Abstract
Anaerobic O-demethylases are inducible multicomponent enzymes which mediate the cleavage of the ether bond of phenyl methyl ethers and the transfer of the methyl group to tetrahydrofolate. The genes of all components (methyltransferases I and II, CP, and activating enzyme [AE]) of the vanillate- and veratrol-O-demethylases of Acetobacterium dehalogenans were sequenced and analyzed. In A. dehalogenans, the genes for methyltransferase I, CP, and methyltransferase II of both O-demethylases are clustered. The single-copy gene for AE is not included in the O-demethylase gene clusters. It was found that AE grouped with COG3894 proteins, the function of which was unknown so far. Genes encoding COG3894 proteins with 20 to 41% amino acid sequence identity with AE are present in numerous genomes of anaerobic microorganisms. Inspection of the domain structure and genetic context of these orthologs predicts that these are also reductive activases for corrinoid enzymes (RACEs), such as carbon monoxide dehydrogenase/acetyl coenzyme A synthases or anaerobic methyltransferases. The genes encoding the O-demethylase components were heterologously expressed with a C-terminal Strep-tag in Escherichia coli, and the recombinant proteins methyltransferase I, CP, and AE were characterized. Gel shift experiments showed that the AE comigrated with the CP. The formation of other protein complexes with the O-demethylase components was not observed under the conditions used. The results point to a strong interaction of the AE with the CP. This is the first report on the functional heterologous expression of acetogenic phenyl methyl ether-cleaving O-demethylases.
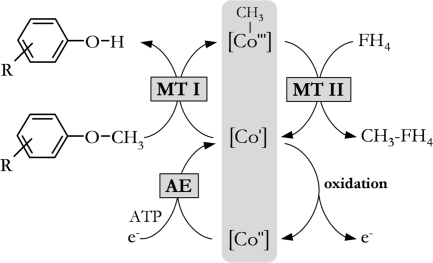
Phenyl methyl ethers are degradation products of lignin, and their methyl groups can be utilized as carbon and energy sources by acetogens such as Acetobacterium dehalogenans (2, 49, 57). In acetogenic bacteria the ether cleavage is mediated by the O-demethylases, which consist of four different proteins: two methyltransferases (MTs), a corrinoid protein (CP), and an activating enzyme (AE) (25). MT I, CP, and MT II are involved in the methyl transfer from the substrate to tetrahydrofolate (FH4) (Fig. 1). MT I binds the substrate and catalyzes the cleavage of the ether bond and the subsequent transfer of the methyl group to the superreduced CP ([CoI]). MT II transfers the methyl group from CP to FH4, yielding methyl-FH4, which is converted to acetate in the acetogenic catabolism. AE is required to reduce CP in an ATP-dependent reaction after inadvertent oxidation of the superreduced corrinoid to the inactive [CoII] form (50).
FIG. 1.
Methyl transfer from phenyl methyl ethers to FH4 catalyzed by anaerobic O-demethylases in Acetobacterium dehalogenans. [CoI], [CoII], and [CoIII], corrinoid protein with cobalt in the respective oxidation state.
Depending on the growth substrate, different O-demethylases are induced by their respective substrates (14, 25, 26). For A. dehalogenans, two O-demethylases have been described so far. The vanillate-O-demethylase (Odm) is induced by syringate or vanillate (Fig. 2). All components of this O-demethylase have been purified (25); however, due to the low yield of the purification procedure for the MTs and especially for AE, a detailed characterization could be performed only for CP, the gene of which has been expressed in Escherichia coli as the cofactor-free apoprotein, which could be reconstituted to a functional form by incubation with hydroxocobalamin (27). The veratrol-O-demethylase (Vdm), which is induced by 3-hydroxyanisole (Fig. 2), has been partially purified, and MT I has been characterized with respect to its substrate spectrum. Hence, the knowledge about bacterial O-demethylases and their reaction mechanism is limited mainly to the biochemical characterization of some isolated components (3, 14, 26, 38) and the CP reduction (50). Besides the two known O-demethylases of A. dehalogenans, a vanillate-O-demethylase of Moorella thermoacetica has been studied in more detail (38). MT I, MT II, and a CP have been isolated and biochemically characterized. For the assay of the latter enzyme system, an AE was not required.
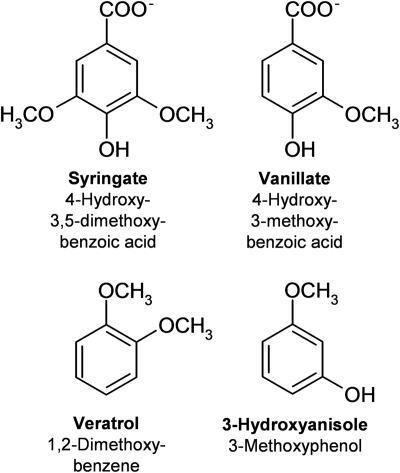
FIG. 2.
Chemical structures of the O-demethylase substrates syringate and vanillate (vanillate-O-demethylase) and veratrol and 3-hydroxyanisole (veratrol-O-demethylase).
Similar corrinoid-dependent methyl transfer reactions are also known for different methanogenic MTs catalyzing methyl transfers from methanol, methylamines, or dimethylsulfide to coenzyme M (7, 16, 48, 55, 56, 59, 62). Recent reports have concentrated on the characteristics of the archaeal MTs, their cofactors (22, 31), the protein structure (20), or the occurrence of isozymes (8, 16, 41, 42) as well as their differential transcription (23, 61). Similar to the case for the O-demethylases of A. dehalogenans, a corrinoid AE, designated either Map or Ram (3, 9), was required for the methanogenic MT reactions. Since the genomes of several methylotrophic methanogens are completely sequenced, the operons encoding the methanogenic MT systems could be partially identified. The genome of M. thermoacetica is also completely sequenced; however, the vanillate-O-demethylase operon is not annotated so far. Meanwhile, the genes encoding MT I, MT II, and CP of the known O-demethylases of A. dehalogenans have been identified and are available in GenBank (accession no. AF087018; AY318856). The gene sequence of the AE was not known. In this communication, the identification and analysis of a gene encoding a corrinoid AE is described for the first time.
We also report on the heterologous expression of MT I, CP, MT II, and AE of the vanillate- and veratrol-O-demethylases from A. dehalogenans. The heterologous expression allowed further biochemical characterization of these enzyme systems as well as studies of the interaction of the O-demethylase components.
MATERIALS AND METHODS
All chemicals, biochemicals, and gases needed for the growth of microorganisms and protein purification were of highest purity and were purchased from Sigma (Taufkirchen, Germany), Fermentas (St. Leon-Roth, Germany), Applichem (Darmstadt, Germany), Roth (Karlsruhe, Germany), and Linde (Leuna, Germany). Enzymes for molecular biology, if not stated otherwise, were purchased from Fermentas (St. Leon-Roth, Germany).
Cultivation of Acetobacterium dehalogenans.
A. dehalogenans was grown on syringate or 3-hydroxyanisole as described earlier (57). The cells were harvested under aerobic conditions by centrifugation for 10 min at 8,000 × g and 10°C.
Identification of the gene encoding the AE. (i) Protein purification.
The AE of A. dehalogenans was purified from cells grown on syringate or 3-hydroxyanisole (57). Cell lysis and the first purification step for AE on Q-Sepharose were performed as described earlier (25). Chromatography was carried out anaerobically at 25°C using a GE Healthcare (Munich, Germany) fast protein liquid chromatography system. Q-Sepharose fractions containing AE were then diluted four times with 50 mM Tris HCl (pH 7.5) containing 0.5 mM dithiothreitol (DTT) (buffer A) and were applied to a MonoQ column (10/10) preequilibrated with buffer A. Proteins were separated applying a linear gradient to buffer A plus 1 M KCl within 5 column volumes (2 ml/min). Fractions containing AE were pooled, and (NH4)2SO4 was added to a final concentration of 1.2 M. After application to a phenyl-Superose column (10/10) preequilibrated with buffer A including 1.2 M (NH4)2SO4, proteins were separated with a linearly decreasing (NH4)2SO4 gradient to buffer A (10 column volumes, 1 ml/min). After addition of (NH4)2SO4 (1.2 M final concentration) to the AE-containing fractions, protein was applied onto a Source-Phenyl column (5/5) preequilibrated with buffer A with 1.2 M (NH4)2SO4. AE was eluted with a linearly decreasing (NH4)2SO4 gradient to buffer A (40 column volumes, 1 ml/min).
(ii) Tryptic digestion.
Internal sequences of AE were obtained after treatment of the 65-kDa protein band from a 12% sodium dodecyl sulfate (SDS) gel with sequencing-grade bovine trypsin (Promega, Madison, WI). Tryptic peptides were separated on an Ettan-μLC high-pressure liquid chromatography system with RP18 columns (Sephasil C18 SC2.1/10) (GE Healthcare, Munich, Germany). Microsequence analysis of the isolated peptides was done using a model 494A amino acid sequencer (Applied Biosystems, Foster City, CA). For N-terminal sequencing, the AE was transferred to a polyvinylidene difluoride blot membrane.
(iii) Isolation of an AE gene fragment by PCR using degenerate primers.
For the isolation of an AE gene fragment, various degenerate primers were derived from the sequence of the N-terminal peptide and the peptides obtained by tryptic digestion of AE. The 1,289-bp AE gene fragment was amplified with the primers GF_AE02 and GF_AE32, derived from the N terminus and the peptide LIYNTQ, respectively (GF_AE02, YTNAAYACNATHCGNGTNTTYTTYCC; GF_AE32, TTYTGGGTRTTATARATCAG). The PCR was performed in 1× PCR buffer, 2.5 mM magnesium chloride, 0.2 mM deoxynucleoside triphosphates (dNTP), 2 ng/μl genomic DNA, 1 μM of each primer, and 40 mU/μl Taq polymerase. After initial denaturation for 2 min at 96°C, 35 cycles of 45 s at 96°C, 45 s at 40°C, and 2 min at 72°C were performed.
(iv) Genome walking.
