Abstract
An outbreak of rubella affected 460 individuals in 2004 and 2005 in the community of Madrid, Spain. Most of the patients were nonvaccinated Latin American immigrants or Spanish males. This study presents the first data on rubella virus genotypes in Spain. Forty selected clinical samples (2 urine, 5 serum, 3 blood, 2 saliva, and 28 pharyngeal exudate samples) from 40 cases were collected. The 739-nucleotide sequence recommended by the World Health Organization obtained from viral RNA in these samples was analyzed by using the MEGA v4.0 software. Seventeen isolates were obtained from 40 clinical samples from the outbreak, including two isolated from congenital rubella syndrome cases. Only viral RNA of genotype 1j was detected in both isolates and clinical specimens. Two variations in amino acids, G253C and T394S, which are involved in neutralization epitopes arose during the outbreak, but apparently there was no positive selection of either of them. The origin of the outbreak remains unknown because of poor virologic surveillance in Latin America and the African countries neighboring Spain. On the other hand, this is the first report of this genotype in Europe. The few published sequences of genotype 1j indicate that it comes from Japan and the Philippines, but there are no epidemiological data supporting this as the origin of the Madrid outbreak.
Rubella virus (RUBV) usually causes a mild exanthematous disease that is frequently accompanied by adenopathy and occasionally by arthralgia. Complications of this infection are rare and include encephalopathy and thrombocytopenia. However, the most severe consequence of this virus is its teratogenicity. It can cause congenital rubella syndrome (CRS) when it occurs in pregnant women, particularly during the first trimester of pregnancy (10).
The direct detection of RUBV RNA in clinical specimens, in addition to the detection of RUBV-specific immunoglobulin M, is a critical factor in the early laboratory diagnosis of recent or congenital infection (18, 27). Currently, the European region of the World Health Organization (WHO) aims to eliminate not only measles but also rubella and to reduce the incidence of CRS to less than one case per 100,000 live births by 2010 (38, 39). For this purpose, epidemiological surveillance based on the laboratory diagnosis of each suspected case and the characterization of the genotype of the circulating strains are included in the WHO's recommendations. In the most recent WHO update, the standard nomenclature for the classification and designation of wild-type RUBV strains recognizes nine definitive and four provisional genotypes (40), expanding the nomenclature established in 2005 (37), which was based on 739 nucleotides (nt) (nt 8731 to 9469) from the E1 gene sequence. This sequence encodes amino acids (aa) 159 to 404 (of the 481 aa) of the E1 glycoprotein. Although our knowledge of the geographic distribution of RUBV genotypes has grown substantially since 2003, the genotypes present in many countries and regions remain unknown (9), even though rubella is still recognized as a globally important disease in a general public health context (41). RUBV is considered monotypic with cross-neutralization among different genotypes.
In Spain, monovalent RUBV vaccine was introduced in the late 1970s, when it was administered in schools to 11-year-old girls (1). In 1981, one dose of the measles-mumps-rubella combined vaccine was introduced in the regular immunization schedule at the age of 15 months for all children. In 1996, a second dose at 11 years of age was introduced (5). In 1999, this second dose was given to 4-year-old children (3). Currently, the seroprevalence of RUBV in the community of Madrid exceeds 95% in all age groups and reaches 98.6% among women of childbearing age (16 to 45 years) (4). Nevertheless, the pattern is very different in other regions around the world and RUBV infection remains endemic in many areas, such as Latin America (15). The rubella vaccine was only introduced in Latin American countries in the late 1990s, so that many adult immigrants to Spain are not immunized. These circumstances led to a small outbreak in Madrid in 2003 (31) and a larger one in 2004 and 2005 (2, 27) among Latin American immigrants. The main aim of this study was to characterize the RUBV strain involved in the latter outbreak, which represents the first data concerning RUBV genotypes in Spain.
MATERIALS AND METHODS
Virus strains.
