Abstract
Intra-amniotic infection and inflammation are major causes of preterm birth (PTB). However, intra-amniotic inflammation is often detected in the absence of infection. This may partly be due to the culturing methods employed in hospital laboratories, which are unable to detect the uncultivated species. In this study, intra-amniotic microbial infections associated with PTB were examined by both culture and 16S rRNA-based culture-independent methods and were corroborated by the presence of intra-amniotic inflammation. Amniotic fluid (AF) specimens from 46 pregnancies complicated by PTB and 16 asymptomatic women were analyzed. No bacterial DNA was amplified in AF collected from the asymptomatic women. Among the 46 samples associated with PTB, bacterial DNA was amplified from all (16/16) of the culture-positive samples and 17% (5/30) of the culture-negative samples. In the culture-positive group, additional species were detected in more than half (9/16) of the cases by PCR and clone analysis. Altogether, approximately two- thirds of the species detected by the culture-independent methods were not isolated by culture. They included both uncultivated and difficult-to-cultivate species, such as Fusobacterium nucleatum, Leptotrichia (Sneathia) spp., a Bergeyella sp., a Peptostreptococcus sp., Bacteroides spp., and a species of the order Clostridiales. To examine intra-amniotic inflammation, an AF proteomic fingerprint (mass-restricted score) was determined by surface-enhanced laser desorption ionization-time-of-flight mass spectrometry. Inflammation was detected in all five samples which were culture negative but PCR positive. Women who were PCR positive more often had elevated interleukin-6 levels in their AF, histological chorioamnionitis, and funisitis and delivered neonates with early-onset neonatal sepsis. Previously unrecognized, uncultivated, or difficult-to-cultivate species may play a key role in the initiation of PTB.
Intrauterine infection has long been recognized to play a role in spontaneous preterm birth (PTB), especially in those occurring at less than 30 to 32 weeks of gestation (18). Bacterial culturing has widely been employed for the detection of microbial invasion of the amniotic cavity, which under normal conditions remains sterile. A variety of microorganisms have been cultivated from amniotic fluid (AF) in pregnancies complicated by PTB, with Ureaplasma urealyticum, Mycoplasma hominis, Fusobacterium nucleatum, Gardnerella vaginalis, and Bacteroides spp. being the most prevalent (18, 22, 23, 38, 52). The current paradigm for infection-induced PTB suggests that the majority of these organisms originate from the lower genital tract and invade the pregnant uterus via an ascending mechanism (44). The bacteria are believed to first infect the decidua and then spread to the fetal membranes, AF, and finally, the fetus (44). A secondary route of infection is hematogenous transmission, with the infectious organism originating from nonreproductive parts of the body (5).
Although most organisms isolated from intra-amniotic infections are of relatively low virulence (18), it is proposed that once they are inside the uterus they stimulate neutrophil infiltration and activation, resulting in increased synthesis and release of proinflammatory cytokines, prostaglandin, and matrix metalloproteases and leading to cervical ripening, membrane rupture, uterine contractions, and PTB (5). This argues for the importance of identifying the etiologic agents of intra-amniotic infections so that targeted antibiotic treatment is initiated early in the pathophysiological chain of events.
Microbial culture of AF is recognized as the “gold standard” for the detection of intra-amniotic infection. However, the disadvantage of using a culture-based approach for bacterial identification is its dependence on growth conditions favorable to a select group of known microorganisms. Thus, uncultivated or difficult-to-cultivate bacteria will not be found if one relies on culture conditions alone (31). For example, by using culture-independent techniques, a previously unrecognized and uncultivated oral species of the genus Bergeyella was found to be implicated in a case of extremely early PTB (19).
The cornerstone of culture-independent detection has been the use of 16S rRNA gene sequencing to identify and differentiate microbial species and to determine the phylogenetic relationship between the microorganisms (53). The 16S rRNA genes, which are usually ∼1,500 bp in length, are highly conserved among bacterial and archaeal species, yet they are sufficiently diverse to allow differentiation. The application of culture-independent techniques led to the striking realization that cultivated organisms represented only a tiny fraction (<1%) of the species in the environment (39). By analogy, it is thus possible that AF cultures are unable to detect the true prevalence of amniotic infection, explaining why discrepancies between the results of tests with AF that assess the host response to infection (i.e., intra-amniotic inflammation) and the presence of bacteria by culture are common in clinical settings (6). In recent years, proteomic profiling of AF has been used to assess intra-amniotic inflammation (6, 9, 11) due to its ability to accurately predict the presence and the severity of histological chorioamniotis (10), funisitis (7), and early-onset neonatal sepsis (6, 7). Yet, the relationship between AF proteomic profiling and bacterial identification has not been fully established, in part due to the incompleteness of bacterial identification by the culturing method. Our hypothesis is that uncultivated microbial species play a previously unrecognized yet significant role in the etiology and pathogenesis of PTB in women who display proteomic biomarkers characteristic for intra-amniotic inflammation. The purpose of our study was to systematically examine the AF of women whose pregnancies were complicated by symptoms of PTB in the presence and absence of intra-amniotic inflammation for the presence of both cultivated and uncultivated bacteria and their prevalence.
