Abstract
Rapid detection and differentiation of Taenia species are required for the control and prevention of taeniasis and cysticercosis in areas where these diseases are endemic. Because of the lower sensitivity and specificity of the conventional diagnosis based on microscopical examination, molecular tools are more reliable for differential diagnosis of these diseases. In this study, we developed and evaluated a loop-mediated isothermal amplification (LAMP) assay for differential diagnosis of infections with Taenia species with cathepsin L-like cysteine peptidase (clp) and cytochrome c oxidase subunit 1 (cox1) genes. LAMP with primer sets to the cox1 gene could differentiate between three species, and LAMP with primer sets to the clp gene could differentiate Taenia solium from Taenia saginata/Taenia asiatica. Restriction enzyme digestion of the LAMP products from primer set Tsag-clp allowed the differentiation of Taenia saginata from Taenia asiatica. We demonstrated the high specificity of LAMP by testing known parasite DNA samples extracted from proglottids (n = 100) and cysticerci (n = 68). LAMP could detect one copy of the target gene or five eggs of T. asiatica and T. saginata per gram of feces, showing sensitivity similar to that of PCR methods. Furthermore, LAMP could detect parasite DNA in all taeniid egg-positive fecal samples (n = 6). Due to the rapid, simple, specific, and sensitive detection of Taenia species, the LAMP assays are valuable tools which might be easily applicable for the control and prevention of taeniasis and cysticercosis in countries where these diseases are endemic.
Cestode parasites Taenia solium, Taenia saginata, and Taenia asiatica are the causative agents of taeniasis. Although taeniasis is relatively innocuous, cysticercosis caused by T. solium larvae is one of the most severe diseases in humans and remains a complicated health problem in many areas around the world, especially in developing countries (4, 15). Therefore, differentiation of Taenia species becomes significant for epidemiological studies and for control of these diseases. Furthermore, it is expected that there are much wider areas in the Asia-Pacific region where the three taeniid species occur sympatrically (3, 5, 23, 24). Diagnosis is mainly performed by microscopic observation of eggs in feces and/or by comparative morphology of proglottids or scolices, but these methods lack both sensitivity and specificity. In order to overcome the lower sensitivity of microscopic diagnosis, various immunological or molecular approaches, including coproantigen and copro-DNA detection methods, have been developed (2, 6, 12, 18, 25). The coproantigen detection method has been shown to be more sensitive than the microscopy method, but it cannot differentiate between Taenia species because of it is genus specific, not species specific (2). By contrast, various copro-DNA detection methods using PCR have been developed for sensitive differential detection of taeniid cestodes (6, 12, 18, 25). Although these techniques provide sensitive and reliable diagnostic results, it is not easy to exploit in the laboratories of developing countries where these diseases are endemic, because PCR requires sophisticated equipment, such as a thermal cycler. Furthermore, Taq DNA polymerase is often inactivated by inhibitors present in biological samples, which sometimes cause problems for sensitivity and reproducibility (1, 13).
Recently, a novel nucleic acid amplification method termed loop-mediated isothermal amplification (LAMP) has been developed (17). LAMP employs a DNA polymerase with strand displacement activity and four primers that recognize six sequences on the target DNA. This method amplifies DNA with high specificity, sensitivity, and rapidity under isothermal conditions. Since LAMP is done under isothermal conditions (60 to 65°C), simple incubators, such as a water bath or a block heater, are sufficient for DNA amplification (17). Moreover, a large amount of white precipitate of magnesium pyrophosphate is produced as a byproduct, which enables the visual judgment of amplification by a naked eye (14). Hence, LAMP is a highly sensitive and specific DNA amplification tool suitable for the rapid diagnosis of infectious diseases, including parasitic diseases (9, 20, 22), in a well-equipped laboratory and/or small-scale clinical laboratories and is expected to be highly useful and feasible in the field.
In the present study, we developed and evaluated the LAMP assay with a cathepsin L-like cysteine peptidase (clp) gene of nuclear DNA and a cytochrome c oxidase subunit 1 (cox1) gene of mitochondrial DNA for differentiation between and rapid diagnosis of infection with Taenia species.
MATERIALS AND METHODS
Parasite materials.
Cysticerci of T. solium, T. saginata, or T. asiatica were obtained from nonobese diabetic/severe combined immunodeficiency (NOD/shi-scid) mice (7, 16) infected by intraperitoneal cavity injection with oncospheres prepared from gravid proglottids of each parasite isolated in Thailand. Genomic DNA was extracted from one cysticercus by using a DNeasy tissue kit (Qiagen, Hilden, Germany).
DNA samples.
