Abstract
Blood isolates of Salmonella enterica serovar Typhi from two recently returned Bangladeshi patients in Kuwait were ciprofloxacin resistant, with ciprofloxacin MICs of 12 mg/liter for both isolates. Both isolates had three novel gyrA mutations (55-Leu→Trp, 87-Asp→Ala, and 106-Gln→Arg) and three novel parC mutations (84-Glu→Lys, 106-Trp→Gly, and 128-Tyr→Asp).
Typhoid fever is a major cause of morbidity and mortality in developing countries (8). In the last two decades, the worldwide emergence of multiresistant salmonellae has led to the withdrawal of chloramphenicol and its replacement with fluoroquinolones and broad-spectrum cephalosporins (2).
However, nalidixic acid-resistant strains (MIC, ≥32 mg/liter) exhibiting reduced susceptibility to ciprofloxacin (MICs, 0.125 to 1 mg/liter) have emerged and become endemic in South and South-East Asia (3, 18, 23). Such strains have also been reported from other parts of the world (26, 27). Consequently, there is treatment failure with ciprofloxacin in patients infected with these organisms (11, 26, 27).
A large expatriate population from South and South-East Asia is employed in the oil-rich Gulf States including Kuwait. We have previously reported enteric fever isolates that are either multiresistant or with reduced susceptibility to ciprofloxacin from expatriate workers of South Asian origin from Kuwait (9, 10, 11). Although there are reports from India (13, 24), Bangladesh (1, 25), and Nepal (5) of Salmonella enterica serovar Typhi strains fully resistant to ciprofloxacin (MICs ≥ 4 mg/liter), infection with such strains is rare. The targets for fluoroquinolones are DNA gyrase and topoisomerase IV, whose subunits are encoded by gyrA and gyrB and by parC and parE genes, respectively (27). Single point mutations in the quinolone resistance-determining region (QRDR) of gyrA gene (spanning amino acids 54 to 171) lead to decreased susceptibility to ciprofloxacin (15). Additional mutations may be required to attain high-level fluoroquinolone resistance (7, 16). There has been only one report of molecular characterization of three serovar Typhi isolates fully resistant to ciprofloxacin from India whose gyrase and topoisomerase genes have been analyzed with mutations in gyrA and parC genes (13). We report here two cases of typhoid fever with full ciprofloxacin resistance. We analyzed these isolates for mutations in both gyrA and parC genes and found novel mutations in both these genes.
Two Bangladeshi male patients, one 29 years old and the other 2 years old, presented with fever at the Infectious Diseases Hospital, Safat, Kuwait, in June 2006 and May 2007, respectively. Both of them returned recently from Bangladesh. Both were treated unsuccessfully at private clinics prior to their visit at our facility. The adult patient received amoxicillin, and the child patient received amoxicillin and cotrimoxazole. Blood cultures were performed on these patients (10) in our facility, and serovar Typhi grew from both blood cultures (laboratory identification numbers of isolates: 1958 from the older patient and 1474 from the child). The isolates were screened for metabolic profiles with API 20E strip (BioMerieux, Marcy l'Etoile, France) and confirmed by slide agglutination with Salmonella antisera (Denka-Seiken, Tokyo, Japan). Antimicrobial susceptibility was performed by disk diffusion method according to Clinical and Laboratory Standards Institute (CLSI) guidelines (6) with disks containing chloramphenicol (30 μg), amoxicillin (10 μg), cotrimoxazole (1.25/23.75 μg), ceftriaxone (30 μg), cefixime (30 μg), nalidixic acid (30 μg), ciprofloxacin (5 μg), moxifloxacin (5 μg), and levofloxacin (5 μg). Escherichia coli ATCC strain 25922 was used as the quality control strain in the susceptibility test.
The MIC of nalidixic acid was determined by the agar dilution method (6) and that of ciprofloxacin by Etest (AB Biodisk, Solna, Sweden) according to the manufacturer's instructions.