For genome walking, the Universal Genome Walker kit (Clontech, Mountain View, CA) was used according to the manufacturer's protocol. DNA libraries were prepared using the restriction enzymes of the kit. Specific PCR products were obtained from the DraI library. The gene-specific primers (upstream, GW_AE44 [CGGTGGTTTCCAGTATTTGCGATAAGG] and GW_AE45 [CCAACCAGAGCGCAGGCTTGAGC]; downstream, GW_AE46 [CGTTCTGCTGGGACTGCCAAAGG] and GW_AE47 [TGACGCCATCGCCTTCCTGTATCGG]) were derived from the DNA fragment obtained with degenerate primers and were used in the first and second PCRs, respectively.
(v) Cloning of PCR products.
The PCR products derived from the protein fragments and obtained by genome walking were extracted from agarose gels using the Qiaquick gel extraction kit (Qiagen, Hilden, Germany) and cloned into the pGEM-T vector (Promega, Mannheim, Germany). Plasmids were transformed in Escherichia coli DH5α by heat shock, and transformants were grown at 37°C in Luria-Bertani (LB) medium (45) containing 100 μg/ml ampicillin. Plasmids were recovered from transformants using the EZNA miniprep kit (peqLab, Erlangen, Germany).
Isolation of genomic DNA.
Cells of Acetobacterium dehalogenans (1 g [wet weight]) were suspended in 5 ml of 10 mM Tris HCl (pH 7.5) containing 1 mM EDTA. Lysozyme was added to a final concentration of 2.5 mg/ml, and the cell suspension was incubated at 37°C for 16 h. SDS, RNase A, and pronase E were then added to final concentrations of 1% (wt/vol), 0.25 mg/ml, and 0.2 mg/ml, respectively, and the mixture was incubated for 2 h at 55°C. Sodium chloride was added to a concentration of 1.6 M and the mixture was extracted twice with chloroform prior to DNA precipitation with an equal volume of isopropanol (45). Genomic DNA was obtained from the mixture using a glass rod and washed in 70% (vol/vol) ethanol. It was then air dried, suspended in water, and stored at 4°C.
Construction of expression cassettes and cloning of the vanillate- and veratrol-O-demethylase genes.
Expression cassettes for the genes for MT I, CP, and MT II of the vanillate- and veratrol-O-demethylases (GenBank accession no. AF087018 and AY318856, respectively) as fusion proteins with a C-terminal Strep-tag II and with restriction sites for cloning in pet11a (Stratagene, Amsterdam, The Netherlands) were constructed from PCR products. Multiple PCR steps with genomic DNA or PCR products as templates were necessary to obtain the inserts. The primer sequences and annealing temperatures for the PCRs are given in Table 1. The PCR mixtures contained 100 ng DNA, 25 pmol of each primer, 10 mM Tris HCl (pH 8.8), 1.5 mM MgCl2, 50 mM KCl, 200 μM dNTP, 2.5 U of Taq polymerase, and 1 U of Pfu polymerase in a final volume of 25 μl. PCR products and vector were digested with NdeI and BamHI according to the manufacturer's protocol. The ligation was performed overnight at 16°C in 1× ligation buffer with 1 U/μl T4 DNA ligase.
TABLE 1.
Oligonucleotides used for cloning of the vanillate- and veratrol-O-demethylase genesa
| Gene | Primer sequence | PCR step (annealing temp, °C) |
|---|---|---|
| odmA | GTTAATCGGCACACACAGCC | 1 (50.0) |
| GACAAAGCGAATTACC | ||
| GGAGATATACATATGTCAAAAATTGAAGAAG | 2 (50.0) | |
| CTGCGGGTGGCTCCAAGCGCTCGCTGTTGCCAG | ||
| GGAGATATACATATGTCAAAAATTGAAGAAG | 3 (50.0) | |
| CAGCCGGATCCTTATTTTTCGAACTGCGGGTGGC | ||
| odmB | GGAGATATACATATGTTAACAAAAAGACAG | 1 (40.0) |
| CTGCGGGTGGCTCCAAGCGCTGAACAATTTCTCTGAC | ||
| GGAGATATACATATGTTAACAAAAAGACAG | 2 (55.0) | |
| CAGCCGGATCCTTATTTTTCGAACTGCGGGTGGC | ||
| odmD | ACGAAGCCTGTGATGAAGC | 1 (60.0) |
| CTGCGGGTGGCTCCAAGCGCTTTTCTTCTGACCG | ||
| GGAGATATACATATGATTATTATCGGAG | 2 (55.0) | |
| CTGCGGGTGCCAAGCGCTTTTTTTTACCGCGC | ||
| GGAGATATACATATGATTATTATCGGAG | 3 (55.0) | |
| CAGCCGGATCCTTATTTTTCGAACTGCGGGTGGC | ||
| vdmA | GCCATCCTTGATCCCACCGATCGG | 1 (57.5) |
| CTCCGGTTTCTTTCCCTCG | ||
| GGAGATATACATATGTCAAAAATTACAGAAGTC | 2 (50.0) | |
| CTGCGGGTGGCTCCAAGCGCTCTTAACCAGGTC | ||
| GGAGATATACATATGTCAAAAATTACAGAAGTC | 3 (50.0) | |
| CAGCCGGATCCTTATTTTTCGAACTGCGGGTGGC | ||
| vdmB | GGAGATATACATATGAATCAGAGAGA | 1 (57.5) |
| CTGCGGGTGGCTCCAAGCGCTAAGCTTGTTTATTTCATTG | ||
| GGAGATATACATATGAATCAGAGAGA | 2 (50.0) | |
| CAGCCGGATCCTTATTTTTCGAACTGCGGGTGGC | ||
| vdmD | TGACCACCATCACCACCCGG | 1 (57.5) |
| TATAAATGTGTGATTAACACG | ||
| GGAGATATACATATGATCATCATCGGA | 2 (50.0) | |
| CTGCGGGTGCCAAGCGCTTTTTTTTACCGCGC | ||
| GGAGATATACATATGATCATCATCGGA | 3 (50.0) | |
| CAGCCGGATCCTTATTTTTCGAACTGCGGGTGGC | ||
| odmC | TTCTTCATATCATACCTG | 1 |
| CCATAGTTGACCATCCTTTCG | ||
| ATGGTAGGTCTCAAATGTCATCTTTGAATACTATTCG | 2 | |
| ATGGTAGGTCTCAGCGCTTTTCATTTCATTTTGACCAAGG |
For all genes except odmC, the PCR was performed as follows: initial denaturation (2 min at 96°C) and 35 cycles of annealing and polymerization (1 min at 96°C, 45 s of annealing at the given temperature, and 2 min at 72°C). For odmC, the PCR conditions used are given in Materials and Methods.
The AE gene (GenBank accession no. EU812561) was cloned as a fusion protein with a C-terminal Strep-tag II using the pASK-IBA3plus expression vector (IBA GmbH, Göttingen, Germany). The insert was amplified using the PCR product obtained in the following reaction mixture with a hot-start protocol: 1× Pfu PCR buffer, 1.1 mM magnesium acetate, 0.2 mM dNTP, genomic DNA (4 ng/μl), 0.4 μM of each primer (Table 1), 60 mU/μl Pfu polymerase (Promega, Mannheim, Germany), and 40 mU/μl Taq polymerase. After initial denaturation for 5 min at 96°C, the reaction was started with the enzyme mix and 5 cycles of 45 s at 96°C, 45 s at 53°C, and 5 min at 72°C were performed, followed by 20 cycles of 45 s at 96°C, 45 s at 50°C, and 5 min at 72°C. The amplified PCR product was used as the template in a second PCR in 1× Pfu PCR buffer, 1.1 mM magnesium acetate, 0.2 mM dNTP, 0.4 μM of each primer (Table 1), 60 mU/μl Pfu polymerase (Promega, Mannheim, Germany), and 40 mU/μl Taq polymerase. After initial denaturation for 5 min at 96°C, the reaction was started with the enzyme mix, and 35 cycles of 45 s at 96°C, 45 s at 55°C, and 5 min at 72°C were performed. After BsaI digestion, the PCR product was ligated to pASK-IBA3plus according to the manufacturer's protocol.
The plasmids were transformed in Escherichia coli DH5α by heat shock and were recovered from transformants using the EZNA miniprep kit (peqLab, Erlangen, Germany). Plasmids containing the insert were then transformed into E. coli expression strains. The properties of the constructed plasmids and expression strains are summarized in Table 2.
TABLE 2.
Plasmids, strains, and conditions for heterologous expression of the components of the vanillate- and veratrol-O-demethylases
| Plasmid | Characteristics | Expression strain | Inducing conditions
|
||
|---|---|---|---|---|---|
| Temp (°C) | IPTG (mM) or AHT (ng/ml) concn | Time (h) | |||
| pCPvanStrep | odmA in pET11a; NdeI, BamHI | E. coli BL21(DE3) | 37 | IPTG, 0.50 | 1.5 |
| pMT1vanStrep | odmB in pET11a; NdeI, BamHI | E. coli BL21(DE3) | 28 | IPTG, 0.25 | 2 |
| pMT2vanStrep | odmD in pET11a; NdeI, BamHI | E. coli BL21(DE3)/pRIL | 28 | IPTG, 0.50 | 2 |
| pCPverStrep | vdmA in pET11a; NdeI, BamHI | E. coli BL21(DE3) | 28 | IPTG, 0.50 | 2 |
| pMT1verStrep | vdmB in pET11a; NdeI, BamHI | E. coli BL21(DE3)/pRIL | 28 | IPTG, 0.50 | 2 |
| pMT2verStrep | vdmD in pET11a; NdeI, BamHI | E. coli BL21(DE3)/pRIL | 28 | IPTG, 0.50 | 2 |
| pAEvanStrep | odmC in pASK IBA3+; BsaI | E. coli BL21(DE3) | 18 | AHT, 50 | 16 |
Heterologous expression of the vanillate- and veratrol-O-demethylase genes in Escherichia coli.
The production of recombinant proteins was performed in LB medium containing the required antibiotics. Gene expression was induced with isopropyl-β-d-thiogalactopyranisde (IPTG) or anhydrotetracycline (AHT) as indicated in Table 2. After induction, the cells were harvested by centrifugation for 10 min at 10,000 × g and stored at −20°C.