MATERIALS AND METHODS
Study population and research design.
We performed a comprehensive bacteriological evaluation of AF in 62 consecutive pregnancies (Fig. 1). This study has been approved by the Human Investigation Committee of Yale University and the Internal Review Board of Case Western Reserve University. Women were enrolled immediately after admission to the Labor and Birth Unit or the High-Risk Antepartum Unit at Yale New Haven Hospital from April 2004 to January 2007. Written informed consent for research purposes was obtained from all participants prior to performance of the procedures. Forty-six women presented with symptoms of either preterm labor or preterm premature rupture of membranes (PPROM) (the study group; mean gestational age ± standard deviation, 27 ± 4 weeks). For obvious ethical reasons, an amniocentesis procedure could not be performed for asymptomatic pregnant women; thus, 16 clinically asymptomatic women undergoing amniocentesis for either genetic testing (n = 7; gestational age, 19 ± 3 weeks) or fetal lung maturity determination (n = 9; gestational age, 36 ± 1 weeks) were recruited as the control group. Control samples were selected in a random fashion with the purpose of providing reassurance that the process of AF sampling and laboratory handling did not cause ex vivo contamination with bacterial DNA.
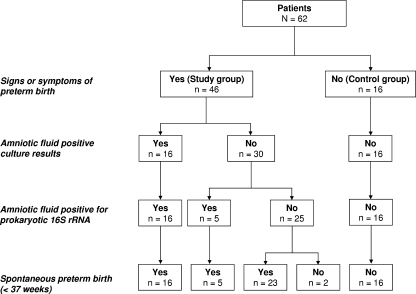
FIG. 1.
Flowchart of patients in the study and control groups, results of culture-dependent and culture-independent methods, and pregnancy outcome.
Subjects eligible for the study group met the following criteria: singleton fetus at <34 weeks of gestational age with a clinically indicated amniocentesis to rule out intra-amniotic infection. Exclusion criteria included anhydramnios, human immunodeficiency virus or hepatitis virus infections, and nonreassuring fetal status. Exclusion criteria for patients in the control group included major fetal structural abnormalities, chromosomal aneuploidy, preeclampsia, gestational or chronic hypertension, cholestasis, multiple gestation, uterine contractions, and fever. Gestational age was established on the basis of the last menstrual period and an ultrasonographic examination prior to 20 weeks of gestation in all instances. Preterm labor was defined as the presence of regular uterine contractions and documented cervical effacement and/or dilatation in patients at <37 weeks of gestation. Cervical dilatation was assessed by speculum examination in all cases. The diagnosis of PPROM was confirmed by vaginal AF pooling, the nitrazine test, or ferning or by a positive amniocentesis dye test result. Digital examinations were not permitted for women with confirmed PPROM. In the absence of signs or symptoms of clinical chorioamnionitis (body temperature of >37.8°C, uterine tenderness, fetal tachycardia), AF laboratory results suggestive of infection, nonreassuring fetal heart tracing, and/or abruption, PPROM was managed expectantly. According to the institutional protocol, patients received corticosteroids for lung maturity if the fetus was at <32 weeks of gestational age and antibiotic therapy, as clinically indicated (1). Fetal monitoring was performed at least twice daily, followed by determination of a biophysical profile if it was so indicated. The induction of labor or a surgical delivery was performed for such clinical indications as AF laboratory test results traditionally considered to indicate intra-amniotic inflammation or infection, fetal lung maturity, prolapsed umbilical cord, and/or a gestational age of ≥34 weeks in the context of PPROM (36).
Microbiological analysis of AF by culture.
AF was retrieved by ultrasound-guided amniocentesis using sterile techniques and was immediately examined in the clinical laboratory for the presence of microorganisms by the traditional culturing method. Briefly, AF was centrifuged and the sediment was resuspended in 0.25 ml of supernatant. The remaining AF was centrifuged at 3,000 × g at 4°C for 20 min, aliquoted, and stored at −80°C until analysis. By using quantitative 0.01- and 0.001-mm loops, the concentrated AF was cultured for aerobic and anaerobic bacteria by using the following media: chocolate, Martin-Lewis, MacConkey, azido benzoic acid, thioglycolate, bacteroides bile esculin/laked blood-kanamycin-vancomycin, and Columbia colistin nalidixic acid agars. To identify any potential anaerobic bacteria, AF was cultured in an anaerobic chamber (Forma anaerobic system; Thermo Electron Co, Waltham, MA) at 37°C. The results of the microbiological tests were available for case management and were reported as final after 5 days of culturing. Presumptive bacterial identification was based on standard microbiological criteria of colonial morphology, medium reaction, and Gram stain and by the use of a Vitek 2 automated card system (bioMérieux, Hazelwood, MO) on the basis of biochemical test results and the results of antibiotic susceptibility testing. To identify Ureaplasma and Mycoplasma, AF was incubated in Ureaplasma broth (Northeast Laboratory, Waterville, ME) for 24 h. If the broth turned pink, the presence of Mycoplasma and/or Ureaplasma was indicated. This was then followed by a subculture to Ureaplasma differential agar (modified U9 agar; Northeast Laboratory). The agar plate was incubated anaerobically for 48 h, followed by 96 h of aerobic incubation at 37°C. The plate was examined under a microscope once a day. If the agar turned pink, Ureaplasma urealyticum was suggested and the identity of the species was confirmed by colony morphology (granular pink-brown urease-positive colonies). Fried egg-type colonies that did not turn pink on the differential agar were classified as Mycoplasma hominis (2).