For amplification of QRDRs of gyrA and parC genes, bacterial DNA template was prepared by the method of Giraud et al. (14). The gyrA gene was amplified by the primers and cycling conditions described by Brown et al. (3), and parC gene was amplified by the primers and cycling conditions described by Giraud et al. (14). The PCR products were photographed under UV transillumination of 1% agarose gel after electrophoresis and staining with ethidium bromide. DNA sequencing was carried out by the dideoxynucleotide chain termination method, using an automated DNA sequencer (ABI Prism 3100 genetic analyzer; Applied Biosystems, Foster City, CA). The sequences were confirmed by sequencing both strands three times. We also included a ciprofloxacin-susceptible strain of serovar Typhi and a serovar Typhi strain with reduced ciprofloxacin susceptibility in sequencing as controls. Sequences were analyzed by the BLAST online search engine (http://www.ncbi.nih.gov/cgi-bin/BLAST) with the sequences of susceptible strains in the database.
Clonal relatedness of the two isolates was studied by pulsed-field gel electrophoresis with XbaI-digested bacterial chromosome using the PulseNet protocol (4).
Both of the serovar Typhi isolates showed the same antimicrobial susceptibility pattern by disk diffusion method. They were resistant to nalidixic acid, ciprofloxacin, moxifloxacin, levofloxacin, chloramphenicol, amoxicillin, and cotrimoxazole but susceptible to ceftriaxone and cefixime. The MICs of nalidixic acid (256 mg/liter) and ciprofloxacin (12 mg/liter) were identical for both of the isolates. Thus, both isolates were deemed multiresistant (being resistant to amoxicillin, chloramphenicol, and cotrimoxazole) and highly resistant to nalidixic acid and ciprofloxacin. Both patients were given a course of ceftriaxone, to which they responded.
Both of the isolates showed the same four point mutations in the QRDR of the gyrA gene. These were 55-TTG (Leu)→TGG (Trp), 83-TCC (Ser)→TTC (Phe), 87-GAC (Asp)→GCC (Ala), and 106-CAG (Gln)→CGG (Arg). Again, both of the isolates showed the same three point mutations in the QRDR (spanning amino acids 12 to 130) of the parC gene (15). These were 84 GAA (Glu)→AAA (Lys), 106 TGG(Trp)→GGG (Gly), and 128 TAC (Tyr)→GAC (Asp). These sequences have been deposited in GenBank (accession no. FJ222660 for gyrA gene and accession no. FJ222661 for parC gene). The fully susceptible control strain did not have mutations, and the strain with reduced susceptibility had a single mutation at Ser-83→Phe in the gyrA gene.
In Salmonella, the more common point mutations found to be associated with resistance to quinolones occur in the gyrA gene and results in substitutions at the Ser-83 position, often to Tyr, Phe, or Ala, and in Asp-87 substitutions to Asn, Gly, or Tyr. The most common amino acid substitution reported in parC gene is Thr-57→Ser, with Thr-66→Ile or Ser-80→Arg being observed as occasional second substitutions (12, 21).