Purification of heterologously expressed CP, MT I, and MT II.
Except for the CPs OdmA and VdmA, recombinant proteins were purified by affinity chromatography in a one-step protocol from cell extracts after lysis with 2 mg/ml of lysozyme and 2 μg/ml DNase I for 1 h at 37°C. The cell debris was removed by centrifugation for 15 min at 10,000 × g and 10°C. About 1 ml of crude extract (5 to 10 mg protein per ml) was mixed with an equal volume of 100 mM Tris HCl (pH 8.0) containing 150 mM NaCl. The cell extract was applied to a 1-ml Strep-Tactin Superflow cartridge (IBA GmbH, Göttingen, Germany) preequilibrated with 10 column volumes of buffer. The purification was performed according to the manufacturers' protocol.
To obtain “active” CP, it was necessary to incorporate the corrinoid cofactor (hydroxocobalamin) into the apoprotein. All purification and reconstitution steps were carried out under anaerobic conditions. OdmA and VdmA were enriched using ion-exchange chromatography. The crude extract (100 to 200 mg protein) was applied to a Q Sepharose column (1.0 cm by 10 cm) preequilibrated with 50 mM Tris HCl (pH 7.5) containing 0.5 mM DTT. Proteins were eluted with a KCl gradient (0 to 0.25 M KCl, 5 column volumes; 0.25 M to 1 M KCl, 5 column volumes). The fractions containing the apoprotein were pooled, and 1 mM phenylmethylsulfonyl fluoride (PMSF) was added. To reconstitute the CP with the cofactor, the protein solution was mixed with a fivefold molar excess of hydroxocobalamin, 3.5 M betaine, and 1 mM DTT. After 16 h of incubation at 4°C, the protein solution was diluted 50-fold by dropwise addition of 50 mM Tris HCl pH (7.5) containing 0.5 mM DTT and 0.1 mM PMSF at 4°C under constant shaking at 100 rpm. After filtration, the protein solution was applied to a Mono Q column (1.0 cm by 10 cm) preequilibrated with 50 mM Tris HCl (pH 7.5) containing 0.5 mM DTT and 0.1 mM PMSF. Fractions containing reconstituted OdmA or VdmA eluted at 0.25 and 0.30 M KCl when a linear gradient from 0 to 0.4 M KCl (5 column volumes) was applied. The corrinoid content of the protein was calculated from the UV/visible spectra using Δɛ386 = 21 mM−1 cm−1 for cob(I)alamin (50) and ɛ475 = 9.2 mM−1 cm−1 for cob(II)alamin (18).
Purification of rAE.
Cells were disrupted in a French pressure cell. Cell debris was removed by centrifugation for 15 min at 10,000 × g and 15°C. Two volumes of 67 mM sodium potassium phosphate buffer (pH 8.5) were added to the cell extract. For purification, 10 ml of the diluted sample was applied to a 5-ml Strep-Tactin Superflow cartridge (IBA GmbH, Göttingen, Germany). After the column was washed, the protein was eluted with 67 mM sodium potassium phosphate buffer (pH 5.0) (5 column volumes). Fractions with recombinant AE (rAE) were pooled. The protein was concentrated to 1 mg/ml using ultrafiltration (10-kDa cutoff, Amicon-Cell; Millipore GmbH, Schwalbach, Germany). Afterwards, the gas atmosphere of the protein solution was replaced by nitrogen. For reconstitution of Fe/S clusters, 2 mM DTT and a 10-fold molar excess of FeCl3 and Na2S were added. The solution was incubated for 16 h at 10°C under N2 as the gas phase. To remove unbound iron and sulfide, the protein sample was applied to a Mono Q 5/5 column preequilibrated with 50 mM Tris HCl (pH 7.5) containing 0.5 mM DTT. After washing, rAE was eluted with a linear KCl gradient (0 to 1 M KCl in 2.5 column volumes) at 0.4 M KCl.
Determination of enzyme activities.
All enzyme activities were determined in a final volume of 100 μl in anaerobic quartz cuvettes with N2 as the gas phase. The assay mixture for AE contained 2 mM ATP, 10 mM MgCl2, 0.5 mM titanium(III) citrate, and 15 μM of CP. The reaction was started by the addition of AE. The activity was calculated from the kinetics of cob(I)alamin formation at 386 nm (Δɛ386 = 21 mM−1 cm−1 [50]).
The MT I activity was determined photometrically by measuring the formation of methylcobalamin from cob(I)alamin at 528 nm (ɛ528 = 7.9 mM−1 cm−1 [18]). The assay mixture contained 50 mM DTT, 2 mM ATP, 10 mM MgCl2, 0.5 mM titanium(III) citrate, 5 mM of vanillate or veratrol, 15 μM CP, and AE (10 μg protein of recombinant E. coli crude extract) in 50 mM Tris HCl (pH 7.5). The reaction was started by the addition of MT I.
MT II activity was measured by following the demethylation of [CoIII-CH3]-OdmA spectrophotometrically at 528 nm. Methylated CP was obtained by incorporation of methylcobalamin instead of hydroxocobalamin during reconstitution of OdmA (see above). The assay mixture contained about 20 μM of methylated OdmA, 3 mM FH4, and 0.5 mM titanium(III) citrate. The reaction was started by the addition of MT II.
With recombinant components, the complete O-demethylase reaction was performed using the coupled-enzyme assay described earlier (35). The slightly modified assay mixture contained 1 mM NAD+, 1 mM NADP+, 1 mM ATP, 1 mM MgCl2, 5 mM DTT, 0.5 mM titanium(III) citrate, 20 mM pyruvate, 3 mM FH4, 40.8 U/ml lactate dehydrogenase, 0.4 U/ml methylene-FH4 reductase, 0.6 U/ml methylene-FH4 dehydrogenase, 5 mM of vanillate or veratrol, 15 μM CP, AE (10 μg recombinant E. coli crude extract), 2 μM MT I, and 2 μM MT II. The test was started by the addition of substrate. The O-demethylase activity was calculated from the kinetics of formation of methyl-FH4 and NADPH observed at 350 nm (ɛ350 = 30.6 mM−1 cm−1 [35]).
Interaction of the O-demethylase components (gel shift experiments).
The following experiments were performed under anoxic conditions in an anaerobic chamber.
(i) Influence of the corrinoid cofactor on the interaction.
Mixtures of the rAE (2 pmol) and the rCP (100 pmol) of the vanillate-O-demethylase (without and with the corrinoid cofactor hydroxocobalamin) were incubated 2 h at 10°C. Afterwards, the protein mixtures were separated using native polyacrylamide gel electrophoresis (PAGE) (8%). For protein detection, specific antibodies against AE and CP were used.
(ii) Stoichiometry of the AE-CP protein complex.
AE-CP mixtures (purified recombinant and reconstituted proteins) with AE/CP ratios (pmol:pmol) of 100:10; 50:10; 10:10; 10:50, and 10:100 were separated by native PAGE (8%) after preincubation (2 h, 10°C). The gels were silver stained.
Analytical methods.
The protein content was determined according to the method of Bradford (5) using bovine serum albumin as a standard protein. Iron was quantified using the method of Fish (17). Acid-labile sulfur was determined according to the method of Gilboa-Garber (19).
The zinc content was determined by a photometric method as described by Zhou et al. (66). Samples of MT I were incubated for 10 min in 50 mM Tris HCl (pH 7.5) containing 1 mM EDTA. EDTA was removed by gel filtration on a Superdex 75 column (1.6 cm by 60 cm) with 50 mM Tris HCl (pH 7.5) at a flow rate of 1 ml/min. Fractions containing MT I were concentrated by ultrafiltration with Vivaspin 50 centrifugation units (Vivascience AG, Hannover, Germany). The protein and zinc contents of the retentate were determined as described above.
EPR spectroscopy.
For a description of the X-band electron paramagnetic resonance (EPR) spectrometer, sample preparation, determination of spin concentration, and simulation for estimation of g values see reference 29.
Determination of the midpoint potential of the Fe/S cluster of AE.
The midpoint potential of the Fe/S cluster of AE was determined by UV/visible potentiometry at pH 7.5 under anoxic conditions. Titanium(III) citrate was used as an electron donor (65) and was added stepwise to the sample in 50 mM Tris HCl buffer (pH 7.5). After the equilibrium was reached, the UV/visible absorption spectrum was recorded and the redox potential of the solution was simultaneously determined with a Pt/Ag/AgCl electrode (Mettler Toledo, Steinbach, Germany). For calculation of the potential versus the standard hydrogen electrode (SHE), 207 mV had to be added to the recorded potential. Redox standard solutions at +220 mV and +468 mV, purchased from Mettler Toledo (Steinbach, Germany), as well as a methyl viologen solution (50 μM in 100 mM Tris HCl buffer [pH 7]; E°′ = −449 mV) titrated with titanium(III) citrate served as controls. The dilution caused by the addition of the titanium(III) citrate solution was taken into account for the calculations.
Bioinformatic analysis.
Analysis of finished and unfinished genomes was performed with the integrated microbial genome system supported by the Department of Energy Joint Genome Institute (33), as well as with the NCBI BLAST server (1). The amino acid sequence of the OdmC of A. dehalogenans was used for BlastP searches, and further searches were then executed with selected hits after inspection of the genetic context in which the hits occurred.
RESULTS
Organization of the O-demethylase genes of Acetobacterium dehalogenans.
At least two O-demethylases have been described to be induced by different substrates in Acetobacterium dehalogenans (14). Due to the substrate preferences, the O-demethylases were designated vanillate-O-demethylase and veratrol-O-demethylase (Odm and Vdm, respectively). The three major components of these enzyme systems mediating the methyl transfer from the phenyl methyl ether to FH4 are MT I (OdmB or VdmB), CP (OdmA or VdmA) as the primary methyl acceptor, and MT II (OdmD or VdmD). The genes of these O-demethylase components are organized in separate operons as odmDAB and vdmDAB. The cotranscription of the three genes could be demonstrated by reverse transcription-PCR in cells grown with the respective inducing substrate (syringate for induction of the vanillate-O-demethylase genes odmDAB and 3-hydroxyanisole for induction of the veratrol-O-demethylase genes vdmDAB) (data not shown). Comparison of the deduced amino acid sequences of the odm and vdm genes showed identities of 80% for MT II, 75% for CP, and 22% for MT I (similarities of 93%, 87%, and 62%, respectively).