Microbial analysis of AF by culture-independent methods. (i) DNA extraction from AF.
DNA was extracted as described previously (19).
(ii) PCRs.
All PCRs were carried out with GoTaq DNA polymerase (Promega, Madison, WI) and an Applied Biosystems 2720 thermal cycler. A total of 2 μl of DNA from each sample was used as the template in a 25-μl reaction volume. The PCR conditions were as follows: an initial denaturation at 94°C for 3 min; 28 to 32 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 2 min; and an additional extension at 72°C for 10 min. Universal primers A17F and 1512R (Table 1) (19, 30) were used for 16S rRNA gene amplification, and the PCR products were observed by 1% agarose gel electrophoresis. Fusobacterium nucleatum 12230, Eikenella corrodens ATCC 23834, and Klebsiella pneumoniae NCIMB8267 were used as positive controls and for determination of the limit of detection. The identities of the control species were verified by DNA sequence analysis of the PCR products. To ensure the validity of our results, negative controls were included with each PCR set to examine the PCR mixtures for environmental or human DNA contamination.
TABLE 1.
Primers used in this study
| Primer | Description or purpose | Sequence (5′-3′) | Reference or source |
|---|---|---|---|
| A17F | Forward primer in 16S region, 17 bases from 5′ end of 16S rRNA | GTTTGATCCTGGCTCAG | 19,30 |
| 1512R | Reverse primer in 16S region 1,512 bases from 5′ end of 16S rRNA | TACCTTGTTACGACTT | 19,30 |
| GW1 | DNA sequencing | GTTGCAACAAATTGATGAGCAATGC | MBCa |
| GW2 | DNA sequencing | GTTGCAACAAATTGATGAGCAATTA | MBC |
MBC, Molecular Biotechnology Core, Lerner Institute, Cleveland, OH.
(iii) Clone analysis and species identification.
To identify the species amplified by PCR and to ensure that the PCR amplicons were indeed bacterial rRNA genes rather than artifacts, a systematic clone analysis was employed. The PCR products were cloned into the pCR8 vector, and at least 20 random transformants were selected for plasmid purification and restriction enzyme digestion. Following electrophoresis, a total of 10 clones with positive 16S rRNA gene inserts were selected for species identification. Plasmids from these 10 clones were extracted by using a WizardPlus SV minipreps DNA purification system (Promega), and their inserts were sequenced (Genomics Core Facility, Case Western Reserve University, Cleveland, OH) by using primers GW1 and GW2 (Table 1), which provided the sequence of the 16S rRNA gene insert from opposite ends. When the DNA sequences obtained with primers GW1 and GW2 were combined, the entire 16S rRNA gene was identified. The sequences were assembled and aligned by using the VectorNTI program (Invitrogen, Carlsbad, CA). The NCBI BLAST nucleotide sequence database (http://www.ncbi.nlm.nih.gov/blast/blast.cgi) was searched for preliminary species identification, regardless of whether the species was cultivated or uncultivated. Phylogenetic analysis was then performed to identify the most closely related species.
(iv) Phylogenetic analysis.
The 16S rRNA gene sequences were aligned by using the ClustalX1.8 program (51). A tree for phylogenetic analysis was constructed by using Molecular Evolutionary Genetics Analysis (version 4) software by calculation of a distance matrix and tree reconstruction by the neighbor-joining method (50). The statistical robustness of the analysis was estimated by bootstrapping with 1,000 replicates. Sequences were considered monophylic when they had a connecting node within a genetic distance of 0.05. This analysis allows the presumptive identification of the bacteria by inferring the evolutionary relationships of homologous sequences. The similarities between the 16S rRNA genes within a species are expected to be >97%, and those between different species within a genus are between 93.3 and 99.9% (13, 48).
AF proteomic profiling and generation of the MR score.
After AF was collected, the fresh AF was used to generate the mass-restricted (MR) score by surface-enhanced laser desorption ionization-time-of-flight mass spectrometry. These results were not available for clinical management. The methodology for the generation of the MR score has been described previously (6, 9, 10, 11). A categorical value of 1 was assigned if a biomarker peak was present, and a value of 0 was assigned if one was absent. The MR score ranges from 0 to 4, depending upon the presence or the absence of each of the four protein biomarkers (9). The MR score provides qualitative information on the presence or the absence of intra-amniotic inflammation, with the scores indicating that intra-amniotic inflammation is absent (MR score, 0), minimal (MR score, 1 or 2), or severe (MR score, 3 or 4) (6, 9).
Immunoassays for IL-6.
