Abstract
We performed a molecular study to determine the occurrence of Candida albicans, Candida africana, and Candida dubliniensis in different clinical samples. The study provides new insights into the epidemiology of candidiasis in hospitalized patients in three hospitals in southern Italy. It also reports the first detailed epidemiological data concerning the occurrence of C. africana in clinical samples.
The incidence of infection caused by Candida spp. has increased steadily over the last two decades, and Candida albicans remains the most common fungal pathogen isolated from clinical samples (14, 15, 19, 24). In 1995, Candida dubliniensis was previously described as being an opportunistic pathogen linked to oral candidiasis in human immunodeficiency virus-infected patients (21), although it has also been recovered from other anatomical sites (7, 11, 12, 25).
Recently, atypical C. albicans strains such as Candida africana have been reported as cause of vaginitis from African, German, Spanish, and Italian patients (2, 5, 17, 23).
Candida africana has been considered to be an atypical C. albicans strain, but in 2001, it was proposed to be a new Candida species that produces a germ tube but not chlamydospores (23). However, comparative studies based on genetic techniques have clearly shown that C. africana cannot be considered as a new species of Candida (2, 5, 17). Therefore, Candida albicans var. africana was suggested to be a more suitable name for these atypical C. albicans strains (17).
More recently, we described the first specific molecular method for discriminating between C. albicans, C. africana, and C. dubliniensis by using a single pair of primers targeting the Hwp1 gene and its homologues in a PCR-based assay (16). Because C. albicans, C. africana, and C. dubliniensis can be identified and differentiated easily and quickly by using this method, we decided to perform a molecular study in order to determine the incidence and distribution of these three important pathogenic yeasts in different clinical samples.
This study provides accurate insights into the epidemiology of candidiasis in hospitalized patients in three hospitals in southern Italy. Moreover, we report here the first detailed epidemiological data concerning the occurrence of C. africana in clinical samples.
Four hundred ninety-eight isolates of Candida spp. obtained from several anatomical sites and clinical samples were examined (Table 1). The strains were obtained from three different hospitals in southern Italy: Riuniti Hospitals, Reggio Calabria (276 strains); Civil Hospital, Vibo Valentia (138 strains); and the University of Messina Medical School (84 strains). In addition, we conducted a retrospective analysis of 52 C. albicans isolates stored in the yeast culture collection at the Department of Life Sciences, University of Messina, Messina, Italy.
TABLE 1.
Clinical samples and Candida isolates examined in this study
| Source | No. of samples | No. of GTPa strains from clinical samples | No. of strains from culture collections | Total no. of GTP strains | No. of strains of other Candida species |
|---|---|---|---|---|---|
| Oral | 121 | 76 | 13 | 89 | 45 |
| Feces | 31 | 23 | 4 | 27 | 8 |
| Vagina | 162 | 136 | 27 | 163 | 26 |
| Urine | 19 | 10 | 0 | 10 | 9 |
| Blood | 16 | 9 | 1 | 10 | 7 |
| Gastric fluid | 29 | 21 | 5 | 26 | 8 |
| Drainage fluid | 67 | 35 | 2 | 37 | 32 |
| Skin lesion | 53 | 14 | 0 | 14 | 39 |
| Total | 498 | 324 | 52 | 376 | 174 |
GTP, germ tube positive.
The germ tube test was the initial screening test. Biochemical identifications were also performed on all isolates by using the Vitek 2 ID-YST system (bioMerieux) in accordance with the manufacturer's instructions. Only germ tube-positive isolates (324 of 498 strains plus 52 strains from a retrospective study) were selected for molecular analysis and were included in this study. In total, 376 isolates were viable and were thus analyzed by PCR.
Molecular identification was performed according to methods described previously by Romeo and Criseo (16).
C. albicans ATCC 10231, C. dubliniensis CD36 (a kind gift from Derek Sullivan, University of Dublin, Republic of Ireland), C. dubliniensis CBS 7987, and C. africana CBS 11016, including strains A1605, A1622, A1653, and M8684 (kind gifts from Hans-Jürgen Tietz, Universitätsklinikum Charité, Berlin, Germany), were used as reference strains.