For evaluation of the LAMP assay, a total of 168 DNA samples extracted from proglottids (n = 100) and cysticerci (n = 68), including 47 T. solium samples, 78 T. saginata samples, and 43 T. asiatica samples, were examined (Table 1). We used the stored DNA samples previously analyzed by multiplex PCR (25).
TABLE 1.
Taenia species DNA samples used in this study
| Taenia species (no. of samples) | Locality | Developmental stage (no. of samples analyzed) |
|---|---|---|
| T. saginata (78) | Cambodia | Proglottid (3) |
| China | Proglottid (5) | |
| Cysticercus (1) | ||
| Indonesia | Proglottid (36) | |
| Cysticercus (12)a | ||
| Nepal | Proglottid (2) | |
| Thailand | Proglottid (9) | |
| Cameroon | Proglottid (1) | |
| Ethiopia | Proglottid (2) | |
| Brazil | Cysticercus (2) | |
| Proglottid (1) | ||
| Ecuador | Proglottid (1) | |
| Belgium | Proglottid (2) | |
| Cysticercus (1)a | ||
| T. asiatica (43) | China | Cysticercus (1)a |
| Proglottid (10) | ||
| Indonesia | Cysticercus (2) | |
| Cysticercus (16)a | ||
| Korea | Cysticercus (2)a | |
| Taiwan | Cysticercus (1)a | |
| Proglottid (1) | ||
| Thailand | Proglottid (6) | |
| Philippines | Proglottid (4) | |
| T. solium (47) | China | Cysticercus (2) |
| Proglottid (11) | ||
| India | Cysticercus (11) | |
| Indonesia | Proglottid (1) | |
| Nepal | Cysticercus (5) | |
| Thailand | Proglottid (5) | |
| Vietnam | Cysticercus (1) | |
| Cameroon | Cysticercus (3) | |
| Mozambique | Cysticercus (2) | |
| South Africa | Cysticercus (1) | |
| Tanzania | Cysticercus (1) | |
| Brazil | Cysticercus (1) | |
| Ecuador | Cysticercus (2) | |
| Mexico | Cysticercus (1) |
The cysticercus was developed in NOD/shi-scid mice.
Cloning and sequencing of clp genes.
The clp gene of each Taenia parasite was cloned by PCR with forward primer 5′-ACATTTTCGTTTCGATCGGTCATG-3′ and reverse primer 5′-TGAACACATGGTTTAAACGTATGG-3′. Primers used were designed from conserved nucleotide sequences between Echinococcus multilocularis (21) and T. solium (10) clp genes to amplify almost the entire gene region. PCR was carried out using high-fidelity polymerase PrimeStar (Takara, Kyoto, Japan) in a final volume of 25 μl reaction mixture containing 0.2 μM of each primer, 200 μM each of deoxynucleoside triphosphate (dNTP), 0.625 units of PrimeStar DNA polymerase, and genomic DNA, as described above. Amplification was performed with 35 cycles of 94°C for 30 s, 60°C for 15 s, and 72°C for 4 min, followed by a final extension at 72°C for 10 min. A single band of approximately 3 kbp was excised from agarose gels by using a NucleoSpin ExTract kit (Macherey-Nagel, Düren, Germany) according to the instruction manual and was cloned into a pGEM-T vector (Promega, Madison, WI) after the addition of adenine to the ends of the PCR products. The plasmid clone was sequenced on an ABI Prism 310 sequencer (Applied Biosystems, Foster City, CA) with BigDye Terminator version 1.1 (AB Applied Biosystems) and was used as a standard plasmid for determining the specificity and sensitivity of LAMP.
Preparation of standard plasmids for cox1 genes.
The cox1 gene of each Taenia parasite was amplified by PCR using forward primer 5′-ATGAATGTCAAATATTTGT-TAAGTT-3′ and reverse primer 5′-CTAAAAGACCATTTCACACGCGAAT-3′ for T. solium, forward primer 5′-ATGAGTGTTAAATTTTTATTAAGTT-3′ and reverse primer 5′-TTAAACTAAAAAACCACGGGCAGGC-3′ for T. saginata, and forward primer 5′-ATGAGTGTTAAATTTTTATTAAGTT-3′ and reverse primer 5′-TTAAACTAAAAAACCACGAGCAAAC-3′ for T. asiatica. PCR was carried out as described above, except the annealing temperature was at 58°C and the elongation time was 90 s. The amplified products of each Taenia species were cloned into a pGEM-T vector and used as a standard plasmid after being confirmed by sequencing.
LAMP primers.