The gene for AE (odmC) was unknown so far, since the amount of purified protein obtained using the method described by Kaufmann et al. (25) was not sufficient for protein sequencing and tryptic digestion. By changing the order of chromatography and omitting the gel filtration step, submilligram quantities of suitable purity for tryptic digestion and N-terminal Edman degradation could be obtained. A comparative matrix-assisted laser desorption ionization-time-of-flight analysis of the peptides obtained by tryptic digestion of AE isolated from syringate- or 3-hydroxyanisole grown cells revealed 100% identity. This finding led to the assumption that only a single AE is responsible for the reduction of different CPs involved in phenyl methyl ether cleavage. In retrospect, about two-thirds of the full AE amino acid sequence was covered by the peptides identified in this procedure. By genome walking with degenerate primers deduced from these peptides, a 4-kb fragment that contained the complete open reading frame (ORF) encoding AE (odmC) was obtained. A putative protein encoded by an additional ORF (divergently transcribed) located upstream of odmC exhibited approximately 45% amino acid sequence identity to the proteins encoded by odmD and vdmD, the CP:FH4 MT II genes of the vanillate- and veratrol-O-demethylases of Acetobacterium dehalogenans. It also showed near 60% amino acid sequence identity to a putative methyl-FH4:corrinoid/iron-sulfur protein MT of Desulfitobacterium hafniense Y51 (40). Another, not completely sequenced ORF assigned to a putative asparaginase or glutamyl-tRNA (Gln) amidotransferase subunit D was found downstream of odmC.
The AE gene is not included in the two known O-demethylase operons of A. dehalogenans. However, by genome walking a new putative O-demethylase operon was identified approximately 3 kb downstream of odmC. The gene organization is similar to that of the known O-demethylase operons. The gene for MT I is located upstream of the MT II gene; downstream of the MT II gene, a gene encoding a CP was identified. The putative gene products showed about 63% (MT I), 98% (MT II), and 92% (CP) amino acid sequence identity to the protein components of the vanillate-O-demethylase of A. dehalogenans.
Southern blot analysis using a gene probe for odmC revealed the presence of just one copy of this gene in the genome of A. dehalogenans. The gene is transcribed independent of the growth substrates used, including fructose, H2-CO2, syringate, and 3-hydroxyanisole (data not shown).
Bioinformatic analysis of the O-demethylase genes.
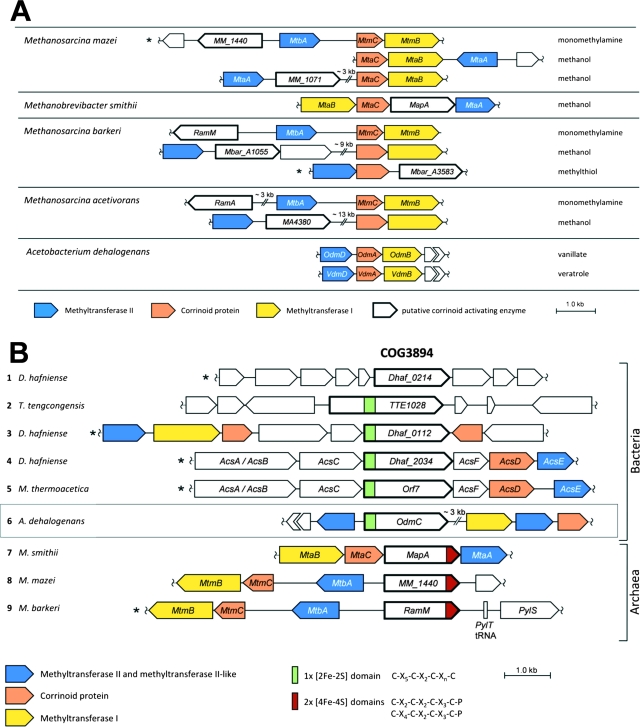
Functionally, the O-demethylases of A. dehalogenans are similar to the methanogenic corrinoid-dependent MTs (3, 7, 10, 15, 48, 59, 60). Therefore, we compared the O-demethylase operons to the operons encoding the methanogenic MT systems mediating the transfer of the methyl groups of methanol, methylamines, or other methyl substrates to coenzyme M. The O-demethylase operons of A. dehalogenans are structured similarly to the operons encoding these methanogenic MTs (Fig. 3A). However, the intergenic region between the MT II gene and the gene encoding the CP is far greater (up to about 15 kb) than that in the O-demethylase genes, where the distance is about 60 to 120 bp. A few examples are shown in Fig. 3A. In most of the methanogenic MT operons, a gene probably encoding an activating protein (e.g., RamA or MapA) is localized either within this intergenic region or upstream of the MT II gene. These proteins belong to the same protein family as the AE of A. dehalogenans (Fig. 3B). Apparently, some of the genes encoding methanogenic AEs are part of the MT operon (Fig. 3A). For A. dehalogenans, it can be excluded that the AE gene odmC is part of a copy of the known O-demethylase genes. The presence of a new O-demethylase operon 3 kb downstream of odmC indicates that the AE gene might be part of another acetogenic MT system, which has not yet been identified.
FIG. 3.
Analysis of the O-demethylase genes. (A) Comparison of the MTII/CP/MTI gene clusters of the vanillate- and veratrol-O-demethylases of Acetobacterium dehalogenans with those encoding some methanogenic MT systems of Methanosarcina barkeri, Methanosarcina acetivorans, Methanosarcina mazei, and Methanobrevibacter smithii. For further explanation, see the text. (B) Comparison of the genetic context of the AE gene with gene clusters with orthologs implied in activation of CPs of anaerobic MTs or CODH/ACS. Nine selected bacterial (1 to 6) and archaeal (7 to 9) gene clusters are shown with the COG3894 genes for the putative protein of Desulfitobacterium hafniense DCB-2 (1), the putative Fe/S protein of Thermoanaerobacter tengcongensis (2), trimethylamine MT of D. hafniense DCB-2 (3), CODH/ACS of D. hafniense DCB-2 (4), CODH/ACS of Moorella thermoacetica (32) (5), AE (OdmC) of Acetobacterium dehalogenans (6), MapA in the methanol:coenzyme M (CoM) MT of Methanobrevibacter smithii (7), monomethylamine:CoM MT of Methanosarcina mazei (8), and RamM in the monomethylamine:CoM MT of Methanosarcina barkeri (9). Map, MT-activating protein; SAM, S-adenosylmethionine binding domain. The organisms whose gene clusters have been oriented in reverse direction in comparison to the orientation in the annotated genome are marked with an asterisk.
The gene product AE of odmC consists of 598 amino acids, accounting for a molecular mass of 65.3 kDa, which is in accordance with the apparent molecular masses determined for the purified AE by SDS-PAGE (25) and for the recombinant enzyme (Fig. 4). Analysis of the protein sequence using ProSite (12) showed a putative [2Fe-2S] cluster binding motif C-X5-C-X2-C-Xn-C (36) in the N-terminal domain. This indicates that C41, C47, C50, and C66 in OdmC could bind a [2Fe-2S] cluster. To preclude [2Fe-2S] ferredoxin-like sequences in database analysis, we used amino acids 100 to 598 of OdmC for a BlastP search. Currently this search yields 163 proteins (410 to 822 amino acids) with amino acid sequence identities to AE of from 20% to 41%. Due to the presence of an N-terminal [2Fe-2S] cluster binding motif (132 hits) or C-terminal [4Fe-4S] motifs (16 hits), the orthologs are usually annotated as metal binding proteins of unknown function. The genes, occurring predominantly in Bacteria (Clostridia; alpha-, gamma-, and deltaproteobacteria) and in methanogenic Archaea, are systematically grouped in the Cluster of Orthologous Groups (COG) database as COG3894. In several cases, multiple genes encoding AE-like proteins were found in completely sequenced genomes of bacteria known to catalyze anaerobic O-demethylation of phenyl methyl ethers, such as M. thermoacetica (11, 63) or Desulfitobacterium hafniense (30, 39). In the genomic sequence of the Y51 strain of the latter organism (40), five paralogs, were found and in the unfinished genome of the deltaproteobacterium MLMS-1, there are seven paralogs. The corresponding genes of numerous proteins which belong to COG3894 are part of known or putative anaerobic MT or carbon monoxide dehydrogenase/acetyl coenzyme A (CoA) synthase (CODH/ACS) gene clusters. Several examples from anaerobic bacteria or archaea are shown in Fig. 3B. In a few methanogens, a corresponding gene has been assigned to the MT-activating protein (Fig. 3B, example 7, MapA). In other methanogens, these proteins are referred to as Ram proteins (T. Ferguson, T. Lienard, G. Gottschalk, and J. Krzycki, unpublished data cited in the legend to Fig. 1A in reference 51) (Fig. 3B, see example 9, RamM). In the anaerobic MTs and the CODH/ACS complexes, protein-bound corrinoids are required in the superreduced state of the cobalt to accept a methyl group. Hence, we believe that the COG3894 genes encode “activases,” which are Fe/S proteins mediating the activation and reduction of protein-bound corrinoids. For future reference, we suggest the term “RACE” (reductive activator of corrinoid-dependent enzymes) for this type of protein.
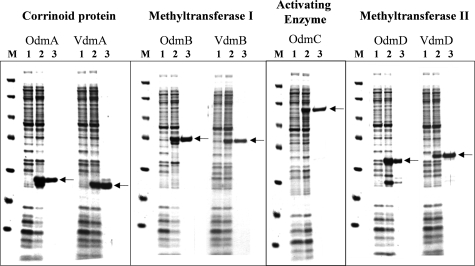
FIG. 4.
SDS-PAGE of heterologously expressed vanillate- and veratrol-O-demethylase components. Proteins were stained with Coomassie blue. The apparent molecular masses of the marker proteins (lanes M) are 116, 67, 45, 35, 25, 18, and 14 kDa, respectively. For each component (CPs, MTs I and II, and AE), 5 μg of crude extract before (lanes 1) or after (lanes 2) induction with IPTG or anhydrotetracycline or 1 μg of purified protein (lanes 3) was loaded.