The RA27/3 RUBV vaccine strain was used for standardization and as a positive control (Beecham, Madrid, Spain). Individual wild isolates of parainfluenza virus types 1, 2, 3, 4A, and 4B; adenovirus type 5; mumps virus; respiratory syncytial virus types A and B; and eastern equine encephalitis virus (from the Instituto de Salud Carlos III collection) were used to evaluate the specificity of the reverse transcription (RT)-PCR assay used in this study.
Clinical samples.
Forty selected clinical samples (2 urine, 5 serum, 3 blood, 2 saliva, and 28 pharyngeal exudate samples) collected from 40 infected people during an RUBV outbreak that occurred in the Madrid community in 2004 and 2005 were studied (see Table S2 in the supplemental material). The outbreak affected 460 people, especially nonvaccinated young Spanish men and Latin Americans, mostly Colombians and Ecuadorians (2). It lasted from week 40 of 2004 to week 35 of 2005. The 40 specimens analyzed were obtained from 10 local Spanish people, 21 immigrants, seven individuals of unknown origin, and two persons with CRS (1313A and 1358A) following the outbreak (GenBank accession numbers EU518617 and EU518618). They had an age range of 13 to 48 years (26.08 ± 6.50 [average ± standard deviation]) between weeks 40 of 2004 and 13 of 2005.
Specimens were collected and processed in accordance with WHO recommendations (39).
Isolation in cell culture.
Isolation was performed as previously described (26), with Vero and fetal lung fibroblast cell lines. Inoculated tubes were monitored for a cytopathic effect (CPE) twice a week. After 7 days without a CPE, the culture supernatant was harvested and used to inoculate fresh monolayers. All tubes showing or not showing a CPE after the second passage (7 days) were monitored for the presence of RUBV by immunofluorescence assay with an RUBV-specific monoclonal antibody (mouse anti-rubella monoclonal antibody; Chemicon International, Inc., CA), followed by final immunostaining with fluorescein-labeled anti-mouse conjugate (anti-mouse immunoglobulin G-fluorescein isothiocyanate conjugate; Sigma-Aldrich Chemie, Steinheim, Germany). Furthermore, cell supernatants were analyzed by multiplex RT-PCR for exanthematic viruses, including RUBV (26).
Primer design.
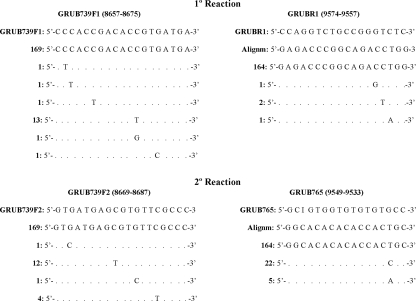
Primers were designed to cover the window of 739 nt for the gene coding for E1 as recommended by the WHO (nt 8731 to 9469) (37). This gene sequence encodes aa 159 to 404 of the E1 glycoprotein. Genomic sequences of the RUBV E1 glycoprotein-encoding gene were taken from GenBank (September 2007) and aligned by the ClustalW method available in the BioEdit 7.0.9 and MEGA v4.0 (32) programs. Alignments were used for primer design (Fig. 1). The forward primer of the first reaction and the forward and reverse primers of the nested reaction were modified from the primer sequences provided by Joe Icenogle (Centers for Disease Control and Prevention Rubella Laboratory team leader), while the reverse primer of the first reaction was designed especially for the present work. Primers were synthesized by a commercial service (Sigma-Aldrich Chemie, Steinheim, Germany).
FIG. 1.
RUBV primers. GRUB739F1 and GRUBR1 are first-reaction primers. GRUB739F2 and GRUB765 are second-reaction primers. The position of each primer following sequence L78917 {Rvi/PA.USA/64VAC[1a] (RA27/3)} is given. The band size obtained with the first reaction was 926 bp, and that for the nested reaction was 875 bp. Sequences used in the alignments were taken from GenBank on 25 September 2007. The number of sequences that are equal to our primer sequences is shown to the left of each illustrated primer.
RT and amplification.