We obtained 324 isolates of germ tube-positive Candida species, which represented 65% of the Candida species found among 498 examined yeast isolates (Table 1). The remaining 174 isolates were identified as being C. glabrata (32.2%), which was the most prevalent one, followed by C. parapsilosis (22%), C. tropicalis (16.6%), C. krusei (12.6%), C. kefyr (10.9%), C. lusitaniae (4%), and C. guilliermondii (1.7%).
Of 376 germ tube-positive isolates, 366 were identified as being C. albicans isolates, whereas 10 isolates were unequivocally identified as being C. dubliniensis isolates by using the Vitek 2 system and the ID-YST card.
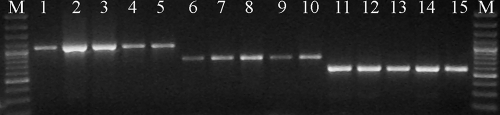
Three hundred thirty-eight isolates of Candida spp. produced a DNA fragment of 941 bp that was identical to that of the reference strain C. albicans ATCC 10231. Twenty-seven isolates produced a DNA fragment of 700 bp, indicating that these strains belong to C. africana, whereas only 11 isolates showed a DNA fragment of 569 bp and were identified as being C. dubliniensis. An overview of the discriminatory power of the PCR primers used is given in Fig. 1.
FIG. 1.
Molecular discrimination between C. albicans (lanes 1 to 5), C. africana (lanes 6 to 10), and C. dubliniensis (lanes 11 to 15) by using a single pair of primers derived from hwp1 genes. M, molecular size markers.
C. albicans was the most common species, representing 89.9% of the isolates, followed by C. africana strains (7.2%) and C. dubliniensis strains (2.9%).
C. albicans was recovered from all different kinds of tested samples. A higher incidence (41%) of this species was found in vaginal samples (139 isolates). Indeed, C. dubliniensis isolates (11 in total) were found primarily in oral specimens (8 isolates [72.7%]), followed by vaginal samples (2 isolates [18.2%]), and one isolate (9.1%) was recovered from gastric fluid.
C. africana was found only in vaginal specimens (27 isolates).
Only 2 of 52 Candida isolates used in the retrospective study were identified as being C. africana isolates.
The remainders of these isolates were all recognized as being C. albicans isolates.
The results obtained by using our molecular method showed that all 27 C. africana strains and 1 isolate of C. dubliniensis were misidentified as being C. albicans isolates and were not resolved by the Vitek 2 system.
The development of improved methods for identifying non-C. albicans species such as the well-known C. dubliniensis during the past years has resulted in a large volume of published data describing the epidemiology of this species (20). However, no rapid and accurate phenotypic methods are available for clinical laboratory applications, and the identification of C. dubliniensis still requires genotyping techniques (9, 10). C. africana, considered to be an atypical chlamydospore-negative C. albicans strain, which has recently been proposed to represent a separate species within the Candida genus, has been added.
Infections due to C. africana in Africa (22) and some European countries (2, 17, 23) have been reported. However, on the basis of some data reported in the literature, we assume that this microorganism has a worldwide distribution.
In 1991, Asakura et al. (3) reported seven atypical C. albicans isolates that shared an unusual assimilation pattern, serotype B, from Japan, and they failed to produce chlamydospores. One of these seven strains (strain TCH23) also showed an atypical electrophoretic karyotype and was classified as a variant of C. albicans with an atypical phenotype (8). Moreover, Al-Hedaithy and Fotedar (1) recovered and studied 25 atypical C. albicans strains from 21 specimens from female patients in Saudi Arabia. The majority of these clinical isolates showed a carbohydrate assimilation profile typical of the C. africana strains reported in other studies (2, 17).
Recently, Odds et al. (13) analyzed data from multilocus sequence typing for a panel of 1,391 C. albicans isolates. Unlike C. africana strains, which have so far been isolated from genital sites (1, 2, 17, 23), one strain from Chile, South America, studied by Odds et al. (13) was isolated from blood culture. Interestingly, this Chilean strain could represent the first isolation of C. africana from a different clinical sample, suggesting that this fungus can be associated with a more wide clinical spectrum.