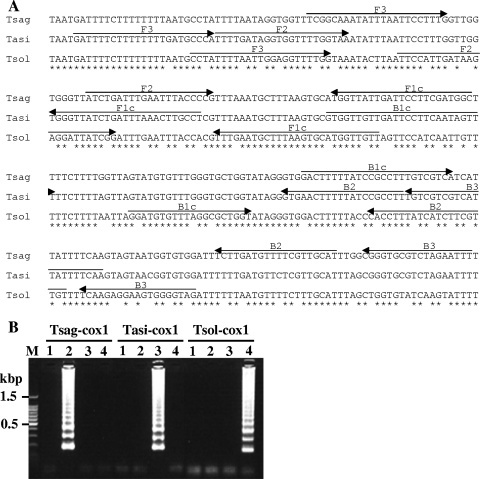
LAMP primers were designed using PrimerExplorer V4 software (http://primerexplorer.jp/). The following four oligonucleotide primers were specifically designed to amplify six distinct regions on the target gene: forward inner primer (FIP), backward inner primer (BIP), and two outer primers, F3 and B3 (Table 2). FIP consists of the sense sequence of F2 at the 3′ end and the F1c region at the 5′ end that is complementary to the F1 region. BIP consists of a B2 region at the 3′ end that is complementary to the B2c region and the same sequence as the B1c region at the 5′ end (Fig. 1A and 2 A).
TABLE 2.
LAMP primer sets
| Species | Primer set | Primer typea | Sequence (5′ to 3′) | Target |
|---|---|---|---|---|
| T. saginata | Tsag-cox1 | F3 | TCGGCAAATATTTAATTCCTTTG | cox1 gene |
| B3 | AAATTCTAGACGCACCCG | |||
| FIP | GCCATCGAAGGAATCAATAACCAATCTGATTTGAATTTACCCCG | |||
| BIP | GACTTTTTATCCGCCTTTGTCGTCAATGCAACGAAAACATCAAGA | |||
| T. asiatica | Tasi-cox1 | F3 | GATTTTCTTTTTTTTGATGCCCA | |
| B3 | TTGAAAATAATGACGACGACA | |||
| FIP | AGGCAAGTTTAAATCAGATAACCCATTTTGATAGGTGGTTTTGGTAA | |||
| BIP | GTGGTTGTTGATTCCTTCAATAGTTTAAGGCGGATAAAAAGTTCAC | |||
| T. solium | Tsol-cox1 | F3 | CCTATTTTAATTGGAGGTTTTGG | |
| B3 | CTACCCCACTTCCTCTTGA | |||
| FIP | CAACCATGCACTTAAAGCATTCAAATTCCATTGATAAGAGGATTATCGG | |||
| BIP | GGATGTGTTTAGGCGCTGGTACAACGAAGATGATAAAGGTG | |||
| T. saginata | Tsag-clp | F3 | GGAAGTCAAAAATCAGGTGAG | clp gene |
| T. asiatica | B3 | CGCTGATAGCTAGGCTAAC | ||
| FIP | CTCAGTCCCACCCAATCCATTTCAAATCTTCATTATGCTGCGTTAC | |||
| BIP | GCACCGTGTCATTGGTAAATTTGGTGGAGCTTACTAAGCTCTATCG | |||
| T. solium | Tsol-clp | F3 | GAGGTCAAAAATCAGGTGAGAT | |
| B3 | AATGCTCCTGACTTGGTT | |||
| FIP | AGGTGCTTTCACAATAGTCCCCTGCGTCATAGGTCTTGC | |||
| BIP | TAGTCGTTGCTTCGATAGAGCTCGCTGATATCTAGGCTAATGCTG |
FIP consists of the sense sequence of F2 at the 3′ end and the F1c region at the 5′ end that is complementary to the F1 region. BIP consists of a B2 region at the 3′ end that is complementary to the B2c region and the same sequence as the B1c region at the 5′ end.
FIG. 1.
Nucleotide sequence alignment of the target region of cox1 genes (A) and LAMP results (B). (A) The locations of the primer recognition sites are indicated by arrows. (B) The LAMP products were run on a 2% agarose gel. Lane M, 100-bp DNA ladder marker (Promega); lane 1, negative control; lane 2, T. saginata genomic DNA; lane 3, T. asiatica genomic DNA; lane 4, T. solium genomic DNA; Tsag-cox1, results of LAMP with primer set Tsag-cox1; Tasi-cox1, results of LAMP with primer set Tasi-cox1; Tsol-cox1, results of LAMP with primer set Tsol-cox1.