In both studies from India (13, 24) and one study from Bangladesh (25) with fully ciprofloxacin-resistant serovar Typhi isolates in which the gyrA gene has been sequenced, all isolates had mutations at Ser-83 and Asp-87. For our isolates, the previously reported codon change at Ser-83→Phe was present. There was also a codon change at 87 with the change of amino acid from Asp to Ala. This amino acid change to alanine has not been reported previously. In addition, there were changes at positions 55 and 106. A BLAST search revealed that these novel mutations have not been reported previously in the gyrA gene of any organism. Site-directed mutagenesis experiments would have to be conducted to determine whether these amino acid changes affect quinolone susceptibilities in a single genetic background. In our previous study using serovar Typhi isolates with decreased ciprofloxacin susceptibility collected during 2000 to 2003 in Kuwait, we found a single mutation in the gyrA gene (Ser-83→Phe) as the mechanism for decreased ciprofloxacin susceptibility (9). A study of three ciprofloxacin-resistant serovar Typhi isolates from India reported mutations in both the gyrA and the parC genes (13). All three isolates had mutations at codon 80 from AGC (Ser)→ATC (Ile) in the parC gene. In contrast, although our isolates lacked this mutation, they carried three novel mutations at Glu-84→Lys, Trp-106→Gly, and Tyr-128→Asp. A BLAST search has revealed that the Trp-106→Gly and the Glu-84→Lys mutations in the parC gene have been previously reported in S. enterica serovar Typhimurium LT2 (20) and S. enterica serovar Paratyphi A (19), respectively. A comparison of mutations previously reported in the QRDR of the gyrA gene of serovar Typhi to those found in our study is shown in Fig. 1. A similar comparison for the QRDR of parC gene is shown in Fig. 2. Although mutations in both gyrA and parC genes have been identified in bacteria highly resistant to fluoroquinolones, the role of parC mutation is less clear (19). The primary target of fluoroquinolones is gyrase rather than topoisomerase IV. Mutation in parC is, however, required to achieve high-level fluoroquinolone resistance.
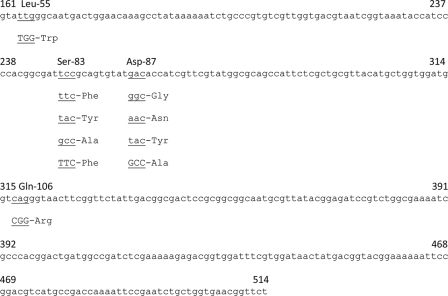
FIG. 1.
QRDR of the gyrA gene of serovar Typhi, with base changes reported previously and now. The QRDR spans nucleotides 161 to 514. The triplet bases with changes are underlined with the amino acids indicated above them. The underlined triplet bases in lowercase letters have been reported previously. The underlined triplet bases in capital letters have been found in the present study.
FIG. 2.
QRDR of the parC gene of serovar Typhi, with base changes reported previously and now. The QRDR spans nucleotides 34 to 390. The triplet bases with changes are underlined with the amino acids indicated above them. The underlined triplet bases in lowercase letters have been reported previously. The underlined triplet bases in capital letters have been found in the present study.
As determined by pulsed-field gel electrophoresis, the two isolates shared 14 bands of identical molecular weights. However, the isolate 1474 had an additional band of approximately 500 bp that was absent in strain 1958 (data not shown).
Thus, the two isolates were closely related. It was not possible to get information on the sources of the infections.
Ours is the second report of molecular characterization of changes in the gyrA and parC genes of fully ciprofloxacin-resistant serovar Typhi strains. However, unlike the first report (13), we have shown novel mutations in both of the genes. Since we have not sequenced gyrB and parE genes, we are not sure of any changes in these genes in our isolates. High-level ciprofloxacin resistance could be due to the cumulative impact of mutations in many genes, decreased membrane permeability, the efflux pump, and plasmid-encoded Qnr genes (17).
The emergence of fully ciprofloxacin-resistant serovar Typhi strains is a worrying development that seriously limits treatment options. It is likely that there will be more reports of fully resistant strains in the future from regions of the world where typhoid fever is endemic. A revision of the MIC breakpoint criteria by the CLSI is also required, since the current reference standards are unable to distinguish between fully susceptible isolates and isolates with reduced susceptibility (22).
Footnotes
Published ahead of print on 29 October 2008.
REFERENCES
- 1.Ahmed, D., L. T. D'Costa, K. Alam, G. B. Nair, and M. A. Hossain. 2006. Multi-resistant Salmonella enterica serovar Typhi isolates with high-level resistance to ciprofloxacin in Dhaka, Bangladesh. Antimicrob. Agents Chemother. 503516-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhan, M. K., R. Bhal, and S. Bhatnagar. 2005. Typhoid and paratyphoid fever. Lancet 366749-762. [DOI] [PubMed] [Google Scholar]
- 3.Brown, J. C., P. M. A. Shanahan, M. V. Jesudason, C. J. Thomson, and S. G. B. Amyes. 1996. Mutations responsible for reduced susceptibility to 4-quinolones in clinical isolates of multi-resistant Salmonella typhi in India. J. Antimicrob. Chemother. 37891-900. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2000. Standardized molecular subtyping of food-borne bacterial pathogen by pulsed-field gel electrophoresis: training manual. Centers for Disease Control and Prevention, Atlanta, GA.