Heterologous expression of O-demethylase genes.
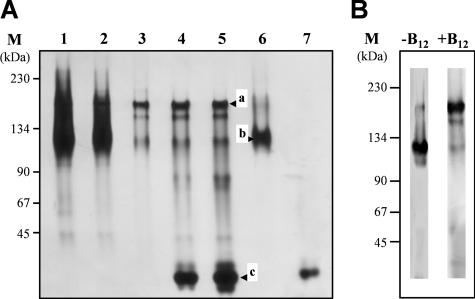
The genes of all vanillate- and veratrol-O-demethylase components were cloned separately in the pet11a or pASK-IBA3plus expression vector (see Materials and Methods). All proteins were produced in Escherichia coli as C-terminal fusions with Strep-tag. Conditions for the heterologous expression of the O-demethylase genes were optimized to obtain a high expression level in the soluble protein fraction of cell extracts of E. coli (Fig. 4, lanes 2). The proteins were purified to apparent homogeneity by affinity chromatography on Strep-Tactin (Fig. 4, lanes 3). The apparent molecular masses of the purified proteins were determined via SDS-PAGE and were identical to the molecular masses calculated from amino acid sequences. Except for the CPs, all components were active in the crude extract of E. coli after induction with IPTG or anhydrotetracycline. Cell extracts of E. coli without any expression vector showed no intrinsic activity similar to that of the O-demethylase components.
Characterization of rMTs I.
MT I catalyzes the cleavage of the substrate ether bond and the transfer of the methyl group to the CP upon formation of methylcobalamin. To assay the MTs OdmB and VdmB, vanillate or veratrol was routinely used as the methyl substrate and reconstituted OdmA (see Materials and Methods) as the methyl acceptor. Previously, the complete O-demethylase reaction (plus accessory substrates and enzymes) was used to test the single components of this enzyme system (35). This assay mixture contained all four O-demethylase components, ATP, Ti(III) citrate, FH4 (which had to be synthesized from folic acid), three accessory enzymes (two of which had to be purified from Peptostreptococcus productus), pyruvate, NAD+, NADP+, and 5 mM DTT. The formation of methenyl-FH4 and NADPH was measured photometrically (ɛ350 = 30.6 mM−1 cm−1 [35]). Here, we developed a simplified assay with a mixture containing MT I, CP, crude extracts of E. coli with rAE, ATP, Ti(III) citrate, and 50 mM DTT. This assay also worked with 5 mM DTT; however, the activity was stimulated fourfold in the presence of 50 mM DTT. The formation of methylcobalamin was followed photometrically by the increase of absorption at 528 nm (ɛ528 = 7.9 mM−1 cm−1 [18]). Using this assay, the activity of MT I could be determined. The results were confirmed with high-pressure liquid chromatography measurements by determination of the phenyl methyl ether and the demethylated reaction product.
The specific activities of the purified rMTs and the substrate spectra were determined and compared to those of the native enzymes (Table 3). The activities of the recombinant proteins were of the same order of magnitude as those of the native enzymes. The slight differences are within the range of the activities of the native enzyme purified from different cell batches. The substrate spectra of the native and recombinant enzymes were identical. Methanol and methylamine did not serve as a substrate for both MTs I. Surprisingly, methyl chloride could be converted by MT Iver. A. dehalogenans was originally isolated with methyl chloride as an energy source. Methyl chloride-grown cells, however, did not contain MT Iver as indicated by Western blot analysis (data not shown), indicating the presence of a separate methyl chloride dehalogenase in A. dehalogenans.
TABLE 3.
Specific activities and substrate spectra of the native and recombinant MTs I of the vanillate- and veratrol-O-demethylases
| Substrate | Sp act (U/g)
|
|||
|---|---|---|---|---|
| MT Ivana
|
MT Iverb
|
|||
| Native | Recombinant | Native | Recombinant | |
| Vanillate | 428 | 146 | 13 | 19 |
| Isovanillate | 647 | 349 | 119 | 125 |
| Syringate | 359 | 146 | 119 | 63 |
| Guajacole | 194 | 76 | 86 | 91 |
| Veratrol | 0 | 0 | 152 | 197 |
| 3-Hydroxyanisole | 0 | 0 | 165 | 236 |
| 2-Chloranisole | 0 | 0 | 86 | 73 |
| 3,4,5-Trimethoxybenzoate | 0 | 0 | 0 | 0 |
| Methyl chloride | 0 | 0 | 115 | 26 |
| Methanol | 0 | 0 | 0 | 0 |
| Methylamine | 0 | 0 | 0 | 0 |
MT I of the vanillate-O-demethylase.
MT I of the veratrol-O-demethylase.
Both MTs I were found to contain zinc. The zinc contents of recombinant and native proteins were quantified with a photometric test (66). For OdmB, 1.02 ± 0.05 mol zinc per mol recombinant protein and 0.94 ± 0.04 mol zinc per mol native protein were determined. VdmB was found to contain 1.03 ± 0.05 mol zinc per mol recombinant protein and 0.93 ± 0.04 mol zinc per mol native protein.
Characterization of rAE.
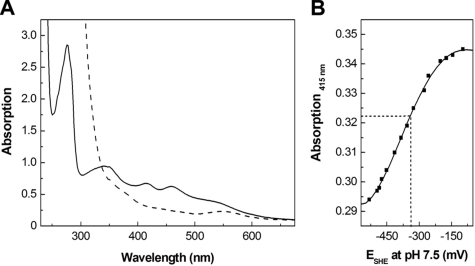
In crude extracts of the expression host, rAE was active and mediated the reduction of the CPs CPvan and CPver of the vanillate- and veratrol-O-demethylases in the presence of ATP. Crude extracts of nontransformed E. coli did not show any corrinoid reduction activity. However, purified rAE was inactive after affinity chromatography. Inactivation upon purification was probably caused by loss or damage of the iron-sulfur cluster, as indicated by the substoichiometric content of about 0.3 mol Fe and 0.6 mol S per mol rAE. The enzyme could be reconstituted to its active form by incubation with FeCl3 and Na2S under anoxic conditions in the presence of DTT. After reconstitution, the enzyme contained approximately 1.8 mol S and 2.0 to 3.0 mol Fe per mol enzyme. If one assumes that the slightly higher Fe content results from binding of adventitious Fe ions, the values are in full accordance with the presence of a [2Fe-2S] cluster in rAE as suggested by the N-terminal cysteine motif. A single [2Fe-2S] cluster was also deduced from the UV/visible spectrum of the reconstituted enzyme (Fig. 5A). Absorbance bands at 345 nm (ɛ345 = 14.5 mM−1 cm−1), 415 nm (ɛ415 = 10.4 mM−1 cm−1), and 458 nm (ɛ458 = 9.6 mM−1 cm−1) and a shoulder at 540 nm (ɛ540 = 5.8 mM−1 cm−1) were detected. The positions and intensities of these absorptions are typical of those observed in the [2Fe-2S]2+ state of proteins with cysteinate-coordinated [2Fe-2S] clusters (58, 64). Upon addition of the reductant Ti(III) citrate, the absorbance of the [2Fe-2S]2+ cluster in rAE diminished due to formation of the [2Fe-2S]1+ state (Fig. 5A). Again, the appearance of the visible spectrum (i.e., bleaching with a residual absorbance band at 550 nm) was typical of [2Fe-2S] proteins (58, 64). By following the bleaching at 415 nm and measuring the solution potential with a redox electrode upon stepwise addition of Ti(III) citrate, a midpoint potential at pH 7.5 of −330 mV ± 20 mV for the [2Fe-2S]2+/1+ redox couple of the rAE was determined (Fig. 5B). This value indicated that sodium dithionite, which was used as a reductant in EPR spectroscopy (see below), should quantitatively reduce the [2Fe-2S]2+ cluster of rAE.
FIG. 5.
UV/visible spectra of rAE and determination of the midpoint potential of the [2Fe-2S]2+/1+ cluster. (A) UV/visible spectra of the isolated (solid line) and reduced (dashed line) forms of AE (65 μM). For reduction of the cluster, titanium(III) citrate was used (final concentration, about 100 μM). (B) Redox titration curve of the determination of the midpoint potential of the [2Fe-2S]2+/1+ cluster measured by its absorbance at 415 nm. Ti(III) was used for the stepwise reduction.
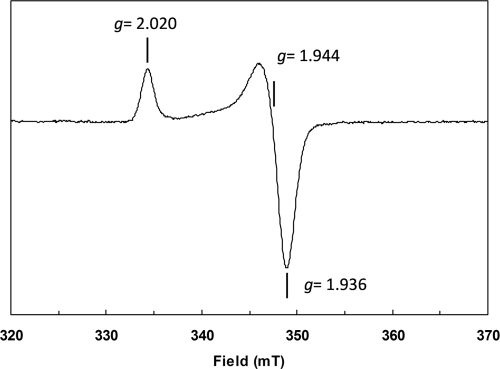
At temperatures of between 4.2 and 60 K, the sample of rAE reduced with 2 mM dithionite exhibited a very intense sharp axial EPR signal with g values of 2.020, 1.944, and 1.936 (Fig. 6). The line shape and g values are in complete agreement with those found for well-characterized cysteinate-coordinated [2Fe-2S]1+ clusters (58, 64) and are different from Rieske-type [2Fe-2S]1+ clusters in proteins (4). Most importantly, the microwave power of half saturation at 10 K of 0.10 ± 0.02 mW, detectability, and absence of broadening at up to 60 K is very typical for [2Fe-2S]1+ clusters and unlike any known [4Fe-4S]1+ cluster (44). Double integration and comparison with a Cu2+ standard showed that 0.85 [2Fe-2S]1+ cluster per rAE was present. Reduction with deazaflavin and light (34) yielded an EPR spectrum identical in shape and g values but with 89% of the intensity of the spectrum obtained upon sodium dithionite reduction (not shown). In conjunction with the UV/visible spectroscopy and Fe/S analysis, the presence of a single [2Fe-2S]2+/1+ cluster is indicated by all experimental data available.
FIG. 6.
