secreted but is defective in fiber assembly (32) (Fig. 6).
We also found that the loss of CsgE, CsgF, CsgA, or CsgB resulted in loss of spatially clustered CsgG foci, with little change in CsgG surface exposure or targeting to the outer membrane (Fig. 4). Stable CsgG oligomers likely form independently of other csg-encoded proteins, since slow-migrating SDS-resistant CsgG species were detected in all strains expressing CsgG (Fig. 5). However, the CsgG complexes formed in WT cells were slower migrating than the CsgG complexes observed in strains lacking CsgE, CsgF, CsgB, or CsgA (Fig. 5). Overexpression of CsgG in the absence of all of the csg proteins did not increase the relative amount of high-molecular-mass CsgG, as overexpressed CsgG migrated predominately as the intermediate-molecular-mass species (see Fig. S1 in the supplemental material). Therefore, some parameter besides CsgG concentration determines formation of the highest-molecular-mass species. Taken collectively, our data suggest two distinct phases of CsgG assembly and organization: (i) CsgG is targeted to the outer membrane and exposes the surface-accessible domain in the absence of any or all of the other csg-encoded proteins, and (ii) spatial restriction of CsgG into microdomains and assembly of the highest-molecular-mass CsgG complexes requires curli fiber polymerization supported by the other csg-encoded proteins. Since CsgG physically interacts with CsgE, CsgF, and CsgA (28), any or all of these proteins may contribute to CsgG spatial restriction. However, genetic analyses to determine exactly which of these CsgG-interacting proteins is required for either the assembly of CsgG multimers or spatial restriction of CsgG were difficult, as deletion of any single csg-encoded protein results in the loss of multiple other csg-encoded proteins from the cell surface (8, 16).
Previous results indicated that the N-terminal cysteine of CsgG was lipidated and that lipidation was required for the transport of CsgG to the outer membrane (21, 28). We showed that CsgG contained a domain exposed to the cell surface (Fig. 2), and a previous study indicated that CsgG had a periplasmic domain (21). Surface-exposed lipoproteins have been identified in several bacterial species, including Escherichia, Klebsiella, and Neisseria spp. Membrane-spanning lipoprotein translocons are not unprecedented. For example, the lipoprotein Wza is an E. coli polysaccharide translocon that spans the outer membrane (11). CsgG and Wza are topologically similar, as both have periplasmic and surface-exposed domains (Fig. 2) (11, 21), and are also functionally similar, as both have been implicated as conduits for secretion across the outer membrane (10, 28). Importantly, CsgG lacks any apparent amino acid sequence similarity with either Wza or any other family of proteins (data not shown), and the architecture of the CsgG membrane-spanning domain(s) remains unclear. Future exploration of the domain architecture of CsgG and definition of the residues governing the contacts between CsgG and its many interacting proteins will help clarify the biology of this unique lipoprotein.
One outstanding question about the biosynthesis of functional amyloids is how cells control amyloid fiber aggregation without any apparent cellular toxicity. In the curli biogenesis system, the activities of nucleation and polymerization are separated into two different proteins, CsgB and CsgA, respectively. This suggests that the cell can avoid unregulated fiber polymerization by keeping CsgA and CsgB separated until they reach the cell surface. The fiber-dependent spatial clustering of CsgG suggests an elegant mechanism to regulate the segregation of CsgA and CsgB: only when the csg-encoded proteins interact with CsgG do spatial restriction and CsgA and CsgB interaction occur. These results support a model where CsgG is the center of a curli assembly platform, although very little is known about the molecular nature of the protein-protein interactions that facilitate CsgG ultrastructural changes. In particular, the mechanisms preventing CsgA and CsgB amyloid assembly on the periplasmic face of the spatially restricted assembly complex remain to be elucidated. Clarification of how the curli assembly platform forms will help further unravel the mechanism of coordinated curli amyloid biogenesis.
Supplementary Material
Acknowledgments
This work was supported by NIH National Research Service Award T32GMO7544 to E.A.E and by NIH grant RO1A1073847 to M.R.C. The transmission electron microscope used in this work was acquired under grant EAR-87-08276 from the NSF.
We thank James Bardwell and members of his laboratory for the anti-DsbA antibody and for the dsbA deletion strain (JP120) used for Fig. 2. Many thanks are also due to Amy Chang and members of her laboratory for their generous and frequent accommodation of our many hours of use of their Olympus microscope. We acknowledge members of the Chapman lab and the lab of Robert Bender for critical evaluation of the manuscript and many helpful discussions, Xuan Wang for technical advice and assistance with electron microscopy, Yizhou Zhou for help with microscopy, and Ryan Frisch for the preimmune rabbit IgG used as a control in IFM and dot blotting.