Total nucleic acids were extracted from samples by using the external lysis protocol on a MagNA Pure LC automatic extractor (Roche, Mannheim, Germany) for clinical specimens. Manual extraction (8) was used for cell culture supernatants. RT-PCR was performed with the Access RT-PCR System kit (Promega, Madison, WI). The extract was added to a PCR mixture composed of 2.5 mM MgSO4, 500 μM each deoxynucleoside triphosphate (dNTP), 0.5 μM each RUBV-specific first-reaction primer (Fig. 1), 10 μl of avian myeloblastosis virus-Tfl 5× reaction buffer, 5 U of avian myeloblastosis virus reverse transcriptase, 10 μl of betaine 5 M (Sigma-Aldrich Chemie, Steinheim, Germany), and 5 U of Thermus flavus DNA polymerase (final volume of 50 μl). After the RT step of 45 min at 48°C and denaturation for 2 min at 94°C, the reaction mixtures were incubated for 30 cycles of 94°C for 1 min, 62°C for 1 min, and 72°C for 1 min, followed by 72°C for 5 min.
For nested reactions, 1 μl of the primary amplification products was added to 49 μl of a fresh PCR mixture containing 3 mM MgCl2, 500 μM each dNTP, 1 μM each nested instead of primary reaction primer (Fig. 1), 5 μl of 10× PCR buffer II (Applied Biosystems, CA), 10 μl of betaine 5 M (Sigma-Aldrich Chemie, Steinheim, Germany), and 0.25 U of Taq DNA polymerase (Applied Biosystems, CA). After denaturation for 2 min at 94°C, the reaction mixtures were incubated for 30 cycles of 94.7°C for 1 min, 57°C for 1 min, and 72°C for 1 min, followed by 72°C for 5 min. The MgCl2, dNTP, and primer concentrations were selected for both primary and nested amplifications on the basis of the results of standardization experiments and hybridization and denaturation temperatures. The PCR products were resolved on a 1% agarose gel and visualized by ethidium bromide staining. The expected band size was 875 bp for RUBV.
Sequencing.
PCR products were purified as described previously (28). Purified products were sequenced in both directions with a Big Dye Terminator v.3.1 cycle sequencing kit (Applied Biosystems, CA) on an automatic sequencer (ABI Prism 3700 DNA sequencer; Applied Biosystems, Foster City, CA). The protocol incorporated betaine 5 M (Sigma-Aldrich Chemie, Steinheim, Germany) to minimize failures associated with the GC-rich template. The nested PCR primers were used as sequencing primers. Sequencing was repeated in cases of nucleotide ambiguity.
Sequence analysis.
Sequences were assembled with the SeqMan tool available in the Lasergene 7.0 program. The nucleotide sequences were aligned by the ClustalW method of BioEdit 7.0.9. Phylogenetic analysis was done with the MEGA v.4.0 program (32), adopting the neighbor-joining Kimura two-parameter distance method for 1,000 replicates. It was based on the 739 nt of the E1 gene sequence, which is the minimum acceptable window defined by the WHO (37). Reference sequences (40) were included in each analysis.
RESULTS
Fifteen viruses from nasopharyngeal exudates and two from urine were isolated and are available for further study (see Table S2 in the supplemental material). Strains were named by following the WHO nomenclature for RUBV (37). The sequences obtained from these isolates were identical to those of the original samples. Sixteen belonged to cluster one, and the remaining one (577 A) belonged to cluster 4 (see below).
Genotyping.
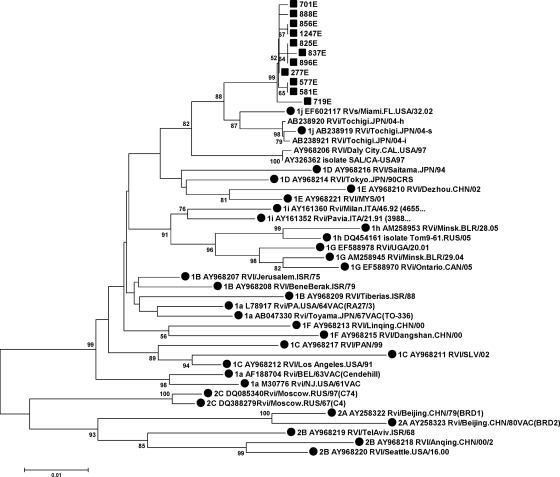
The homology observed among all of the sequences from the outbreak and the reference strains ranged from 97.8 to 98.2% for 1j and from 89.5% (2a) to 96.8% (1b) for the other genotypes (see Table S3 in the supplemental material). All of the sequences of the outbreak strains formed a well-supported cluster in the distance tree (Fig. 2) and grouped with the 1j reference strains with a significant bootstrap value of 88. These results together allow the strain causing the outbreak to be assigned to genotype 1j.