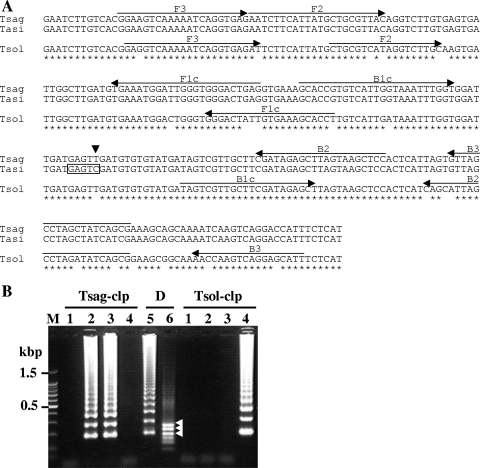
FIG. 2.
Nucleotide sequence alignment of the target region of clp genes (A) and LAMP results (B). (A) The locations of the primer recognition sites are indicated by arrows and the restriction enzyme HinfI recognition site in the LAMP products of T. asiatica clp gene is boxed. The single nucleotide substitution, C to T, in the T. saginata sequence is indicated by an arrowhead. (B) The LAMP products and HinfI digestion products were run on a 2% agarose gel. Lane M, 100-bp DNA ladder marker (Promega); lane 1, negative control; lane 2, T. saginata genomic DNA; lane 3, T. asiatica genomic DNA; lane 4, T. solium genomic DNA; lane 5, HinfI digestion of LAMP products from T. saginata genomic DNA with primer set Tsag-clp; lane 6, HinfI digestion of LAMP products from T. asiatica genomic DNA with primer set Tsag-clp; Tsag-clp, results of LAMP with primer set Tsag-clp; D, results of HinfI digestion; Tsol-clp, results of LAMP with primer set Tsol-clp. The DNA fragments generated after digestion with HinfI are indicated by arrowheads.
LAMP reaction.
LAMP was carried out in a 25-μl volume of reaction mixture containing 40 pmol each of FIP and BIP, 5 pmol each of F3 and B3, an 8-U large fragment of Bst DNA polymerase (New England Biolabs, Beverly, MA), 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 8 mM MgSO4, 10 mM (NH4)2SO4, 0.1% Tween 20, 0.8 M betaine (Sigma, St. Louis, MO), and 1.4 mM each of dNTP and the template DNA. The reaction mixture was incubated for 60 min at 63°C for the clp gene and at 60°C for the cox1 gene and heated at 80°C for 5 min. The LAMP products from primer set Tsag-clp were digested with the HinfI restriction enzyme (New England Biolabs) for 2 h at 37°C. The LAMP products and restriction enzyme-digested products were electrophoresed on a 2.0% agarose gel and detected by staining with ethidium bromide.
Sensitivity analyses.
The sensitivities of the LAMP and PCR methods were assessed using a standard plasmid diluted from 104 copies/reaction to 1. LAMP was performed as previously mentioned. PCR was carried out by using F3 and B3 primers of each LAMP primer set. PCRs were performed in a 25-μl volume of reaction mixture containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.001% gelatin, 0.2 μM of each primer, 0.2 mM each of dNTP, diluted standard plasmid, and 0.5 units of Taq DNA polymerase (ExTaq; Takara), and cycling conditions were 30 s at 94°C (first cycle, 2 min at 94°C), 30 s at 60°C (clp gene) and 58°C (cox1 gene), and 30 s at 72°C for 30 cycles. The PCR products were electrophoresed on a 2.0% agarose gel and detected by staining with ethidium bromide.
DNA extraction from fecal samples.
In order to evaluate the detection limit of taeniid DNA in fecal samples, 1 gram of feces from a noninfected volunteer was mixed with 5, 10, 20, 30, 40, or 50 T. asiatica or T. saginata eggs, and DNA was extracted by using a QIAamp DNA stool mini kit (Qiagen) after egg disruption treatment with glass beads (19). In addition, taeniid egg-positive or -negative fecal samples obtained in Indonesia and Thailand were used to assess LAMP, and DNA samples were extracted by the same procedure. LAMP was performed as previously described except that the reaction was performed for 90 min.
Nucleotide sequence accession numbers.
The nucleotide sequence data of the clp genes cloned in this study are available in the GenBank, EMBL, and DDBJ databases under accession numbers AB441815 (T. solium), AB441816 (T. saginata), and AB441817 (T. asiatica).