- 5.Chau, T. T., J. I. Campbell, C. M. Galindo, N. V. M. Hoang, T. S. Diep, T. T. T. Nga, N. V. V. Chau, P. Q. Tuan, A. L. Page, R. L. Ochiai, C. Schultsz, J. Wain, Z. A. Bhutta, C. M. Parry, S. K. Bhattacharya, S. Dutta, M. Agtini, B. Dong, Y. Honghui, D. D. Anh, D. G. Canh, A. Naheed, M. J. Albert, R. Phetsouvanh, P. N. Newton, B. Basnyat, A. Arjyal, T. T. P. La, N. N. Rang, L. T. Phuong, P. V. B. Bay, L. von Seidlein, G. Dougan, J. D. Clemens, H. Vinh, T. T. Hien, N. T. Chinh, C. J. Costa, J. Farrar, and C. Dolecek. 2007. Antimicrobial drug resistance of Salmonella enterica serovar Typhi in Asia and molecular mechanism of reduced susceptibility to the fluoroquinolones. Antimicrob. Agents Chemother. 514315-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement M100-S16, vol. 26, no. 3. CLSI, Wayne, PA.
- 7.Cloeckaert, A., and E. Chaslus-Dancla. 2001. Mechanism of quinolone resistance in Salmonella. Vet. Res. 32291-300. [DOI] [PubMed] [Google Scholar]
- 8.Crump, J. A., S. P. Lubsy, and E. D. Mintz. 2004. The global burden of enteric fever. Bull. W. H. O. 82346-353. [PMC free article] [PubMed] [Google Scholar]
- 9.Dashti, A. A., M. M. Jadaon, F. Habeeb, P. W. West, D. Panigrahi, and S. G. B. Amyes. 2008. Salmonella enterica serotype Typhi in Kuwait and its reduced susceptibility to ciprofloxacin. J. Chemother. 2038-43. [DOI] [PubMed] [Google Scholar]
- 10.Dimitrov, T., E. E. Udo, O. Albaksami, S. Al-Shehab, A. Kilani, M. Shehab, and A. Al-Nakkas. 2007. Clinical and microbiological investigations of typhoid fever in an infectious disease hospital in Kuwait. J. Med. Microbiol. 56538-544. [DOI] [PubMed] [Google Scholar]
- 11.Dimitrov, T., E. E. Udo, O. Albaksami, A. A. Kilani, and E. M. R. Shehab. 2007. Ciprofloxacin treatment failure in a case of typhoid fever caused by Salmonella enterica serotype Paratyphi A with reduced susceptibility to ciprofloxacin. J. Med. Microbiol. 56277-279. [DOI] [PubMed] [Google Scholar]
- 12.Eaves, D. J., L. Randall, D. T. Gray, A. Buckley, M. J. Woodward, A. P. White, and L. J. Piddock. 2004. Prevalence of mutations with the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica. Antimicrob. Agents Chemother. 484012-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaind, R., B. Paglietti, M. Murgia, R. Dawar, S. Uzzau, P. Cappuccinelli, M. Deb, P. Aggarwal, and S. Rubino. 2006. Molecular characterization of ciprofloxacin-resistant Salmonella enterica serovar Typhi and Paratyphi A causing enteric fever in India. J. Antimicrob. Chemother. 581139-1144. [DOI] [PubMed] [Google Scholar]
- 14.Giraud, E., A. Brisabois, J. L. Martel, and E. Chaslus-Dancla. 1999. Comparative studies of mutations in animal isolates and experimental in vitro- and in vivo-selected mutants of Salmonella spp. suggest a counterselection of highly fluoroquinolone-resistant strains in the field. Antimicrob. Agents Chemother. 432131-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirose, K., A. Hashimoto, K. Tamura, Y. Kawamura, T. Ezaki, H. Sagara, and H. Watanabe. 