FIG. 2.
Phylogenetic tree of the minimum acceptable window recommended by the WHO in the E1 gene. It shows the RUBV outbreak isolates and samples (▪) in 2005 and all accepted reference strains, as well as sequences of the three provisional genotypes 1 h, 1i, and 1j (•). Furthermore, it includes the other sequences from genotype 1j. The 277E sequence represents 75.0% (30 of 40) of the samples and isolates analyzed.
Sequence analysis.
Four clusters and three sequences that did not fall within any group were identified within the outbreak (Fig. 2). Identical groups were obtained by the minimum-evolution method and the unweighted-pair group method using average linkages with the same Mega v. 4.0 program, as well as by Bayesian inference with the Mr. Bayes program (data not shown). Patients in cluster 4 (577A, 581E) lived in the same area, first exhibited symptoms in the same week, and had the same maternal family name (although we have no direct evidence that they were related). No other significant correlations with epidemiological characteristics were found in other clusters.
The sequences in this study showed 28 variable positions with respect to vaccine strain RA27/3, 5 of them nonsynonymous. Three of these variations were present in all of the sequences, i.e., Y211H, V378L, and L339S, the last one located in a region that could be involved in the induction of proliferative responses of T-cell lines (29). Sequence 277E was present in 30 (75.0%) of the 40 sequences analyzed (Table 1; Fig. 2) that formed cluster 1. This strain seems to be the originally imported one since it was present in the first detected case, and no other sequence was found until week 8 (Table 1). Interestingly, the two sequences from the CRS cases also contained this strain. Cluster 2 (sequences 856E and 1247E) had one additional variable nucleotide at position 178, the third base of the codon, and remained silent. Cluster 3 (sequences 825E, 837E, and 896E) had one additional variable nucleotide at position 328, which remained silent, and sequence 837E had one additional variable nucleotide at position 704 which affected the first base of the codon, causing a change in amino acid T394S. This amino acid maps within an immunoreactive region (11, 14, 25, 33). Cluster 4 (577A and 581E) contained one additional variable nucleotide at position 247, which remained silent. Finally, sequences 701E, 719E, and 888E had particular nucleotide variations, but only sequence 701E showed the G253C amino acid alteration. This amino acid is also located in an immunoreactive region (11, 14, 25, 33).
TABLE 1.
Differences in the nucleotide and predicted amino acid sequences of the four clusters and the individual sequence differences found compared with the reference sequence RVs/Miami.FL.USA/32.02[1j] in the 739-nt window from the E1 gene as recommended by the WHO
| Cluster | Sequence | GenBank accession no. | Wk/yr | Nucleotide change (amino acid change[s]) in 739 nt from E1 gene with respect to:
|
|
|---|---|---|---|---|---|
| RA27/3 vaccine sequence L78917 | 1j prototype sequence EF602117 | ||||
| 1 | 277Ec | EU518607 | 40/2004-13/2005 | 28 (Y211H,a L339S,b V378Lb) | 13 (L339Sb) |
| 2 | 856E | EU518614 | 10/2005 | 29 (Y211H, L339S, V378L) | 14 (L339S) |
| 1247E | EU518606 | 13/2005 | 29 (Y211H, L339S, V378L) | 14 (L339S) | |
| 3 | 825E | EU518612 | 10/2005 | 29 (Y211H, L339S, V378L) | 14 (L339S) |
| 837E | EU518613 | 10/2005 | 30 (Y211H, L339S, V378L, T394Sa) | 15 (L339S, T394Sa) | |
| 896E | EU518616 | 10/2005 | 29 (Y211H, L339S, V378L) | 14 (L339S) | |
| 4 | 577A | EU518608 | 8/2005 | 29 (Y211H, L339S, V378L) | 14 (L339S) |
| 581E | EU518609 | 8/2005 | 29 (Y211H, L339S, V378L) | 14 (L339S) | |
| None | 701E | EU518610 | 9/2005 | 29 (Y211H, G253C,a L339S, V378L) | 13 (G253C,a L339S) |
| 719E | EU518611 | 9/2005 | 31 (Y211H, L339S, V378L) | 16 (L339S) | |
| 888E | EU518615 | 10/2005 | 29 (Y211H, L339S, V378L) | 14 (L339S) | |
Change is located in an immunoreactive region.