An enzyme-linked immunosorbent assay for human interleukin-6 (IL-6; Pierce-Endogen, Rockford, IL) was performed in duplicate, according to the manufacturer's instructions. The minimal detectable concentration was 1 pg/ml, and the inter- and intra-assay coefficients of variation were <10%.
Histological evaluation of the placenta and diagnosis of inflammation.
Histological examination of the placenta was performed by a perinatal pathologist who was unaware of the results of the microbial cultures, the MR scores, the AF IL-6 levels, and the results of the clone analysis and species identification. Hematoxylin-eosin-stained sections of the chorionic plate, extraplacental membranes, and umbilical cord were available for histological examination for 42/46 cases. The remaining four cases represented either deliveries at outside hospitals and/or deliveries at >37 weeks of gestation. Neutrophil infiltration of the chorionic plate and amniochorion membranes was used to establish the presence of histological chorioamnionitis, and the severity was established by using well-recognized histological stages and grading systems (37, 45).
Evaluation of early-onset neonatal sepsis.
Neonatal hematological indices and sepsis categorization were assessed by the use of blood specimens and cultures of samples obtained after delivery, as previously described (6, 40, 41). Neonatal sepsis was defined as the presence of confirmed or suspected sepsis at ≤3 days after birth. A diagnosis of early-onset neonatal sepsis was based on clinical symptoms corroborated by hematological laboratory test results and was dichotomized into either present (when sepsis was either confirmed or suspected) or absent. All neonates with confirmed or suspected sepsis received antibiotic therapy.
Statistical analysis.
The Kolmogorov-Smirnov test was used for the testing of data normality. The data are presented as either means with standard deviations (if they were normally distributed) or as the medians and interquartile ranges if they were nonnormally distributed. Statistical analyses were performed with Sigma Stat statistical software (version 2.03; SPSS Inc., Chicago, IL). The data were compared by the Student t test, the Mann-Whitney test, or the Kruskal-Wallis analysis of variance on ranks followed by Dunn's tests to adjust for multiple comparisons as appropriate. Comparisons between proportions were performed by the chi-square or Fisher's exact test. A P value of <0.05 was considered significant.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of the species identified in this study have been deposited in the GenBank database (http://www.ncbi.nlm.nih.gov), and the accession numbers are listed in Table 4.
TABLE 4.
Results of 16S rRNA-based identification of the prokaryotic species found in AF of patients with signs and/or symptoms of PTB
| Sample source | Species | GenBank accession no. | No. of clones | Size (bp) of available sequence |
|---|---|---|---|---|
| Case 1 | Leptotrichia sanguinegensa | EU644451 | 6 | 1,470 |
| Case 1 | Bacteroides ureolyticusb | EU644452 | 4 | 1,467 |
| Case 2 | Bacteroides fragilisb | EU644453 | 7 | 1,484 |
| Case 2 | Citrobacter koseri | EU644454 | 3 | 1,497 |
| Case 3 | Fusobacterium nucleatumb | EU644455 | 10 | 1,473 |
| Case 4 | Prevotella bivia | EU644456 | 10 | 1,484 |
| Case 5 | Shigella sp. | EU644457 | 5 | 1,497 |
| Case 5 | Shigella sp. | EU644458 | 4 | 1,497 |
| Case 5 | Shigella sp. | EU644459 | 1 | 1,497 |
| Case 6 | Bacteroides ureolyticusb | EU644460 | 9 | 1,467 |
| Case 6 | Fusobacterium nucleatumb | EU644461 | 1 | 1,474 |
| Case 7 | Clostridiales bacteriuma | EU644462 | 7 | 1,487 |
| Case 7 | Fusobacterium nucleatumb | EU644463 | 3 | 1,474 |
| Case 8 | Fusobacterium nucleatumb | EU644464 | 10 | 1,473 |
| Case 9 | Bergeyella sp.a | EU644465 | 8 | 1,483 |
| Case 9 | Fusobacterium nucleatumb | EU644466 | 2 | 1,474 |
| Case 10 | Ureaplasma parvum serovar | EU644473 | 10 | 1,472 |
| Case 11 | Ureaplasma parvum serovar | EU644474 | 10 | 1,472 |
| Case 12 | Streptococcus agalactiae | EU644467 | 10 | 1,504 |
| Case 13 | Streptococcus agalactiae | EU644468 | 10 | 1,504 |
| Case 14 | Leptotrichia amnioniia | EU644469 | 10 | 1,468 |
| Case 15 | Shigella sp. | EU644470 | 5 | 1,497 |
| Case 15 | Shigella sp. | EU644471 | 3 | 1,497 |
| Case 15 | Shigella sp. | EU644472 | 2 | 1,497 |
| Case 16 | Mycoplasma hominisb | EU644475 | 10 | 1,473 |
| Case 17 | Peptostreptococcus sp.b | EU644476 | 8 | 1,603 |
| Case 17 | Mycoplasma hominisb | EU644477 | 2 | 1,473 |
| Case 18 | Fusobacterium nucleatumb | EU644478 | 10 | 1,476 |
| Case 19 | Sneathia sanguinegensa | EU644479 | 10 | 1,470 |
| Case 20 | Fusobacterium nucleatumb | EU644480 | 10 | 1,474 |
| Case 21 | Sneathia sanguinegensa | EU644481 | 10 | 1,470 |
Uncultivable species.