RESULTS AND DISCUSSION
The standard plasmid of each gene was constructed to facilitate initial evaluation and optimization of LAMP. The LAMP products are detected as a ladder of multiple bands on the gel due to the formation of a mixture of stem-loop DNAs with various stem lengths and cauliflower-like structures, with multiple loops formed by annealing between alternately inverted repeats of the target sequence in the same strand (17). Application of the LAMP assay with each specific primer set to each Taenia cox1 gene resulted in successful amplification of the target gene from the respective parasite genomic DNA (Fig. 1B). On the other hand, because of high nucleotide sequence similarity between the clp genes of T. saginata and T. asiatica, we could not design a specific primer set to differentiate between these two Taenia species but could design specific primer sets Tsol-clp and Tsag-clp to differentiate T. solium from T. saginata and T. asiatica and differentiate T. saginata/T. asiatica from T. solium, respectively. However, the recognition site GAGTC for the restriction enzyme HinfI in the amplified region of the T. asiatica clp gene (Fig. 2A) enables us to differentiate T. asiatica from T. saginata. In this case, restriction enzyme digestion of the LAMP products from T. asiatica genomic DNA with primer set Tsag-clp produces three bands with the predicted sizes of 179, 217, and 255 bp, because, unlike the PCR products, the LAMP products are characteristic structures with inverted repeats of the target sequence. In fact, three bands that agreed with the predicted size in the restriction enzyme digestion of the LAMP products from T. asiatica genomic DNA with primer set Tsag-clp were detected (Fig. 2B, lane 6).
Analytical specificity of the LAMP assays.
In order to evaluate the specificity of the LAMP assays, the known parasite DNA samples prepared from proglottids and cysticerci were examined. Because LAMP requires four primers that recognize six different sequences on a target sequence, the target sequence specificity of the LAMP reaction appears to be high. Indeed, LAMP with primer sets to the cox1 or clp gene specifically amplified each respective target gene, with a species-specific detection. The results obtained by LAMP with cox1 primer sets were consistent with those obtained by multiplex PCR with the cox1 gene, whereas two DNA samples extracted from proglottids, which were identified as being T. saginata by both LAMP and multiplex PCR with cox1 primer sets, were detected as T. asiatica by LAMP with clp primer sets (Table 3). The size of bands produced by restriction enzyme digestion indicated that these two LAMP products were specifically derived from the T. asiatica clp gene (data not shown). After cloning of the clp gene from these two DNA samples by PCR with primer set Tsag-clp F3 and B3 and sequencing, we found a single nucleotide substitution, T to C, as indicated in Fig. 2A, which leads to the appearance of the HinfI recognition site in the region between B1c and B2 and is identical to the nucleotide substitution in T. asiatica. Thus, the nucleotide substitution in the clp gene of these two T. saginata samples may lead to the different result from the diagnosis based on the cox1 gene. Alternatively, the possibility that these two DNA samples were obtained from a hybrid parasite having T. asiatica nuclear DNA and T. saginata mitochondrial DNA could not be ruled out, because samples of these parasite materials were collected from areas where both parasites exist sympatrically in China and Thailand, respectively, and these materials were identified by only mitochondrial DNA (3, 8). Exactly two tapeworms obtained in Thailand where three human Taenia species were confirmed to be sympatrically occurring (3) are concluded to be the hybrids of T. saginata and T. asiatica (M. Okamoto, M. Nakao, M. T. Anantaphruti, J. Waikagul, and A. Ito, unpublished data). More-detailed characterizations of these two DNA samples must be performed using another nuclear DNA marker, in addition to analyses of longer nucleotide sequences of the clp genes. These results demonstrated that the LAMP methods with each primer set designed in this study can specifically amplify the target gene and are applicable to a differential diagnosis of infections with Taenia species.
TABLE 3.
Analytical specificity of the LAMP assaysa
| Taenia species (no. of samples) | No. of samples (%) detected by LAMP with indicated primer set
|
|
|---|---|---|
| cox1 | clp | |
| T. saginata (78) | 78 (100) | 76 (97.4)b |
| T. asiatica (43) | 43 (100) | 43 (100) |
| T. solium (47) | 47 (100) | 47 (100) |
Taenia species were confirmed by multiplex PCR with cox1 genes (25).
Two samples were detected as T. asiatica by LAMP.
Analytical sensitivity of the LAMP assay.
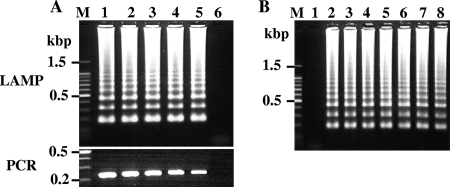
Tenfold serial dilution of each standard plasmid was used to determine the lower detection limits for LAMP and PCR, and the sensitivities of the two methods were compared. In this study, F3 and B3 primers of each LAMP primer set were utilized for PCR. Figure 3A shows results of LAMP and PCR with primer set Tsol-clp. Both LAMP and PCR detected up to one copy of target gene/reaction, which indicated no difference in sensitivity between the two methods. LAMP and PCR with other primer sets provided the same results as LAMP with primer set Tsol-clp (data not shown).