Change is not located in an immunoreactive region.
Present in 30 sequences.
In summary, 18 (85.7%) of the 21 specific mutations occurred at codon position 3 and remained silent. Of the three nonsynonymous mutations, two occurred at position 2 and one occurred at position 1 of the codon, leading to changes in the amino acid sequence.
DISCUSSION
In this report, we present the first data on RUBV genotypes in Spain in the context of an outbreak that involved a mainly nonvaccinated population from Latin America, as well as Spanish males (2) born before the introduction of the measles-mumps-rubella vaccine in the early 1980s (2). As the index case is unknown, the geographical origin of the outbreak remains unknown. It is unlikely that the origin was Latin American because the only information about the circulation of RUBV at the time of the outbreak corresponded to genotype 1C (36). Data concerning genotype circulation in Europe during these years showed genotypes 1E, 1G, and 1D (36). Genotypes 1E and 1G circulated in Belarus in 2004 and 2005 (17), and genotype 1E circulated in Poland in 2007 (22). Furthermore, recent findings about RUBV circulation in 2007 in Africa corresponded to genotypes 1E in Morocco, 1G in Uganda and Cote d'Ivoire, and 2B in South Africa (7). All of the published sequences of genotype 1j came from Japan and the Philippines (40), but we do not have any epidemiological evidence linking the outbreak with the Far East. Consequently, this is the first report, to our knowledge, of the detection and isolation of genotype 1j in Europe. Considerable additional effort in RUBV genotyping is needed worldwide to obtain enough data and available sequences to reach consistent conclusions about global RUBV circulation, as is the case for measles virus in Europe (21).
Our results indicate that only one genotype circulated during this outbreak, in contrast to the three (1E, 1G, and 1D) that were circulating in the city of Minsk during the outbreak in Belarus (17). This can be explained by the difference in the lengths of the vaccination programs in Minsk, where rubella vaccination was introduced in 1996 (17), and Madrid, where the universal program started in 1981 (1). The earlier introduction of the vaccine in Madrid could account for a smaller susceptible population, which would make the simultaneous establishment of three genotypes unlikely. Additional studies of RUBV genotype circulation in areas with low or no vaccine coverage are needed to clarify this matter.
The strain causing this outbreak showed a characteristic amino acid change (L339S) with respect to vaccine strain RA27/3 that could be involved in the induction of proliferative responses of T-cell lines (29); however, vaccine failure was not observed. The nucleotide sequence of this strain seemed to remain invariable in the region studied during the first 19 weeks of the outbreak. However, two additional mutations in amino acids in immunoreactive regions (6, 23, 24, 29, 35) of the E1 glycoprotein arose subsequently, although signs of positive selection events were not observed. The E1 glycoprotein has an important role in attachment to the cell and contains important neutralization epitopes (10). Further studies of the biological properties and especially of the degree of neutralization of these strains by vaccine-induced antibodies are required. The proportions of synonymous and nonsynonymous mutations were similar to those reported by other authors (6, 16, 17, 19, 30), confirming that RUBV is very stable compared with some alphaviruses and other RNA viruses, such as poliovirus and human immunodeficiency virus (12, 13, 20, 34). Additional research into the short-term evolution of RUBV in the context of outbreaks seems necessary in the light of these results.