Difficult-to-cultivate species.
RESULTS
A flowchart of the patients assigned to the study and control groups, detection of intra-amniotic infection by the traditional culturing method and 16S rRNA gene-based culture-independent methods, and pregnancy outcomes is presented in Fig. 1. Briefly, of the 46 pregnant women included in the study group, 32 delivered newborns who were admitted to the Newborn Special Care Unit. Of the remainder, nine women delivered fetuses below the limit of viability (<24 weeks of gestation), three had term or near-term newborns who did not require intensive special care, and two delivered at outside hospitals. The rate of PTB occurring at <34 weeks of gestation was 89% (41/46). The high prevalence of spontaneous early PTB in the study group supports the clinical relevance of this cohort.
Table 2 summarizes the demographic, clinical, and outcome characteristics of the study group stratified by the status of the fetal membranes at the time of amniocentesis. Of note was that women with intact membranes were presenting with more advanced cervical dilatation and were of lower gestational ages at the times of both amniocentesis and delivery than women presenting with PPROM. Of the viable infants requiring admission in the Newborn Special Care Unit (n = 32), those delivered by mothers with intact membranes had lower birth weights and more often had early-onset neonatal sepsis, as confirmed by positive blood culture results at birth.
TABLE 2.
Demographic, clinical, and outcome characteristics of the study group
| Variable | Study group
|
P valuea | ||
|---|---|---|---|---|
| All | Intact membranes | PPROM | ||
| Mothers (no. of patients) | 46 | 26 | 20 | |
| Maternal age (yr)b | 28 ± 6 | 28 ± 6 | 29 ± 6 | 0.792 |
| Non-Caucasian racec | 30 (65) | 16 (62) | 14 (70) | 0.776 |
| Gravidityd | 3 (2-4) | 3 (2-3) | 3 (2-5) | 0.956 |
| Parityd | 1 (0-1) | 1 (0-1) | 1 (0-1) | 0.556 |
| History of PTBc | 12 (26) | 6 (23) | 6 (30) | 0.848 |
| Gestational age at amniocentesis (wk)b | 27 ± 4 | 25 ± 4 | 29 ± 4 | 0.002 |
| Uterine contractionsc | 23 (50) | 16 (62) | 7 (35) | 0.137 |
| Cervical dilatationd | 1 (0-2) | 2 (1-3) | 0 (0-1) | <0.001 |
| Clinical chorioamnionitise | 6 (13) | 4 (15) | 2 (10) | 0.684 |
| Steroid exposurec | 33 (72) | 18 (69) | 15 (75) | 0.920 |
| Antibiotic treatmentc | 35 (76) | 16 (62) | 19 (95) | 0.013 |
| Tocolytic treatmentb | 16 (35) | 11 (42) | 5 (25) | 0.363 |
| Time from amniocentesis to delivery (h)d | 19 (10-155) | 17 (5-293) | 19 (11-89) | 0.883 |
| Gestational age at delivery (wk)b | 28 ± 5 | 27 ± 6 | 30 ± 4 | 0.033 |
| Term delivery (≥37 wk of gestation)e | 2 (4) | 2 (8) | 0 (0) | 0.498 |
| Cesarean deliveryc | 15 (33) | 8 (31) | 7 (35) | 0.989 |
| Placentas (no.) | 42 | 22 | 20 | |
| Histological chorioamnionitis stages II and IIIc | 31 (74) | 18 (82) | 13 (65) | 0.375 |
| Funisitis grades 1 to 4c | 25 (60) | 16 (73) | 9 (45) | 0.130 |
| Neonates (no. admitted to NBSCUf) | 32 | 13 | 19 | |
| Birth weight (g)b | 1,432 ± 658 | 1,036 ± 380 | 1,702 ± 678 | 0.003 |
| Apgar score at 1 min of <7c | 15 (47) | 9 (69) | 8 (42) | 0.166 |
| Apgar score at 5 min of <7e | 9 (28) | 5 (38) | 4 (21) | 0.427 |
| Early-onset neonatal sepsisd | 19 (60) | 7 (54) | 6 (32) | 0.372 |
| Positive blood cultures at birthe | 4 (8) | 4 (31) | 0 (0) | 0.020 |
P value of statistical comparison between groups with intact membranes and PPROM.
Data are presented as means ± standard deviations and were analyzed by Student's t test.
Data are presented as the number (percent) of patients and were analyzed by the chi-square test.
Data are presented as the median (interquartile range) and were analyzed by the Mann-Whitney test.
Data are presented as the number (percent) of patients and were analyzed by Fisher's exact test.
NBSCU, newborn special care unit.