Our data clearly indicate that the incidence of C. albicans in clinical samples could be overestimated if the variants of this species were placed into a separate group from typical C. albicans isolates. However, until now, none of the epidemiological studies took into account the incidence of C. africana. This is because most clinical laboratories use automated identification systems that do not always allow discrimination between closely related Candida species and/or variants (4, 6). In addition, only a few clinical laboratories perform extensive mycological investigations, including chlamydospore production on cornmeal agar at 24°C for up to 5 days. Therefore, as still happens for some C. dubliniensis strains, C. africana strains are often misidentified as being typical C. albicans strains. In fact, in this study, the Vitek 2 system identified all C. africana isolates as being typical C. albicans strains (with 85% probability, which is an acceptable identification), indicating that the biochemical reactions of the Vitek ID-YST card are not appropriate for distinguishing between these two closely related yeasts.
On the other hand, infections due to C. dubliniensis and/or C. africana are underestimated and not well known because although some discriminative methods for these pathogenic yeasts are described in literature, they are not routinely used in many clinical diagnostic laboratories.
To our knowledge, this is the first well-documented report regarding the occurrence of C. africana in clinical samples.
Acknowledgments
We thank Giuseppe Teti, Department of Pathology and Experimental Microbiology, University of Messina Medical School, Messina, Italy; the staff of the Department of Microbiology and Virology, Civil Hospital, Vibo Valentia, Italy; and Giuseppe Bolignano and his staff of the Department of Microbiology, Riuniti Hospitals (OORR), Reggio Calabria, Italy, for providing Candida sp. strains. We are grateful to Derek. J. Sullivan, Dublin Dental School and Hospital, University of Dublin, Dublin, Ireland, and Hans-Jürgen Tietz, Dermatologische Universitätsklinik und Poliklinik, Universitätsklinikum Charité, Berlin, Germany, for supplying C. dubliniensis CD36 and C. africana A1605, A1622, A1653, and M8684. We are also very grateful to Frank C. Odds, Medical Mycology Aberdeen Fungus Group, Institute of Medical Sciences, Aberdeen, United Kingdom, for critical reading and comments during the realization of the manuscript.
Footnotes
Published ahead of print on 5 November 2008.
REFERENCES
- 1.Al-Hedaithy, S. S. A., and R. Fotedar. 2002. Recovery and studies on chlamydospores-negative Candida albicans isolated from clinical specimens. Med. Mycol. 40301-306. [DOI] [PubMed] [Google Scholar]
- 2.Alonso-Vargas, R., L. Elorduy, E. Eraso, J. F. Cano, J. Guarro, J. Pontón, and G. Quindòs. 2008. Isolation of Candida africana, probable atypical strains of Candida albicans, from a patients with vaginitis. Med. Mycol. 46167-170. [DOI] [PubMed] [Google Scholar]
- 3.Asakura, K., S. Iwaguchi, M. Homma, T. Sukai, K. Higashide, and K. Tanaka. 1991. Electrophoretic karyotypes of clinically isolated yeasts of Candida albicans and C. glabrata. J. Gen. Microbiol. 1372531-2538. [DOI] [PubMed] [Google Scholar]
- 4.Campanha, N. H., K. H. Neppelenbroek, D. M. Spolidorio, L. C. Spolidorio, and A. C. Pavarina. 2005. Phenotypic methods and commercial systems for the discrimination between C. albicans and C. dubliniensis. Oral Dis. 11392-398. [DOI] [PubMed] [Google Scholar]
- 5.Forche, A., G. Schönian, Y. Gräser, R. Vilgalys, and T. G. Mitchell. 1999. Genetic structure of typical and atypical populations of Candida albicans from Africa. Fungal Genet. Biol. 28107-125. [DOI] [PubMed] [Google Scholar]
- 6.Freydiere, A. M., R. Guinet, and P. Boiron. 2001. Yeast identification in the clinical microbiology laboratory: phenotypical methods. Med. Mycol. 399-33. [DOI] [PubMed] [Google Scholar]