In conclusion, this is the first characterization of an RUBV genotype causing an outbreak in Spain that has involved the circulation of a single genotype (1j). However, it could not be linked to any other concomitant circulating strain in the world due to the paucity of available data on RUBV genotypes. Further studies like this are necessary to obtain a more accurate picture of the global distribution of RUBV genotypes. Such information would allow outbreaks to be managed better and enable the effectiveness of the elimination effort to be monitored, as has been done with measles virus.
Supplementary Material
Acknowledgments
We acknowledge Joseph P. Icenogle, Emily Abernathy, and Paul A. Rota (Centers for Disease Control and Prevention, Atlanta, GA) for providing the primer sequences and the protocol for amplifying RUBV from isolates and clinical samples with the minimal acceptable window recommended by the WHO. We thank the Genomic Unit of the Instituto de Salud Carlos III for carrying out all of the automated sequencing. We also thank Fernando de Ory, Instituto de Salud Carlos III, for his careful review of an earlier draft of our manuscript.
This work received financial support from the fellowship for Ph.D. study by the Republic of Panama and Acuerdo de Encomienda de Gestión entre la Dirección General de Salud Pública del Ministerio de Sanidad y Consumo and the Instituto de Salud Carlos III (dossier DGVI-1429/05-3).
Footnotes
Published ahead of print on 19 November 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Amela, C., and I. Pachón. 2000. La Vigilancia Epidemiológica del sarampión en el contexto del “Plan de acción para eliminar el sarampión en España.” Bol. Epidemiol. Semanal. 8169-172. [Google Scholar]
- 2.Anonymous. 2005. Brote Comunitario de Rubéola en la Población Residente en la Comunidad de Madrid. Bol. Epidemiol. Comunidad Madrid 1133-53. [Google Scholar]
- 3.Anonymous. 1999. Calendario vacunal 1999. Bol. Epidemiol. Comunidad Madrid 450-59. [Google Scholar]
- 4.Anonymous. 2002. III Encuesta de serovigilancia de la Comunidad de Madrid. Bol. Epidemiol. Comunidad Madrid 83-43. [Google Scholar]
- 5.Anonymous. 1996. Nuevo calendario vacunal. Bol. Epidemiol. Comunidad Madrid 1925-27. [Google Scholar]
- 6.Bosma, T. J., J. M. Best, K. M. Corbett, J. E. Banatvala, and W. G. Starkey. 1996. Nucleotide sequence analysis of a major antigenic domain of the E1 glycoprotein of 22 rubella virus isolates. J. Gen. Virol. 772523-2530. [DOI] [PubMed] [Google Scholar]
- 7.Caidi, H., E. S. Abernathy, A. Benjouad, S. Smit, J. Bwogi, M. Nanyunja, R. El Aouad, and J. Icenogle. 2008. Phylogenetic analysis of rubella viruses found in Morocco, Uganda, Cote d'Ivoire and South Africa from 2001 to 2007. J. Clin. Virol. 4286-90. [DOI] [PubMed] [Google Scholar]
- 8.Casas, I., L. Powell, P. E. Klapper, and G. M. Cleator. 1995. New method for the extraction of viral RNA and DNA from cerebrospinal fluid for use in the polymerase chain reaction assay. J. Virol. Methods 5325-36. [DOI] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2005. Global Measles and Rubella Laboratory Network, January 2004-June 2005. MMWR Morb. Mortal. Wkly. Rep. 541100-1104. [PubMed] [Google Scholar]
- 10.Chantler, J., J. S. Wolinsky, and A. Tingle. 2001. Rubella virus, p. 963-990. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, PA. [Google Scholar]
- 11.Chaye, H., P. Chong, B. Tripet, B. Brush, and S. Gillam. 1992. Localization of the virus neutralizing and hemagglutinin epitopes of E1 glycoprotein of rubella virus. Virology 189483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coffin, J. M. 1992. Genetic diversity and evolution of retroviruses. Curr. Top. Microbiol. Immunol. 176143-164. [DOI] [PubMed] [Google Scholar]