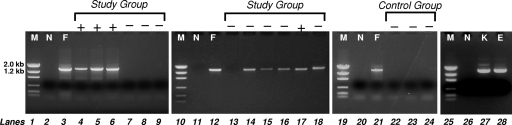
Of the 46 AF samples included in the study group, microbial species were identified in 16 specimens by culture (Fig. 1). By using 16S rRNA gene-based PCR analysis, bacteria were detected in all (16/16) samples which were culture positive (as exemplified in Fig. 2, lanes 4 to 6). Bacterial DNA was not detected in 83% (25/30) of the study group samples which tested culture negative (as exemplified in Fig. 2, lanes 7 to 9 and 13). Conversely, 17% (5/30) of the study group samples which were culture negative tested positive by PCR (as exemplified in Fig. 2, lanes 14 to 16 and 18). Therefore, bacterial 16S rRNA genes were detected in a total of 21 AF specimens. For the control group, all bacterial culture results were negative and no bacterial DNA was amplified by PCR (as exemplified in Fig. 2, lanes 22 to 24).
FIG. 2.
PCR examination of AF samples with universal primers A17F and 1512R. The PCR products from representative samples of AF were examined on 1% agarose gels. Lanes + and −, AF samples with positive and negative microbial culture results, respectively; lanes M, DNA size markers; lanes N, negative control consisting of phosphate-buffered saline; lanes F, Fusobacterium nucleatum 12230; lanes K, Klebsiella pneumoniae NCIMB8267; lane E, Eikenella corrodens ATCC 23834.
The bacterial species identified by culture were the following: Ureaplasma urealyticum (five cases), Fusobacterium spp. (four cases), Prevotella bivia (two cases), Escherichia coli (two cases), group B streptococci (two cases), Citrobacter koseri (1 case), a viridans group streptococcus (one case), Klebsiella pneumoniae (one case), and Eikenella corrodens (one case) (Table 3). Mixed infections, in which more than one microorganism was isolated, were identified by culture in six AF samples (from cases 2, 3, 4, 6, 13, and 14) (Table 3).
TABLE 3.
Summary of prokaryotic species detected by culture and culture-independent techniques
| Test result and case no. | Membrane status | Gestational age (wk) at:
|
MR score | Cultured species | DNA-based identification | |
|---|---|---|---|---|---|---|
| Amniocentesis | Birth | |||||
| AF culture positive and PCR positive | ||||||
| 1 | Intact | 27 6/7 | 28 1/7 | 4 | Prevotella biviaa | Leptotrichia sanguinegens,bBacteroides ureolyticusb |
| 2 | Intact | 21 5/7 | 21 6/7 | 4 | Citrobacter koseri, viridans group streptococcusa | Citrobacter koseri, Bacteroides fragilisb |
| 3 | Intact | 21 1/7 | 21 1/7 | 4 | Ureaplasma urealyticum,aFusobacterium spp. | Fusobacterium nucleatumb |
| 4 | PPROM | 32 6/7 | 32 6/7 | 3 | Ureaplasma urealyticum,aKlebsiella pneumoniae,aPrevotella bivia | Prevotella bivia |
| 5 | Intact | 24 1/7 | 24 1/7 | 3 | Escherichia colia | Shigella spp.b |
| 6 | Intact | 20 6/7 | 21 1/7 | 4 | Mixed anaerobes | Fusobacterium nucleatum,bBacteroides ureolyticusb |
| 7 | Intact | 27 6/7 | 27 6/7 | 4 | Fusobacterium nucleatum | Fusobacterium nucleatum, Clostridiales bacteriumb |
| 8 | Intact | 27 4/7 | 27 4/7 | 4 | Fusobacterium spp. | Fusobacterium nucleatum |
| 9 | Intact | 23 3/7 | 23 4/7 | 3 | Fusobacterium nucleatum | Fusobacterium nucleatum, Bergeyella sp.b |
| 10 | Intact | 19 5/7 | 20 1/7 | 4 | Ureaplasma urealyticum | Ureaplasma parvum serovar |
| 11 | PPROM | 22 2/7 | 22 3/7 | 4 | Ureaplasma urealyticum | Ureaplasma parvum serovar |
| 12 | PPROM | 23 3/7 | 23 3/7 | 4 | Group B streptococcus | Streptococcus agalactiae |
| 13 | PPROM | 27 4/7 | 27 4/7 | 4 | Mixed anaerobes | Streptococcus agalactiae |
| 14 | PPROM | 24 6/7 | 25 0/7 | 3 | Group B streptococcus,aEikenella corrodensa | Leptotrichia amnioniib |
| 15 | Intact | 24 1/7 | 24 4/7 | 4 | Escherichia colia | Shigella spp.b |
| 16 | PPROM | 24 1/7 | 24 1/7 | 4 | Ureaplasma urealyticuma | Mycoplasma hominisb |
| AF culture negative and PCR positive | ||||||
| 17 | Intact | 27 1/7 | 27 1/7 | 3 | Peptostreptococcus sp.,bMycoplasma hominisb | |
| 18 | PPROM | 29 1/7 | 29 2/7 | 3 | Fusobacterium nucleatumb | |
| 19 | PPROM | 32 1/7 | 32 1/7 | 2 | Sneathia sanguinegensb | |
| 20 | Intact | 25 3/7 | 25 3/7 | 3 | Fusobacterium nucleatumb | |
| 21 | Intact | 23 6/7 | 24 1/7 | 3 | Sneathia sanguinegensb | |
Species identified by culture but not by the culture-independent methods.