2002. DNA sequence analysis of DNA gyrase and DNA topoisomerase IV quinolone resistance-determining regions of Salmonella enterica serovar Typhi and serovar Paratyphi A. Antimicrob. Agents Chemother. 463249-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooper, D. C. 2001. Mechanisms of fluoroquinolone resistance. Emerg. Infect. Dis. 7337-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacoby, G. A. 2005. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 41(Suppl. 2)S120-S126. [DOI] [PubMed] [Google Scholar]
- 18.Jesudason, M. V., B. Malathy, and T. J. John. 1996. Trend of increasing minimum inhibitory concentration of ciprofloxacin to Salmonella typhi. Indian J. Med. Res. 103247-249. [PubMed] [Google Scholar]
- 19.McClelland, M., K. E. Sanderson, S. W. Clifton, P. Latreille, S. Porwollik, A. Sabo, R. Meyer, T. Bieri, P. Ozersky, M. McLellan, C. R. Harkins, C. Wang, C. Nguyen, A. Berghoff, G. Elliot, S. Kohlberg, C. Strong, F. Du, J. Carter, C. Kremizki, D. Laymen, S. Leonard, H. Sun, L. Fulton, W. Nash, T. Miner, P. Minx, K. Delehaunty, C. Fronick, V. Magrini, M. Nhan, W. Warren, L. Florea, J. Spieth, and R. K. Wilson. 2004. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 361268-1274. [DOI] [PubMed] [Google Scholar]
- 20.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413852-856. [DOI] [PubMed] [Google Scholar]
- 21.Piddock, L. J., V. Ricci, I. McLaren, and D. J. Griggs. 1998. Role of mutation in gyrA and parC genes in nalidixic-acid-resistant Salmonella serotypes isolated from animals in the United Kingdom. J. Antimicrob. Chemother. 41635-641. [DOI] [PubMed] [Google Scholar]
- 22.Poutanen, S., and D. E. Low. 2003. Is it time to change fluoroquinolones MIC breakpoints for Salmonella spp? Clin. Microbiol. Newsl. 2597-102. [Google Scholar]
- 23.Rahman, M. M., J. A. Haq, M. A. Morshed, and M. A. Rahman. 2005. Salmonella enterica serovar Typhi with decreased susceptibility to ciprofloxacin-an emerging problem in Bangladesh. Int. J. Antimicrob. Agents 25345-346. [DOI] [PubMed] [Google Scholar]
- 24.Renuka, K., S. sood, B. K. Das, and A. Kapil. 2005. High-level ciprofloxacin resistance in Salmonella enterica serotype Typhi in India. J. Med. Microbiol. 54999-1000. [DOI] [PubMed] [Google Scholar]
- 25.Saha, S. K., G. L. Darmstadt, A. H. Baqui, D. W. Crook, M. N. Islam, M. Islam, M. Hossain, S. El Arifeen, M. Santosham, and R. E. Black. 2006. Molecular basis of resistance displayed by highly ciprofloxacin-resistant Salmonella enterica serovar Typhi in Bangladesh. J. Clin. Microbiol. 443811-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Threlfall, E. J., and L. R. Ward. 2001. Decreased susceptibility to ciprofloxacin in Salmonella enterica serotype Typhi, United Kingdom. Emerg. Infect. Dis. 7448-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wain, J., N. T. T. Hoa, N. T. Chinh, H. Vinh, M. J. Everett, T. S. diep, N. P. J. Day, T. Solomon, N. J. White, L. J. Piddock, and C. M. Parry. 1997. Quinolone-resistant Salmonella typhi in Viet Nam: molecular basis of resistance and clinical response to treatment. Clin. Infect. Dis. 251404-1410. [DOI] [PubMed] [Google Scholar]