- 13.Fitch, W. M. 1996. The variety of human virus evolution. Mol. Phylogenet. Evol. 5247-258. [DOI] [PubMed] [Google Scholar]
- 14.Giessauf, A., T. Letschka, G. Walder, M. P. Dierich, and R. Wurzner. 2004. A synthetic peptide ELISA for the screening of rubella virus neutralizing antibodies in order to ascertain immunity. J. Immunol. Methods 2871-11. [DOI] [PubMed] [Google Scholar]
- 15.Hinman, A. R., B. S. Hersh, and C. A. de Quadros. 1998. Rational use of rubella vaccine for prevention of congenital rubella syndrome in the Americas. Rev. Panam. Salud Publica 4156-160. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann, J., M. Renz, S. Meyer, A. von Haeseler, and U. G. Liebert. 2003. Phylogenetic analysis of rubella virus including new genotype I isolates. Virus Res. 96123-128. [DOI] [PubMed] [Google Scholar]
- 17.Hübschen, J. M., M. Yermalovich, G. Semeiko, E. Samoilovich, E. Blatun, S. De Landtsheer, and C. P. Muller. 2007. Co-circulation of multiple rubella virus strains in Belarus forming novel genetic groups within clade 1. J. Gen. Virol. 881960-1966. [DOI] [PubMed] [Google Scholar]
- 18.Jin, L., and B. Thomas. 2007. Application of molecular and serological assays to case based investigations of rubella and congenital rubella syndrome. J. Med. Virol. 791017-1024. [DOI] [PubMed] [Google Scholar]
- 19.Katow, S., H. Minahara, M. Fukushima, and Y. Yamaguchi. 1997. Molecular epidemiology of rubella by nucleotide sequences of the rubella virus E1 gene in three East Asian countries. J. Infect. Dis. 176602-616. [DOI] [PubMed] [Google Scholar]
- 20.Kinnunen, L., T. Poyry, and T. Hovi. 1992. Genetic diversity and rapid evolution of poliovirus in human hosts. Curr. Top. Microbiol. Immunol. 17649-61. [DOI] [PubMed] [Google Scholar]
- 21.Kremer, J. R., K. E. Brown, L. Jin, S. Santibanez, S. V. Shulga, Y. Aboudy, I. V. Demchyshyna, S. Djemileva, J. E. Echevarria, D. F. Featherstone, M. Hukic, K. Johansen, B. Litwinska, E. Lopareva, E. Lupulescu, A. Mentis, Z. Mihneva, M. M. Mosquera, M. Muscat, M. A. Naumova, J. Nedeljkovic, L. S. Nekrasova, F. Magurano, C. Fortuna, H. R. de Andrade, J. L. Richard, A. Robo, P. A. Rota, E. O. Samoilovich, I. Sarv, G. V. Semeiko, N. Shugayev, E. S. Utegenova, R. van Binnendijk, L. Vinner, D. Waku-Kouomou, T. F. Wild, D. W. Brown, A. Mankertz, C. P. Muller, and M. N. Mulders. 2008. High genetic diversity of measles virus, World Health Organization European region, 2005-2006. Emerg. Infect. Dis. 14107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makowka, A., W. Gut, B. Litwinska, S. Santibanez, and A. Mankertz. 2007. Genotyping of measles and rubella virus strains circulating in Poland in 2007. Euro Surveill. 12E071025 2. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=3295. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell, L. A., D. Decarie, A. J. Tingle, M. Zrein, and M. Lacroix. 1993. Identification of immunoreactive regions of rubella virus E1 and E2 envelope proteins by using synthetic peptides. Virus Res. 2933-57. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell, L. A., A. J. Tingle, D. Decarie, and R. Shukin. 1999. Identification of rubella virus T-cell epitopes recognized in anamnestic response to RA27/3 vaccine: associations with boost in neutralizing antibody titer. Vaccine 172356-2365. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell, L. A., T. Zhang, M. Ho, D. Decarie, A. J. Tingle, M. Zrein, and M. Lacroix. 1992. Characterization of rubella virus-specific antibody responses by using a new synthetic peptide-based enzyme-linked immunosorbent assay. J. Clin. Microbiol. 