- 7.Gutiérrez, J., P. Morales, M. A. González, and G. Quindós. 2002. Candida dubliniensis, a new fungal pathogen. J. Basic Microbiol. 42207-227. [DOI] [PubMed] [Google Scholar]
- 8.Iwaguchi, S. I., M. Sato, B. B. Magee, P. T. Magee, K. Makimura, and T. Suzuki. 2001. Extensive chromosome translocation in a clinical isolate showing the distinctive carbohydrate assimilation profile from a candidiasis patient. Yeast 181035-1046. [DOI] [PubMed] [Google Scholar]
- 9.Khlif, M., H. Sellami, A. Sellami, H. Chelly, F. Makni, M. Bouaziz, and A. Ayadi. Candida dubliniensis: first identification in Sfax hospital, Tunisia. Mycoses, in press. [DOI] [PubMed]
- 10.Loreto, E. S., A. R. Bolzan, C. E. Linares, E. Boff, J. M. Santurio, and S. H. Alves. 2006. Evaluation of 5 new media containing extracts of seeds applied to Candida dubliniensis screening. Diagn. Microbiol. Infect. Dis. 55191-193. [DOI] [PubMed] [Google Scholar]
- 11.Marriott, D., M. Laxton, and J. Harkness. 2001. Candida dubliniensis candidemia in Australia. Emerg. Infect. Dis. 7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odds, F. C., L. Van Nuffel, and G. Dams. 1998. Prevalence of Candida dubliniensis isolates in a yeast stock collection. J. Clin. Microbiol. 62869-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Odds, F. C., M. E. Bougnoux, D. J. Shaw, J. M. Bain, A. D. Davidson, D. Diogo, M. D. Jacobsen, M. Lecomte, S. Y. Li, A. Tavanti, M. C. Maiden, N. A. Gow, and C. d'Enfert. 2007. Molecular phylogenetics of Candida albicans. Eukaryot. Cell 61041-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfaller, M. A., and D. J. Diekema. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20133-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfaller, M. A., P. G. Pappas, and J. R. Wingard. 2006. Invasive fungal pathogens: current epidemiological trends. Clin. Infect. Dis. 43S3-14. [Google Scholar]
- 16.Romeo, O., and G. Criseo. 2008. First molecular method for discriminating between Candida africana, Candida albicans and Candida dubliniensis by using hwp1 gene. Diagn. Microbiol. Infect. Dis. 62230-233. [DOI] [PubMed] [Google Scholar]
- 17.Romeo, O., and G. Criseo. Morphological, biochemical and molecular characterisation of the first Italian Candida africana isolate. Mycoses, in press. [DOI] [PubMed]
- 18.Reference deleted.
- 19.Shao, P. L., L. M. Huang, and P. R. Hsueh. 2007. Recent advances and challenges in the treatment of invasive fungal infections. Int. J. Antimicrob. Agents 30487-495. [DOI] [PubMed] [Google Scholar]
- 20.Sullivan, D. J., G. P. Moran, E. Pinjon, A. Al-Mosaid, C. Stokes, C. Vaughan, and D. C. Coleman. 2004. Comparison of the epidemiology, drug resistance mechanisms, and virulence of Candida dubliniensis and Candida albicans. FEMS Yeast Res. 4369-376. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan, D. J., T. J. Westerneng, K. A. Haynes, D. E. Bennett, and D. C. Coleman. 1995. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology 1411507-1521. [DOI] [PubMed] [Google Scholar]
- 22.Tietz, H. J., A. Küssner, M. Thanos, M. Pinto De Andreade, W. Presber, and G. Schönian. 1995. Phenotypic and genotypic characterization of unusual vaginal isolates of Candida albicans from Africa. J. Clin. Microbiol. 332462-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tietz, H. J., M. Hopp, A. Schmalreck, W. Sterry, and V. Czaika. 2001. Candida africana sp. nov., a new human pathogen or a variant of Candida albicans? Mycoses 44437-445. [DOI] [PubMed] [Google Scholar]
- 24.Tortorano, A. M., C. Kibbler, J. Peman, H. Bernhardt, L. Klingspor, and R. Grillot. 2006. Candidaemia in Europe: epidemiology and resistance. Int. J. Antimicrob. Agents 27359-366. [DOI] [PubMed] [Google Scholar]
- 25.van Hal, S. J., D. Stark, J. Harkness, and D. Marriott. 2008. Candida dubliniensis meningitis as delayed sequela of treated C. dubliniensis fungemia. Emerg. Infect. Dis. 14327-329. [DOI] [PMC free article] [PubMed] [Google Scholar]