Footnotes
Published ahead of print on 14 November 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arnqvist, A., A. Olsen, J. Pfeifer, D. G. Russell, and S. Normark. 1992. The Crl protein activates cryptic genes for curli formation and fibronectin binding in Escherichia coli HB101. Mol. Microbiol. 62443-2452. [DOI] [PubMed] [Google Scholar]
- 2.Barnhart, M. M., and M. R. Chapman. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60131-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bian, Z., A. Brauner, Y. Li, and S. Normark. 2000. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J. Infect. Dis. 181602-612. [DOI] [PubMed] [Google Scholar]
- 4.Bian, Z., and S. Normark. 1997. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. EMBO J. 165827-5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandon, L. D., N. Goehring, A. Janakiraman, A. W. Yan, T. Wu, J. Beckwith, and M. B. Goldberg. 2003. IcsA, a polarly localized autotransporter with an atypical signal peptide, uses the Sec apparatus for secretion, although the Sec apparatus is circumferentially distributed. Mol. Microbiol. 5045-60. [DOI] [PubMed] [Google Scholar]
- 6.Buddelmeijer, N., M. E. Aarsman, A. H. Kolk, M. Vicente, and N. Nanninga. 1998. Localization of cell division protein FtsQ by immunofluorescence microscopy in dividing and nondividing cells of Escherichia coli. J. Bacteriol. 1806107-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campo, N., H. Tjalsma, G. Buist, D. Stepniak, M. Meijer, M. Veenhuis, M. Westermann, J. P. Muller, S. Bron, J. Kok, O. P. Kuipers, and J. D. Jongbloed. 2004. Subcellular sites for bacterial protein export. Mol. Microbiol. 531583-1599. [DOI] [PubMed] [Google Scholar]
- 7a.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. Mol. Microbiol. 104541-555. [DOI] [PubMed] [Google Scholar]
- 8.Chapman, M. R., L. S. Robinson, J. S. Pinkner, R. Roth, J. Heuser, M. Hammar, S. Normark, and S. J. Hultgren. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295851-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collinson, S. K., S. C. Clouthier, J. L. Doran, P. A. Banser, and W. W. Kay. 1996. Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J. Bacteriol. 178662-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, C., K. Beis, J. Nesper, A. L. Brunkan-Lamontagne, B. R. Clarke, C. Whitfield, and J. H. Naismith. 2006. Wza, the translocon for E. coli capsular polysaccharides, defines a new class of membrane protein. Nature 444226-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drummelsmith, J., and C. Whitfield. 2000. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane. EMBO J. 1957-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fowler, D. M., A. V. Koulov, W. E. Balch, and J. W. Kelly. 2007. Functional amyloid—from bacteria to humans. Trends Biochem. Sci. 32217-224. [DOI] [PubMed] [Google Scholar]
- 13.Gibson, D. L., A. P. White, C. M. Rajotte, and W. W. Kay. 2007. AgfC and AgfE facilitate extracellular thin aggregative fimbriae synthesis in Salmonella Enteritidis. Microbiology 1531131-1140. [DOI] [PubMed] [Google Scholar]
- 14.Hammar, M., A. Arnqvist, Z. Bian, A. Olsen, and S. Normark. 1995. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18661-670. [DOI] [PubMed] [Google Scholar]
- 15.Hammar, M., Z. Bian, and S. Normark. 1996. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc. Natl. Acad. Sci. USAU S A 936562-6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammer, N. D., J. C. Schmidt, and M. R. Chapman. 2007. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc. Natl. Acad. Sci. USA 10412494-12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardie, K. R., S. Lory, and A. P. Pugsley. 1996. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 15978-988. [PMC free article] [PubMed] [Google Scholar]
- 18.Herwald, H., M. Morgelin, A. Olsen, M. Rhen, B. Dahlback, W. Muller-Esterl, and L. Bjorck. 1998. Activation of the contact-phase system on bacterial surfaces—a clue to serious complications in infectious diseases. Nat. Med. 4298-302. [DOI] [PubMed] [Google Scholar]
- 19.Hwang, J., D. Bieber, S. W. Ramer, C. Y. Wu, and G. K. Schoolnik. 2003. Structural and topographical studies of the type IV bundle-forming pilus assembly complex of enteropathogenic Escherichia coli. J. Bacteriol. 1856695-6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kayed, R., E. Head, J. L. Thompson, T. M. McIntire, S. C. Milton, C. W. Cotman, and C. G. Glabe. 2003. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 300486-489. [DOI] [PubMed] [Google Scholar]
- 21.Loferer, H., M. Hammar, and S. Normark. 1997. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol. Microbiol. 2611-23. [DOI] [PubMed] [Google Scholar]
- 22.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 2591717-1723. [DOI] [PubMed] [Google Scholar]
- 23.Nudleman, E., D. Wall, and D. Kaiser. 2006. Polar assembly of the type IV pilus secretin in Myxococcus xanthus. Mol. Microbiol. 6016-29. [DOI] [PubMed] [Google Scholar]
- 24.Olsen, A., A. Jonsson, and S. Normark. 1989. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338652-655. [DOI] [PubMed] [Google Scholar]
- 25.Olsen, A., M. J. Wick, M. Morgelin, and L. Bjorck. 1998. Curli, fibrous surface proteins of Escherichia coli, interact with major histocompatibility complex class I molecules. Infect. Immun. 66944-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prigent-Combaret, C., G. Prensier, T. T. Le Thi, O. Vidal, P. Lejeune, and C. Dorel. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2450-464. [DOI] [PubMed] [Google Scholar]
- 27.Reithmeier, R. A., and P. D. Bragg. 1974. Purification and characterization of heat-modifiable protein from the outer membrane of Escherichia coli. FEBS Lett. 41195-198. [DOI] [PubMed] [Google Scholar]
- 28.Robinson, L. S., E. M. Ashman, S. J. Hultgren, and M. R. Chapman. 2006. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 59870-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosch, J. W., and M. G. Caparon. 2005. The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol. Microbiol. 58959-968. [DOI] [PubMed] [Google Scholar]
- 30.Sauvonnet, N., P. Gounon, and A. P. Pugsley. 2000. PpdD type IV pilin of Escherichia coli K-12 can be assembled into pili in Pseudomonas aeruginosa. J. Bacteriol. 182848-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, X., and M. R. Chapman. 2008. Sequence determinants of bacterial amyloid formation. J. Mol. Biol. 380570-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, X., N. D. Hammer, and M. R. Chapman. 2008. The molecular basis of functional bacterial amyloid polymerization and nucleation. J. Biol. Chem. 28321530-21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, X., D. R. Smith, J. W. Jones, and M. R. Chapman. 2007. In vitro polymerization of a functional Escherichia coli amyloid protein. J. Biol. Chem. 2823713-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.