X-band EPR spectrum of dithionite-reduced rAE. Freshly reconstituted rAE (0.3 mg/ml in 100 mM Tris Cl, pH 8.0) was reduced with 2 mM sodium dithionite (final concentration). The sample was shock frozen after 2 min by immersion in liquid nitrogen. Measurement conditions: temperature, 10 K; microwave power, 3.2 μW; modulation frequency, 100 kHz; modulation amplitude, 0.8 mT; microwave frequency, 9.459 GHz.
The activity of the rAE was measured by following the reduction of the CP from the cob(II)alamin to the cob(I)alamin form (50). Both native and rCPs of the two O-demethylases could serve as substrates for AE. rAE catalyzed the ATP-dependent reduction of the corrinoid cofactor of the CP with Ti(III) as an artificial electron donor up to 0.6 U per mg protein (data not shown). Free cobalamin was not reduced. An increase of the midpoint potential of the CP of at least 200 mV compared to conditions in the absence of AE has earlier been shown with redox titrations of the CP of the veratrol-O-demethylase in the presence of native AE (50). This effect was confirmed with rAE, indicating functionality of the enzyme. In repeated redox titrations the midpoint potentials for the reduction of the corrinoid cofactor to cob(I)alamin of either native or rOdmA were increased to −320 mV ± 30 mV by the AE. In the absence of the AE and ATP, native as well as rOdmA could not be reduced to [CoI]-CP with titanium(III) citrate at potentials of as low as −545 mV (data not shown).
Complete O-demethylase reaction and interaction of the components.
The complete O-demethylase reaction (methyl transfer from the phenyl methyl ether to FH4) was measured using the coupled-enzyme assay described earlier by Messmer et al. (35). In this assay, the formation of methyl-FH4 mediated by the protein components of the O-demethylases is monitored by the conversion of methyl-FH4 via methylene-FH4 to methenyl-FH4 upon concomitant reduction of NADP+ to NADPH. The reaction was started by addition of the respective substrate or of one of the four heterologously expressed protein components. After the start of the reaction, an increase of the absorption at 350 nm, indicating the formation of methenyl-FH4 and NADPH, could be observed (data not shown). Hence, the heterologously expressed protein components mediated the complete vanillate- and veratrol-O-demethylase reactions.
The complete O-demethylase reaction requires the interaction of at least three of the four components involved. In addition, an interaction of AE with its substrate, CP, should occur. Preliminary indications for an interaction of AE and CP were based on affinity chromatography using cell extracts of A. dehalogenans with attached AE as bait as well as a copurification of both proteins (data not shown). Native PAGE was used to investigate the interaction of recombinant vanillate-O-demethylase components. In native PAGE with rAE and the rCP of the vanillate-O-demethylase (rCPvan) using different stoichiometries of AE and CP (10:1 to 1:10), three bands were observed, corresponding to about 180 kDa (Fig. 7A, band a), 130 kDa (Fig. 7A, band b), and 25 kDa (Fig. 7A, band c). The AE alone appears to migrate as a 130-kDa homodimer (2 × 65 kDa; see also reference 28). CP is a monomeric protein of 25 kDa. The 180-kDa band is most probably a protein complex formed from two AEs and two CPs. A stoichiometry of 1:1 of AE and CP is supported by the finding that the 1:1 mixture of AE and CP yields only one major band at 180 kDa; almost no silver stain was observed at 25 or 130 kDa (Fig. 7A, lane 3). Using Western blot analysis with antibodies directed against AE or CP, both proteins were detected in the 180-kDa band (shown for AE in Fig. 7B). The formation of this band was observed only in the presence of vitamin B12. When AE was preincubated with the recombinant nonreconstituted CP in the absence of vitamin B12, no gel shift was observed (Fig. 7B, lane −B12), indicating that the holo-CP was required for the interaction. Under the same experimental conditions, all four components of the vanillate-O-demethylase were mixed in a 1:1:1:1 stoichiometry. The only bands observed were those of the single components and the AE-CP complex. This result points to a strong interaction of AE and CP.
FIG. 7.
Gel shift experiments to demonstrate interaction of AE and CP (recombinant and reconstituted). (A) Native PAGE of rAE-rCP mixtures. The purified components (20 pmol each of rAE [lane 6] or rCP [lane 7]) or rAE-rCP mixtures with AE/CP ratios (pmol:pmol) of 100:10 (lane 1), 50:10 (lane 2), 10:10 (lane 3), 10:50 (lane 4), and 10:100 (lane 5) were separated by native PAGE (8%) after preincubation (2 h, 10°C) under anoxic conditions. The gel was silver stained. Band a, AE-CP complex; band b, AE; band c, CP. The molecular masses of the marker proteins used are indicated on the left. (B) Influence of the corrinoid cofactor on the interaction. Mixtures of rAE (2 pmol) and rCP (100 pmol; without [−B12] and with [+B12] the corrinoid cofactor hydroxocobalamin) were separated by native PAGE (8%) after preincubation (2 h, 10°C) under anoxic conditions. For protein detection, specific antibodies against AE were used.
DISCUSSION
The O-demethylases of homoacetogenic bacteria are corrinoid-dependent MTs which are key enzymes in the anaerobic metabolism of phenyl methyl ethers. Two different O-demethylases have been purified from Acetobacterium dehalogenans and were partially characterized (14, 25). Both exhibit a rather broad substrate spectrum, which is useful for the organism when utilizing phenyl methyl ethers with different substituents resulting from lignin degradation. From experiments with growing cells and cell extracts of these organism, it is known that A. dehalogenans has the ability to demethylate a wide range of phenyl methyl ethers in addition to the growth substrates used (14, 25). Similar observations were described for other homoacetogenic bacteria, such as Sporomusa ovata (52, 53) and Moorella thermoacetica (11, 63). The O-demethylases of A. dehalogenans are organized in different operons containing the genes for MT II, CP, and MT I. The protein sequences of the two CPs and MTs II seem to be rather conserved, whereas the sequences of the ether-cleaving MTs I have only short regions of homology. These findings are not surprising, since it must be assumed that MT I is responsible for the substrate specificity with respect to the phenyl methyl ether, whereas MT II mediates the conversion of structurally similar substrates (methylated CP). The high identity of the two CPs may be explained by the finding that only one AE mediates the reduction of the CPs and therefore may require high structural similarity of these substrates. The similarity of the two MTs II and the CP is likely the reason why these components can be combined with each of the two MTs I in the O-demethylase assay.
Assuming a similar genetic structure for other homoacetogenic O-demethylases, the analysis of the genome sequence of M. thermoacetica (GenBank accession no. NC_007644) showed at least four gene clusters with genes coding for proteins similar to OdmD and OdmA. The putative gene products of M. thermoacetica share about 56% (OdmD) and 60 to 70% (OdmA) similarity to the corresponding protein sequences of A. dehalogenans. Thus, the broad substrate range for growth of M. thermoacetica on phenyl methyl ethers (11) seems to be accomplished by only a few O-demethylases.
In this communication, a “reductive activator for corrinoid enzymes” (RACE) was identified for the first time and assigned to the COG3894 protein family. Most of these COG3894-harboring proteins seem to be involved in anaerobic MT reactions. In the acetogenic bacterium M. thermoacetica, a gene encoding the AE-like COG3894-harboring protein with an N-terminal binding motif for a [2Fe-2S] cluster (Fig. 3B, example 5) is located within the CODH/ACS operon (acs operon). It exhibits 30% identity (50% similarity) to OdmC of A. dehalogenans. A role of this putative protein in the acetyl-CoA pathway is unknown so far (32). In this pathway, an MT mediates the transfer of the methyl group from methyl-FH4 to the ACS via a corrinoid iron-sulfur protein (CFeSP) (37). It has been proposed that for the reduction of inadvertently oxidized corrinoid in CFeSP, the electrons are transferred from an unknown electron donor to cobalt via the [4Fe-4S] cluster of CFeSP (43). To our knowledge, an ATP requirement for the CFeSP reduction has not been reported. Here we propose that a RACE-type protein is involved in the reduction and/or the activation of the corrinoid in CFeSP. The acetyl-CoA pathway is the most important CO2 fixation pathway in nonphototrophic anaerobic bacteria. Therefore, it is expected that anaerobes using this pathway for carbon assimilation should involve RACEs. An example of such an organism is Carboxydothermus hydrogenoformans (54), where the acs operon also was found to contain a RACE-encoding gene.
Various domain architectures for COG3894 proteins were encountered in database searches (Fig. 3B). The “minimal” version of a protein similar to AE has 521 to 537 amino acids and lacks the ∼110-amino-acid N-terminal domain proposed to bind the [2Fe-2S] cluster in the A. dehalogenans AE (Fig. 3B, example 1). The genetic context was heterogeneous and did not allow derivation of a function. In other proteins (Fig. 3B, example 2), an N-terminal domain of, e.g., 220 amino acids was fused to the [2Fe-2S] and COG3894 domains (25% identity with A. dehalogenans AE). This N-terminal domain has 25% amino acid sequence identity with a methionine synthase-associated S-adenosylmethionine binding protein from Thermotoga maritima (Tm0269, PDB 1J6R) (24). Another domain organization was encountered in 16 COG3894-harboring proteins of methanogenic Archaea encoded by genes of which the majority (13 hits) lie in the vicinity of gene clusters known to involve methanol, N-MT, or S-MT systems. In these COG3894 proteins no N-terminal extension with a proposed [2Fe-2S] cluster binding motif was seen. In contrast, two C-X2/4-C-X2-C-X3-C-P patterns typical of 2x [4Fe-4S] ferredoxins were present in the approximately 80 amino acids fused to the C terminus (Fig. 3B). Apparently, all Archaea appear to contain the latter type of COG3894-harboring proteins, whereas all Bacteria seem to possess COG3894 proteins with an N-terminal [2Fe-2S] cluster.