FIG. 3.
(A) Comparison of detection sensitivities of LAMP and PCR with Tsol-clp primer set. The standard plasmids were serially diluted from 104 copies per reaction to 1 copy per reaction and amplified by LAMP (upper panel) and PCR (lower panel). The F3 and B3 primers of the Tsol-clp primer set were used in the PCR. The LAMP reactions were carried out for 60 min. Lane M, 100-bp DNA ladder marker (Promega); lanes 1 to 5 represent 104, 103, 102, 10, and 1 copy(ies)/reaction, respectively; lane 6, negative control. (B) Detection limits of target genes by LAMP with Tasi-cox1 primer set against DNA samples prepared from feces containing various numbers of T. asiatica eggs. The LAMP reactions were carried out for 90 min. Lane M, 100-bp DNA ladder marker; lane 1, negative control; lane 2, T. asiatica genomic DNA as a positive control; lanes 3 to 8 represent 5, 10, 20, 30, 40, and 50 T. asiatica eggs/g of feces, respectively.
Next, the detection limit of taeniid eggs in feces was evaluated using T. asiatica and T. saginata eggs (Fig. 3B). At least five eggs per gram of feces was sufficient for taeniid eggs to be detected by LAMP with primer sets Tasi-cox1 and Tsag-cox1, whereas more than 10 eggs/g of feces was needed to be detected by LAMP with primer set Tsag-clp (data not shown). The differences between the cox1 gene and the clp gene in the detection sensitivity may be responsible for the number of copies of each target gene within the samples, since a large number of mitochondrial DNA exists in a cell, one feature to be selected as a target DNA for detection. Several detection methods for Taenia species in feces based on PCR techniques have been reported. The multiplex PCR method with mitochondrial DNA (25), the PCR-restriction fragment length polymorphism method with mitochondrial DNA (18), and the nested-PCR method with the Tso31 gene encoding the T. solium oncosphere-specific protein (12) have been reported to show detection limits of 5 eggs/g of feces, 17 eggs/g of feces, and 40 eggs/g of feces, respectively. It seems that more eggs are needed for detection when the nuclear gene is chosen as a diagnostic gene marker, although the amplification efficiency of each PCR method varies. In this study, T. solium eggs were not available, but a similar sensitivity might be expected because all LAMP primer sets could amplify one copy of the target gene when using pure plasmid DNA samples.
Differential detection of Taenia species in fecal samples.
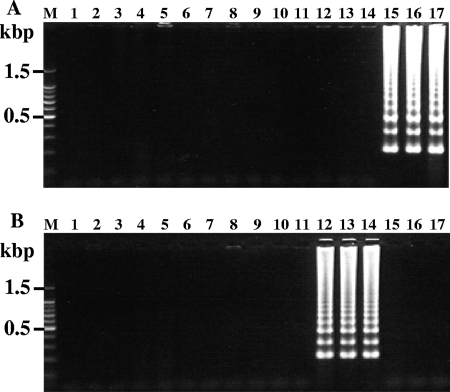
Furthermore, in order to assess the LAMP method for the detection of copro-DNA, we investigated fecal samples that were positive (n = 6) or negative (n = 10) for taeniid eggs by microscopy and collected in Indonesia and Thailand (Fig. 4). Out of six taeniid egg-positive fecal samples, three specimens from Indonesia tested positive with primer sets Tsag-cox1 (Fig. 4A, lanes 15 to 17) and Tsag-clp (data not shown), and the remaining specimens from Thailand were positive with primer sets Tasi-cox1 (Fig. 4B, lanes 12 to 14) and Tsag-clp (data not shown). No differences in detection between nuclear DNA and mitochondrial DNA were observed. The results obtained by LAMP were consistent with those of multiplex PCR. No positive results were observed with the taeniid egg-negative samples.
FIG. 4.
Differential detection of Taenia species in fecal samples. Each genomic DNA extracted from taeniid egg-negative fecal samples collected in Indonesia (lanes 2 to 11) and taeniid egg-positive fecal samples collected in Thailand (lanes 12 to 14) and Indonesia (lanes 15 to 17) was examined by LAMP. The LAMP reactions were carried out for 90 min. The LAMP results with primer sets Tsag-cox1 (A) and Tasi-cox1 (B) are shown. Lane M, 100-bp DNA ladder marker (Promega); lane 1, negative control.