Species identified by the culture-independent methods but not by culture.
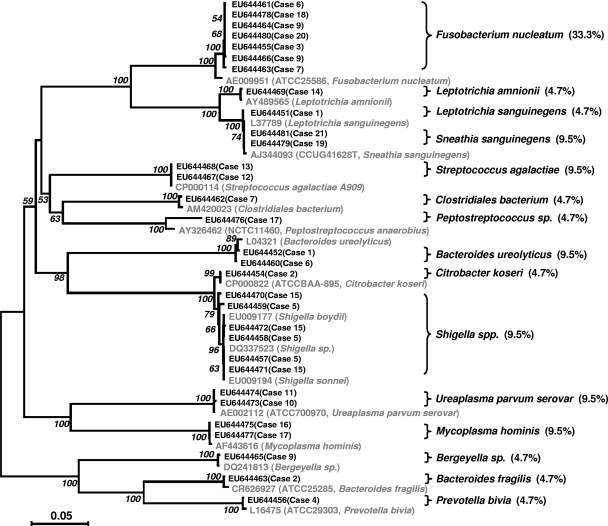
To identify the microorganisms from the study group detected by PCR, a clone library was generated for each AF sample. Ten random clones were selected from each library for DNA sequence analysis (Table 3 and Table 4). The species were identified preliminarily by a BLAST search by use of the 16S rRNA gene sequences. The sequences were deposited in the GenBank database, and the results are presented in Table 4. Furthermore, to confirm the species identities, phylogenetic analysis was performed (Fig. 3). A total of 15 different species were identified (Table 3). In decreasing order of prevalence, they were Fusobacterium nucleatum (seven cases; 33.3% prevalence); Sneathia (Leptotrichia) sanguinegens, Streptococcus agalactiae (group B streptococcus), Bacteroides ureolyticus, Shigella spp., Ureaplasma parvum, and Mycoplasmsa hominis (two cases each; 9.5% prevalence); and Leptotrichia (Sneathia) amnionii, Leptotrichia (Sneathia) sanguinegens, a bacterium of the order Clostridiales, a Peptostreptococcus sp., Citrobacter koseri, a Bergeyella sp., Bacteroides fragilis, and Prevotella bivia (one case each; 4.7% prevalence). Among these, 10 species were not identified by culture, including S. sanguinegens, B. ureolyticus, M. hominis, L. sanguinegens, L. amnionii, the bacterium of the order Clostridiales, the Peptostreptococcus sp., the Shigella spp., the Bergeyella sp., and B. fragilis. With the exception of the Shigella spp., which we think were probably mistakenly identified by the clinical laboratory as its close relative, E. coli (cases 5 and 15), the remaining nine species which were not identified by culture were either uncultivated or difficult to cultivate. These nine species were present in 48% (10/21) of the AF samples found to be positive by PCR. In addition, although F. nucleatum was identified by culture, it was detected in only four cases, whereas it was detected in seven cases by PCR and clone analysis (Table 3). Mixed infections were detected in eight AF samples by the culture-independent methods, whereas mixed infections were detected in six AF samples by culture (Table 3). Altogether, the uncultivated or difficult-to-cultivate species were detected in two-thirds (14/21) of the PCR-positive samples (Table 4). Of the 14 pregnancies from which these PCR-positive samples were obtained, 10 resulted in viable premature neonates, of whom 60% (6/10) were diagnosed with early-onset neonatal sepsis.
FIG. 3.
Phylogenic analysis of total 16S rRNA gene sequences amplified from AF samples. The 16S rRNA sequences were aligned by using the ClustalX1.8 program and were edited manually to remove ambiguities. Molecular Evolutionary Genetics Analysis (version 4) software was used to estimate the evolutionary distance (the number of nucleotide substitutions per site). The statistical robustness of the neighbor-joining tree was confirmed by bootstrapping (1,000 replicates). The reference strains are shown in gray. The prevalence of each species in the 21 positive samples is shown in parentheses as a percentage.
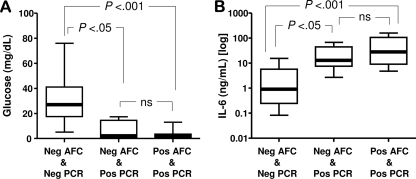
Using AF proteomic analysis, we determined that a total of 65% (30/46) of the specimens from the study group demonstrated evidence of severe intra-amniotic inflammation (i.e., MR scores, 3 to 4). None of the samples with an MR score of 0 (n = 11) tested positive by either culture or PCR. All samples positive by both culture (n = 16) and PCR had MR scores of either 3 or 4 (Table 3). However, by PCR, five additional AF samples (four with MR scores of 3 and one with an MR score of 2) were classified as infected (Table 3). To further demonstrate that the culture-independent method detects intra-amniotic infection of biological relevance, we analyzed the relationship between the microbial detection techniques and the AF glucose and IL-6 levels. The women with negative culture results but positive PCR results had lower AF glucose levels (Fig. 4A) and higher IL-6 levels (Fig. 4B) than those who tested negative by both culture and PCR. When microbial DNA was identified by PCR, there was no significant difference in the AF glucose or IL-6 levels, regardless of the microbial cultures results. Similarly, 100% (five of five) of the placentas of women with negative culture results but positive PCR results had histological chorioamnionitis of stage II or III and 80% (four of five) had funisitis of grade 2 or 3. Women with positive PCR results more often had histological chorioamnionitis, funisitis, and neonates diagnosed with early-onset neonatal sepsis (Table 5).