Other soluble Fe/S enzymes which catalyze a thermodynamically unfavorable reduction driven by ATP are known. Such enzymes have been referred to as “archerases” (6). Two examples are the 2-hydroxyglutaryl-CoA dehydratase (21) and the 2-hydroxyisocaproyl-CoA dehydratase (28) of clostridia. These enzymes do not belong to the COG3894 protein family and neither reduce corrinoid cofactors nor show significant similarity with the RACE proteins. In the reaction mediated by these archerases, an electron is transferred to an Fe/S cluster, and upon hydrolysis of two ATPs, the redox potential of this reduced Fe/S cluster is decreased considerably so that the electrons can easily be transferred to the dehydratase subunit. In contrast, the RACE of the O-demethylase of A. dehalogenans increases the redox potential of its target, the corrinoid cofactor of CP, so that the electrons of the Fe/S cluster of AE (midpoint potential of ≈−330 mV at pH 7.5) can be transferred to the CP with an increased midpoint potential (≈−320 mV at pH 7.5). These findings lead to the conclusion that the corrinoid-independent archerases differ in structure, function, and gene sequence from the RACE proteins.
The functional expression of the O-demethylase components now allows further studies of the structure and function of these proteins. In principle, all of the recombinant components have properties similar to those of their native counterparts with respect to, e.g., the specific activities and substrate spectrum of MT I, the increase of the midpoint potential of CP in the presence of AE, and the inability of MT I and MT II to methylate/demethylate free cobalamin/methylcobalamin, respectively. The latter characteristic appears to be a unique property of the Acetobacterium O-demethylases, since the methanogenic methanol:coenzyme M MT of Methanosarcina barkeri and the vanillate-O-demethylase of Moorella thermoacetica were reported to use free cobalamin as a methyl acceptor (38, 47).
Novel findings derived from studies of the recombinant proteins were the detection of cofactors of the AE (Fe/S) and the MT I (Zn) as well as the interaction of AE and CP. Both MTs I contained 1 mol zinc per mol protein. In this respect the enzymes are similar to the methanogenic MTs, which also were found to contain zinc (46). In contrast, zinc was not detected in stoichiometric amounts in the MT I of the vanillate-O-demethylase of M. thermoacetica (38).
A possible interaction between the O-demethylase components was studied in gel shift experiments. These studies revealed the formation of a protein complex consisting of two AEs and two CPs. Since CP is the substrate for AE, this is not surprising. The presence of the corrinoid cofactor is essentially required for the interaction (Fig. 7B). This may reflect either an exposed surface position of the corrinoid in CP and a direct involvement of the corrinoid in the interaction or a correct CP folding, which might occur exclusively in the presence of the cofactor, leading to a tertiary structure that enables interaction. An exposed position of the corrinoid cofactor has been deduced from crystal structures of the corrinoid-containing methionine synthase of E. coli (13) and of the methanol:coenzyme M MT of M. barkeri (20). Since CP of vanillate-O-demethylase has a high homology to methionine synthase with respect to the corrinoid binding domain, a surface position of the corrinoid in CP is feasible. On the other hand, a correct folding only in the presence of the corrinoid is also very likely. Hence, both explanations seem reasonable, and both may apply. Other interactions of O-demethylase components were not observed, although an interaction in vivo is very likely. It is feasible that the interaction is too weak to be monitored by gel shift experiments. Further studies of the interaction of all O-demethylase components using a different experimental approach are currently under way in our laboratory.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft and the EU.
Rolf K. Thauer (Max Planck Institut für Terrestrische Mikrobiologie, Marburg, Germany) is gratefully acknowledged for the use of his EPR spectrometer. We thank Dirk Merten (Institute of Applied Geology, Friedrich-Schiller-University, Jena) for the metal analysis, Bernhard Schlott (Leibniz Institute for Age Research Fritz Lipmann, Jena) for the protein sequencing of AE, and Yvonne Greiser for skillful technical assistance.
Footnotes
Published ahead of print on 14 November 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bache, R., and N. Pfennig. 1981. Selective isolation of Acetobacterium woodii on methoxylated aromatic acids and determination of growth yields. Arch. Microbiol. 130255-261. [Google Scholar]
- 3.Banerjee, R., and S. W. Ragsdale. 2003. The many faces of vitamin B12: catalysis by cobalamin-dependent enzymes. Annu. Rev. Biochem. 72209-247. [DOI] [PubMed] [Google Scholar]
- 4.Bertrand, P., B. Guigliarelli, J.-P. Gayda, P. Beardwood, and J. F. Gibson. 1985. A ligand-field model to describe a new class of 2Fe-2S clusters in proteins and their synthetic analogues. Biochim. Biophys. Acta 831261-266. [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilising the principle of protein dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 6.Buckel, W., B. M. Martins, A. Messerschmidt, and B. T. Golding. 2005. Radical-mediated dehydration reactions in anaerobic bacteria. Biol. Chem. 386951-959. [DOI] [PubMed] [Google Scholar]
- 7.Burke, S. A., and J. A. Krzycki. 1997. Reconstitution of monomethylamin:coenzyme M methyl transfer with a corrinoid protein and two methyltransferases purified from Methanosarcina barkeri. J. Bacteriol. 27216570-16577. [DOI] [PubMed] [Google Scholar]
- 8.Burke, S. A., S. L. Lo, and J. A. Krzycki. 1998. Clustered genes encoding the methyltransferases of methanogenesis from monomethylamine. J. Bacteriol. 1803432-3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daas, P. H. J., K. A. A. Gerrits, J. T. Keltjens, C. van der Drift, and G. D. Vogels. 1993. Involvement of an activation protein in the methanol:2-mercaptoethanesulfonic acid methyltransferase reaction in Methanosarcina barkeri. J. Bacteriol. 1751278-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daas, P. H. J., W. R. Hagen, J. T. Keltjens, C. van der Drift, and G. D. Vogels. 1996. Purification and properties of an enzyme involved in ATP-dependent activation of the methanol:5-hydroxybenzimidazolylcobamide methyltransferase from Methanosarcina barkeri. J. Biol. Chem. 27122339-22345. [DOI] [PubMed] [Google Scholar]
- 11.Daniel, S. L., E. S. Keith, H. Yang, Y. Lin, and H. L. Drake. 1991. Utilization of methoxylated aromatic compounds by the acetogen Clostridium thermoaceticum: expression and specificity of the CO-dependent O-demethylating activity. Biochem. Biophys. Res. Commun. 180416-422. [DOI] [PubMed] [Google Scholar]
- 12.De Castro, E., C. J. A. Sigrist, A. Gattiker, V. Bulliard, P. S. Langendijk-Genevaux, E. Gasteiger, A. Bairoch, and N. Hulo. 2006. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34W362-W365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drennan, C. L., S. Huang, J. T. Drummond, R. G. Matthews, and M. L. Ludwig. 1994. How a protein binds B12: a 3.0 Å X-ray structure of B12-binding domains of methionine synthase. Science 2661669-1674. [DOI] [PubMed] [Google Scholar]
- 14.Engelmann, T., F. Kaufmann, and G. Diekert. 2001. Isolation and characterization of a veratrol:corrinoid protein methyltransferase from Acetobacterium dehalogenans. Arch. Microbiol. 175376-383. [DOI] [PubMed] [Google Scholar]
- 15.Ferguson, D. J., and J. A. Krzycki. 1997. Reconstitution of trimethylamine-dependent coenzyme M methylation with the trimethylamine corrinoid protein and the isozymes of methyltransferase II from Methanosarcina barkeri. J. Bacteriol. 179846-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferguson, D. J., J. A. Krzycki, and D. A. Grahame. 1996. Specific roles of methylcobamide:coenzyme M methyltransferase isozymes in metabolism of methanol and methylamines in Methanosarcina barkeri. J. Biol. Chem. 2715189-5194. [DOI] [PubMed] [Google Scholar]
- 17.Fish, W. W. 1988. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Methods Enzymol. 158357-364. [DOI] [PubMed] [Google Scholar]
- 18.Friedrich, W. 1975. Vitamin B12 und verwandte Corrinoide, p. 47-55. In R. Ammon and W. Discherl (ed.), Fermente, Hormone, Vitamine. Georg Thieme Verlag, Stuttgart, Germany.