Our results demonstrated that the LAMP method has high sensitivity and specificity for differential detection of Taenia species. Compared to PCR, LAMP has the advantages of reaction simplicity and cost-effectiveness. LAMP does not need sophisticated and expensive equipments, and a simple water bath or a heat block is sufficient to furnish a constant temperature for reactions requiring only 1 to 2 h. Another useful feature of LAMP is that a white precipitate of magnesium pyrophosphate leading to turbidity of reaction mixtures as a byproduct of gene amplification makes it easy to distinguish positive samples from negative samples. In fact, we could discriminate between positive and negative samples by a naked eye in all cases (data not shown). Although limited numbers of clinical specimens were analyzed in the present study, it has been shown that the LAMP method has the potential for use in the differential diagnoses of infections with Taenia species that could be easily applicable in countries where taeniasis/cysticercosis are endemic for better control and prevention of these diseases.
Acknowledgments
Fecal and/or parasite samples used for this study were obtained through joint projects on epidemiological studies of taeniasis/cysticercosis in Indonesia, Thailand, and China (3, 11, 24). We sincerely thank T. Wandra, J. Waikagul, and T. Y. Li for their great help with these samplings.
This work was supported by the leadership in science and technology in Asia from the Ministry of Education, Culture, Sports, Science and Technology, Japan; International Collaboration Research Fund from the Japan Society for the Promotion of Science (JSPS) (17256002); and JSPS-Asia/Africa Science Platform Fund to A.I.
Footnotes
Published ahead of print on 12 November 2008.
REFERENCES
- 1.Abu Al-Soud, W., and P. Rådström. 2000. Effects of amplification facilitators on diagnostic PCR in the presence of blood, feces, and meat. J. Clin. Microbiol. 384463-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allan, J. C., G. Avila, J. Garcia-Noval, A. Flisser, and P. S. Craig. 1990. Immunodiagnosis of taeniasis by coproantigen detection. Parasitology 101473-477. [DOI] [PubMed] [Google Scholar]
- 3.Anantaphruti, M. T., H. Yamasaki, M. Nakao, J. Waikagul, D. Watthanakulpanich, S. Nuamtanong, W. Maipanich, S. Pubampen, S. Sanguankiat, C. Muennoo, K. Nakaya, M. O. Sato, Y. Sako, M. Okamoto, and A. Ito. 2007. Sympatric occurrence of Taenia solium, T. saginata, and T. asiatica, Thailand. Emerg. Infect. Dis. 131413-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bern, C., H. H. Garcia, C. Evans, A. E Gonzalez, M. Verastegui, V. C. W. Tsang, and R. H. Gilman. 1996. Magnitude of the disease burden from neurocysticercosis in a developing country. Curr. Infect. Dis. 291203-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eom, K. S., H. K. Jeon, Y. Kong, U. W. Hwang, Y. Yang, X. Li, L. Xu, Z. Feng, Z. S. Pawlowski, and H. J. Rim. 2002. Identification of Taenia asiatica in China: molecular, morphological, and epidemiological analysis of a Luzhai isolate. J. Parasitol. 88758-764. [DOI] [PubMed] [Google Scholar]
- 6.González, L. M., E. Montero, L. J. S. Harrison, R. M. E. Parkhouse, and T. Garate. 2000. Differential diagnosis of Taenia saginata and Taenia solium infection by PCR. J. Clin. Microbiol. 38737-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ito, A., K. Nakaya, Y. Sako, M. Nakao, and M. Ito. 2001. NOD-scid mouse as an experimental animal model for cysticercosis. Southeast Asian J. Trop. Med. Public Health 3285-89. [PubMed] [Google Scholar]
- 8.Ito, A., M. Nakao, T. Wandra, T. Suroso, M. Okamoto, H. Yamasaki, Y. Sako, and K. Nakaya. 2005. Taeniasis and cysticercosis in Asia and the Pacific: present state of knowledge and perspectives. Southeast Asian J. Trop. Med. Public Health 36123-130. [PubMed] [Google Scholar]
- 9.Kuboki, N., N. Inoue, T. Sakurai, F. D. Cello, D. J. Grab, H. Suzuki, C. Sugimoto, and I. Igarashi. 2003. Loop-mediated isothermal amplification for detection of African trypanosomes. J. Clin. Microbiol. 415517-5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, A. H., S. U. Moon, Y. K. Park, B. K. Na, M. G. Hwang, C. M. Oh, S. H. Cho, Y. Kong, T. S. Kim, and P. R. Chung. 2006. Identification and characterization of a cathepsin L-like cysteine protease from Taenia solium metacestode. Vet. Parasitol. 141251-259. [DOI] [PubMed] [Google Scholar]
- 11.Li, T., P. S. Craig, A. Ito, X. Chen, D. Qiu, J. Qiu, M. O. Sato, T. Wandra, H. Bradshaw, L. Li, Y. Yang, and Q. Wang. 2006. Taeniasis/cysticercosis in a Tibetan population in Sichuan province, China. Acta Trop. 100223-231. [DOI] [PubMed] [Google Scholar]
- 12.Mayta, H., R. H. Gilman, E. Prendergast, J. P. Castillo, Y. O. Tinoco, H. H. Garcia, A. E. Gonzalez, and C. R. Sterling. 2008. Nested PCR for the specific diagnosis of Taenia solium taeniasis. J. Clin. Microbiol. 46286-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monteiro, L., D. Bonnemaison, A. Vekris, K. G. Petry, J. Bonnet, R. Vidal, J. Cabrita, and F. Mégraud. 1997. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J. Clin. Microbiol. 35995-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori, Y., K. Nagamine, N. Tomita, and T. Notomi. 2001. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289150-154. [DOI] [PubMed] [Google Scholar]
- 15.Murell, K. D. 2005. Epidemiology of taeniasis and cysticercosis, p. 32-44. In K. D. Murell (ed.), WHO/FAO/OIE guidelines for the surveillance, prevention and control of taeniasis/cysticercosis. OIE, Paris, France.