301841-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mosquera Mdel, M., F. de Ory, M. Moreno, and J. E. Echevarria. 2002. Simultaneous detection of measles virus, rubella virus, and parvovirus B19 by using multiplex PCR. J. Clin. Microbiol. 40111-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mosquera Mdel, M., J. C. Sanz, J. E. Echevarria, N. Herranz, M. Fernandez, and F. de Ory. 2006. Diagnostic performance of specific IgM detection and genomic amplification in rubella. Enferm. Infecc. Microbiol. Clin. 24251-253. [DOI] [PubMed] [Google Scholar]
- 28.Mosquera, M. M., F. Ory, and J. E. Echevarria. 2005. Measles virus genotype circulation in Spain after implementation of the national measles elimination plan 2001-2003. J. Med. Virol. 75137-146. [DOI] [PubMed] [Google Scholar]
- 29.Ou, D., P. Chong, A. J. Tingle, and S. Gillam. 1993. Mapping T-cell epitopes of rubella virus structural proteins E1, E2, and C recognized by T-cell lines and clones derived from infected and immunized populations. J. Med. Virol. 40175-183. [DOI] [PubMed] [Google Scholar]
- 30.Saitoh, M., N. Shinkawa, S. Shimada, Y. Segawa, K. Sadamasu, M. Hasegawa, M. Kato, K. Kozawa, T. Kuramoto, O. Nishio, and H. Kimura. 2006. Phylogenetic analysis of envelope glycoprotein (E1) gene of rubella viruses prevalent in Japan in 2004. Microbiol. Immunol. 50179-185. [DOI] [PubMed] [Google Scholar]
- 31.Sanz, J. C., C. Lemos, D. Herrera, and R. Ramirez-Fernandez. 2004. Brote de rubéola en población inmigrante de origen latinoamericano. Enferm. Infecc. Microbiol. Clin. 22197. [DOI] [PubMed] [Google Scholar]
- 32.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 33.Terry, G. M., L. Ho-Terry, P. Londesborough, and K. R. Rees. 1988. Localization of the rubella E1 epitopes. Arch. Virol. 98189-197. [DOI] [PubMed] [Google Scholar]
- 34.Wain-Hobson, S. 1993. The fastest genome evolution ever described: HIV variation in situ. Curr. Opin. Genet. Dev. 3878-883. [DOI] [PubMed] [Google Scholar]
- 35.Wolinsky, J. S., M. McCarthy, O. Allen-Cannady, W. T. Moore, R. Jin, S. N. Cao, A. Lovett, and D. Simmons. 1991. Monoclonal antibody-defined epitope map of expressed rubella virus protein domains. J. Virol. 653986-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.World Health Organization. 2006. Global distribution of measles and rubella genotypes—update. Wkly. Epidemiol. Rec. 81474-479. [PubMed] [Google Scholar]
- 37.World Health Organization. 2005. Standardization of the nomenclature for genetic characteristics of wild-type rubella viruses. Wkly. Epidemiol. Rec. 80126-132. [PubMed] [Google Scholar]
- 38.World Health Organization. 25 September 2007, accession date. Strategic plan for measles and congenital rubella infection in the European region of WHO. World Health Organization Regional Office for Europe, Copenhagen, Denmark. [Online.] http://www.euro.who.int/document/e81567.pdf.
- 39.World Health Organization. 25 September 2007, accession date. Surveillance guidelines for measles and congenital rubella infections in the WHO European region. World Health Organization Regional Office for Europe, Copenhagen, Denmark. [Online.] http://www.euro.who.int/document/e82183.pdf.
- 40.World Health Organization. 2007. Update of standard nomenclature for wild-type rubella viruses, 2007. Wkly. Epidemiol. Rec. 82216-222. [PubMed] [Google Scholar]
- 41.Zheng, D. P., T. K. Frey, J. Icenogle, S. Katow, E. S. Abernathy, K. J. Song, W. B. Xu, V. Yarulin, R. G. Desjatskova, Y. Aboudy, G. Enders, and M. Croxson. 2003. Global distribution of rubella virus genotypes. Emerg. Infect. Dis. 91523-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.