- 19.Gilboa-Garber, N. 1971. Direct spectrophotometric determination of inorganic sulfide in biological material and in other complex mixtures. Anal. Biochem. 43129-133. [DOI] [PubMed] [Google Scholar]
- 20.Hagemeier, C. H., M. Krüer, R. K. Thauer, E. Warkentin, and U. Ermler. 2006. Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex. Proc. Natl. Acad. Sci. USA 10318917-18922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hans, M., J. Sievers, U. Müller, E. Bill, J. A. Vorholt, D. Linder, and W. Buckel. 1999. 2-Hydroxyglutaryl-CoA dehydratase from Clostridium symbosium. Eur. J. Biochem. 265404-414. [DOI] [PubMed] [Google Scholar]
- 22.Hao, B., W. Gong, T. K. Ferguson, C. M. James, J. A. Krzycki, and M. K. Chan. 2002. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science 2961462-1466. [DOI] [PubMed] [Google Scholar]
- 23.Hovey, R., S. Lentes, A. Ehrenreich, K. Salmon, K. Saba, G. Gottschalk, R. P. Gunsalus, and U. Deppenmeier. 2005. DNA microarray analysis of Methanosarcina mazei Gö1 reveals adaptation to different methanogenic substrates. Mol. Gen. Genomics 273225-239. [DOI] [PubMed] [Google Scholar]
- 24.Huang, S., G. Romanchuk, K. Pattridge, S. A. Lesley, I. A. Wilson, R. G. Matthews, and M. Ludwig. 2007. Reactivation of methionine synthase from Thermotoga maritima (TM0268) requires the downstream gene product TM0269. Protein Sci. 161588-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaufmann, F., G. Wohlfarth, and G. Diekert. 1997. Isolation of O-demethylase, an ether-cleaving enzyme system of the homoacetogenic strain MC. Arch. Microbiol. 166136-142. [DOI] [PubMed] [Google Scholar]
- 26.Kaufmann, F., G. Wohlfarth, and G. Diekert. 1998. O-Demethylase from Acetobacterium dehalogenans. Substrate specificity and function of the participating proteins. Eur. J. Biochem. 253706-711. [DOI] [PubMed] [Google Scholar]
- 27.Kaufmann, F., G. Wohlfarth, and G. Diekert. 1998. O-Demethylase from Acetobacterium dehalogenans—cloning, sequencing and active expression of the gene encoding the corrinoid protein. Eur. J. Biochem. 257515-521. [DOI] [PubMed] [Google Scholar]
- 28.Kim, J., D. Darley, and W. Buckel. 2005. 2-Hydroxyisocaproyl-CoA dehydratase and its activator from Clostridium difficile. FEBS J. 272550-561. [DOI] [PubMed] [Google Scholar]
- 29.Kim, J., D. J. Darley, W. Buckel, and A. J. Pierik. 2008. An allylic ketyl radical intermediate in clostridial amino-acid fermentation. Nature 452239-242. [DOI] [PubMed] [Google Scholar]
- 30.Kreher, S., A. Schilhabel, and G. Diekert. 2008. Enzymes involved in the anoxic utilization of phenyl methyl ethers by Desulfitobacterium hafniense DCB-2 and Desulfitobacterium hafniense PCE-S. Arch. Microbiol. 190489-495. [DOI] [PubMed] [Google Scholar]
- 31.Krzycki, J. A. 2004. Function of genetically encoded pyrrolysine in corrinoid dependent methylamine methyltransferases. Curr. Opin. Chem. Biol. 8484-491. [DOI] [PubMed] [Google Scholar]
- 32.Loke, H.-K., and P. A. Lindahl. 2003. Identification and preliminary characterization of AcsF, a putative Ni-insertase used in the biosynthesis of acetyl-CoA synthase from Clostridium thermoaceticum. J. Inorg. Biochem. 9333-40. [DOI] [PubMed] [Google Scholar]
- 33.Markowitz, V. M., and N. C. Kyrpides. 2007. Comparative genome analysis in the integrated microbial genomes (IMG) system. Methods Mol. Biol. 39535-56. [DOI] [PubMed] [Google Scholar]
- 34.Massey, V., and P. Hemmerich. 1978. Photoreduction of flavoproteins and other biological compounds catalyzed by deazaflavin. Biochemistry 179-16. [DOI] [PubMed] [Google Scholar]
- 35.Messmer, M., S. Reinhardt, G. Wohlfarth, and G. Diekert. 1996. Studies on methyl chloride dehalogenase and O-demethylase in cell extracts of the homoacetogen strain MC based on a newly developed coupled enzyme assay. Arch. Microbiol. 16518-25. [Google Scholar]
- 36.Meyer, J. 1988. The evolution of ferredoxins. Trends Ecol. Evol. 3222-226. [DOI] [PubMed] [Google Scholar]
- 37.Müller, V. 2003. Energy conservation in acetogenic bacteria. Appl. Environ. Microbiol. 696345-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naidu, D., and S. W. Ragsdale. 2001. Characterization of a three-component vanillate O-demethylase from Moorella thermoacetica. J. Bacteriol. 1833276-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neumann, A., T. Engelman, R. Schmitz, Y. Greiser, A. Orthaus, and G. Diekert. 2004. Phenyl methyl ethers: novel electron donors for respiratory growth of Desulfitobacterium hafniense and Desulfitobacterium sp. strain PCE-S. Arch. Microbiol. 181245-249. [DOI] [PubMed] [Google Scholar]
- 40.Nonaka, H., G. Keresztes, Y. Shinoda, Y. Ikenaga, M. Abe, K. Naito, K. Inatomi, K. Furukawa, M. Inui, and H. Yukawa. 2006. Complete genome sequence of the dehalorespiring bacterium Desulfitobacterium hafniense Y51 and comparison with Dehalococcoides ethenogenes 195. J. Bacteriol. 1882262-2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul, L., D. J. Ferguson, and J. A. Krzycki. 2000. The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons. J. Bacteriol. 1822520-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pritchett, M. A., and M. W. Metcalf. 2005. Genetic, physiological and biochemical characterization of multiple methanol methyltransferase isozymes in Methanosarcina acetivorans C2A. Mol. Microbiol. 561183-1194. [DOI] [PubMed] [Google Scholar]
- 43.Ragsdale, S. W. 2004. Life with carbon monoxide. Crit. Rev. Biochem. Mol. Biol. 39165-195. [DOI] [PubMed] [Google Scholar]
- 44.Rupp, H., K. K. Rao, D. O. Hall, and R. Cammack. 1978. Electron spin relaxation of iron-sulphur proteins studied by microwave power saturation. Biochim. Biophys. Acta 537255-269. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Sauer, K., and R. K. Thauer. 1997. Methanol:coenzyme M methyltransferase from Methanosarcina barkeri. Zinc dependence and thermodynamics of the methanol:cob(I)alamin methyltransferase reaction. Eur. J. Biochem. 249280-285. [DOI] [PubMed] [Google Scholar]
- 47.Sauer, K., and R. K. Thauer. 1999. Methanol:coenzyme M methyltransferase from Methanosarcina barkeri—substitution of the corrinoid harbouring subunit MtaC by free cob(I)alamin. Eur. J. Biochem. 261674-681. [DOI] [PubMed] [Google Scholar]
- 48.Sauer, K., U. Harms, and R. K. Thauer. 1997. Methanol:coenzyme M methyltransferase from Methanosarcina barkeri. Purification, properties and encoding genes of the corrinoid protein MT1. Eur. J. Biochem. 243670-677. [DOI] [PubMed] [Google Scholar]
- 49.Schink, B., A. Brune, and S. Schnell. 1992. Anaerobic degradation of aromatic compounds, p. 219-242. In G. Winkelmann (ed.), Microbial degradation of natural products. VCH, New York, NY.
- 50.Siebert, A., T. Schubert, T. Engelmann, S. Studenik, and G. Diekert. 2005. Veratrol-O-demethylase of Acetobacterium dehalogenans: ATP dependent reduction of the corrinoid protein. Arch. Microbiol. 183378-384. [DOI] [PubMed] [Google Scholar]
- 51.Srinivasan, G., C. M. James, and J. A. Krzycki. 2002. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science 2961459-1462. [DOI] [PubMed] [Google Scholar]
- 52.Stupperich, E., and R. Konle. 1993. Corrinoid-dependent methyl transfer reactions are involved in methanol and 3,4-dimethoxybenzoate metabolism by Sporomusa ovata. Appl. Environ. Microbiol. 593110-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stupperich, E., R. Konle, and C. Eckerskorn. 1996. Anaerobic O-demethylations of methoxynaphthols, methoxyfuran, and fluoroanisols by Sporomusa ovata. Biochem. Biophys. Res. Commun. 223770-777. [DOI] [PubMed] [Google Scholar]
- 54.Svetlitchnaia, T., V. Svetlitchnyi, O. Meyer, and H. Dobbek. 2006. Structural insights into methyltransfer reactions of a corrinoid iron-sulfur protein involved in acetyl-CoA synthesis. Proc. Natl. Acad. Sci. USA 10314331-14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tallant, T. C., and J. A. Krzycki. 1997. Methylthiol:coenzyme M methyltranferase from Methanosarcina barkeri, an enzyme of methanogenesis from dimethylsulfide and methylmercaptopropionate. J. Bacteriol. 1796902-6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tallant, T. C., L. Paul, and J. A. Krzycki. 2001. The MtsA subunit of the methylthiol:coenzyme M methyltransferase of Methanosarcina barkeri catalyzes both half-reactions of corrinoid-dependent dimethylsulfide:coenzyme M methyl transfer. J. Biol. Chem. 2764485-4493. [DOI] [PubMed] [Google Scholar]
- 57.Traunecker, J., A. Preuss, and G. Diekert. 1991. Isolation and characterization of a methyl chloride utilizing, strictly anaerobic bacterium. Arch. Microbiol. 156416-421. [Google Scholar]
- 58.Türk, K., A. Puhar, F. Neese, E. Bill, G. Fritz, and J. Steuber. 2004. NADH oxidation by the Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae: functional role of the NqrF subunit. J. Biol. Chem. 27921349-21355. [DOI] [PubMed] [Google Scholar]
- 59.van der Meijden, P., B. W. Brommelstroet, C. M. Poirot, C. van der Drift, and G. D. Vogels. 1984. Purification and properties of methanol:5-hydroxybenzimidazolylcobamide methyltransferase from Methanosarcina barkeri. J. Bacteriol. 160629-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van de Wijngaard, W. M. H., R. L. Lugtigheid, and C. van der Drift. 1991. Reductive activation of the corrinoid-containing enzyme involved in methyl group transfer between methyl-tetrahydromethanopterin and coenzyme M in Methanosarcina barkeri. Antonie van Leeuwenhoek. 601-6. [DOI] [PubMed] [Google Scholar]
- 61.Veit, K., C. Ehlers, and R. A. Schmitz. 2005. Effects of nitrogen and carbon sources on transcription of soluble methyltransferases in Methanosarcina mazei strain Gö1. J. Bacteriol. 1876147-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wassenaar, R. W., J. T. Keltjens, C. van der Drift, and G. D. Vogels. 1998. Purification and characterization of dimethylamine:5-hydroxybenzimidazolylcobamide methyltransferase from Methanosarcina barkeri Fusaro. Eur. J. Biochem. 253692-697. [DOI] [PubMed] [Google Scholar]
- 63.Wu, Z., S. L. Daniel, and H. L. Drake. 1988. Characterization of a CO-dependent O-demethylating enzyme system from the acetogen Clostridium thermoaceticum. J. Bacteriol. 1705747-5750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xia, B., H. Cheng, V. Bandarian, G. H. Reed, and J. L. Markley. 1996. Human ferredoxin: overproduction in Escherichia coli, reconstitution in vitro, and spectroscopic studies of iron-sulfur cluster ligand cysteine-to-serine mutants. Biochemistry 359488-9495. [DOI] [PubMed] [Google Scholar]
- 65.Zehnder A. J., and K. Wuhrmann. 1976. Titanium (III) citrate as a nontoxic oxidation-reduction buffering system for the culture of obligate anaerobes. Science 194:1165-1166. [DOI] [PubMed] [Google Scholar]
- 66.Zhou, Z. S., K. Peariso, J. E. Penner-Hahn, and R. G. Matthews. 1999. Identification of zinc ligands in cobalamin-independent methionine synthase (MetE) from Escherichia coli. Biochemistry 3815915-15926. [DOI] [PubMed] [Google Scholar]