- 16.Nakaya, K., W. Mamuti, N. Xiao, M. O. Sato, T. Wandra, M. Nakao, Y. Sako, H. Yamasaki, Y. Ishikawa, P. S. Craig, P. M. Schantz, and A. Ito. 2006. Usefulness of severe combined immunodeficiency (scid) and inbred mice for studies of cysticercosis and echinococcosis. Parasitol. Int. 55(Suppl.)S91-S97. [DOI] [PubMed] [Google Scholar]
- 17.Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nunes, C. M., A. K. K. Dias, F. E. F. Dias, S. M. Aoki, H. B. D. Paula, L. G. F. Lima, and J. F. Garcia. 2005. Taenia saginata: differential diagnosis of human taeniasis by polymerase chain reaction-restriction fragment length polymorphism assay. Exp. Parasitol. 110412-415. [DOI] [PubMed] [Google Scholar]
- 19.Nunes, C. M., L. G. F. Lima, C. S. Manoel, R. N. Pereira, M. M. Nakano, and J. F. Garcia. 2006. Technical report: fecal specimens preparation methods for PCR diagnosis of human taeniosis. Rev. Inst. Med. Trop. Sao Paulo 4845-47. [DOI] [PubMed] [Google Scholar]
- 20.Poon, L. L. M., B. W. Y. Wong, E. H. T. Ma, K. H. Chan, L. M. C. Chow, W. Abeyewickreme, N. Tangpukdee, K. Y. Yuen, Y. Guan, S. Looareesuwan, and J. S. M. Peiris. 2006. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin. Chem. 52303-306. [DOI] [PubMed] [Google Scholar]
- 21.Sako, Y., H. Yamasaki, K. Nakaya, M. Nakao, and A. Ito. 2007. Cloning and characterization of cathepsin L-like peptidases of Echinococcus multilocularis metacestodes. Mol. Biochem. Parasitol. 154181-189. [DOI] [PubMed] [Google Scholar]
- 22.Savan, R., T. Kono, T. Itami, and M. Sakai. 2005. Loop-mediated isothermal amplification: an emerging technology for detection of fish and shellfish pathogens. J. Fish Dis. 28573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sudewi, A. A., T. Wandra, A. Artha, A. Nkouawa, and A. Ito. 2008. Taenia solium cysticercosis in Bali, Indonesia: serology and mtDNA analysis. Trans. R. Soc. Trop. Med. Hyg. 10296-98. [DOI] [PubMed] [Google Scholar]
- 24.Wandra, T., P. Sutisna, N. S. Dharmawan, S. S. Margono, R. Sudewi, T. Suroso, P. S. Craig, and A. Ito. 2006. High prevalence of Taenia saginata taeniasis and status of Taenia solium cysticercosis in Bali, Indonesia, 2002-2004. Trans. R. Soc. Trop. Med. Hyg. 100346-353. [DOI] [PubMed] [Google Scholar]
- 25.Yamasaki, H., J. C. Allan, M. O. Sato, M. Nakao, Y. Sako, K. Nakaya, D. Qiu, W. Mamuti, P. S. Craig, and A. Ito. 2004. DNA differential diagnosis of taeniasis and cysticercosis by multiplex PCR. J. Clin. Microbiol. 42548-553. [DOI] [PMC free article] [PubMed] [Google Scholar]