FIG. 4.
Biological relevance of intra-amniotic infection as assessed by culture-dependent and culture-independent methods. The glucose (A) and interleukin-6 (B) concentrations in AF samples positive by culture and/or by culture-independent methods are presented as percentiles with medians. The ends of the boxes define the 25th and 75th percentiles, the line inside the box defines the median, and the whiskers show the highest and the lowest values. Comparisons among groups were done by Kruskal-Wallis analysis of variance, followed by post-hoc Dunn's tests. AFC, AF culture; PCR, analysis of AF for 16S rRNA by PCR; Neg, negative result; Pos, positive result; ns, not significant.
TABLE 5.
Clinical relevance of intra-amniotic infection, as determined by culture-independent methods
| Variable | No. (%) of patients with the following PCR result:
|
P value | |
|---|---|---|---|
| Negative (n = 25) | Positive (n = 21) | ||
| Positive AF culture resulta | 0 (0) | 16 (76) | <0.001 |
| Early-onset neonatal sepsisa | 2 (12)b | 11 (73)c | <0.001 |
| Clinical chorioamnionitisa | 2 (8) | 4 (19) | 0.390 |
| Histological chorioamnionitis stages II and IIIa | 10 (48)d | 21 (100) | <0.001 |
| Funisitis grades 1 to 4a | 8 (38)d | 17 (81) | 0.011 |
Data were analyzed by Fisher's exact test.
Data were unavailable for seven previable cases.
Data were unavailable for six previable cases.
Data were unavailable for four cases.
DISCUSSION
By employing 16S rRNA gene-based PCR followed by clone analysis, we stand to determine the identity of the bacteria involved in intra-amniotic bacterial infections and the true relationship between intra-amniotic bacterial infection and PTB. We first determined that the true prevalence of intra-amniotic infection is higher than the one determined and accepted by traditional culture techniques. This conclusion is based on our analysis indicating that bacterial DNA is identified in AF in the absence of a traditional positive microbial culture result. This observation elucidates the incompleteness of the culturing method. We further determined that perhaps as many as two-thirds of the organisms associated with intra-amniotic infection are uncultivated or difficult-to-cultivate bacteria. Similar to other studies, we have confirmed that inflammation, as determined by the occurrence of proteomic biomarkers characteristic for intra-amniotic inflammation, is associated with the presence of bacterial DNA in AF yet, in several instances, the absence of a positive microbiological culture result (17). By analyzing multiple clones, we identified the presence of mixed infections and the relative abundance of different species in the sample. Therefore, this approach has the potential to determine the diversity of the microbial species most likely acting as etiologic agents involved in triggering intra-amniotic inflammation and, hence, PTB.
Unlike other prior studies, our investigation employed clone analysis, which is by far the most effective means of identifying multiple species in an infection. Previous studies (3, 17, 24, 26) used either direct sequencing of the amplified 16S rRNA genes or primers for one or two specific species, and therefore, only a few species were anecdotally identified by PCR. In contrast, by sequencing multiple clones in the same sample, our study provides, for the first time, a comprehensive list of the microbial species identified in the intra-amniotic infection. Also, while in some of the previous studies the presence of bacteria was correlated with IL-6 and glucose levels, a much more extensive panel of inflammation and infection markers was correlated with the presence of bacteria in our study. This includes not only the traditional markers of IL-6 and glucose but also early-onset neonatal sepsis, histological chorioamnionitis, funisitis, and the novel intra-amniotic inflammatory marker (the proteomic MR score). The discovery that bacteria can be identified in the AF of women with negative bacterial cultures and high levels of intra-amniotic inflammation is thus of significant clinical relevance.
Phylogenetic classification of the AF species associated with PTB found that the most prevalent taxa were Fusobacterium nucleatum, followed by Shigella spp. and Leptotrichia spp. Fusobacterium nucleatum, a gram-negative anaerobe, is an opportunistic oral pathogen (20). Intrauterine F. nucleatum has been suspected to originate from the oral cavity, where the species is ubiquitous (22, 23, 34). It was previously proposed that after transient bacteremia, oral F. nucleatum translocates to the pregnant uterus and possibly the fetus hematogenously (20). Such a hypothesis was supported by animal studies in which hematogenous injection of orally related F. nucleatum resulted in its preferential localization to placental blood vessels, from which it crossed the endothelium and finally spread to AF to induce premature delivery and stillbirths in a pattern similar to that observed in humans (20). Further studies showed that F. nucleatum causes adverse pregnancy outcomes in mice through activation of Toll-like receptor 4-mediated placental inflammatory responses (33). Although rare cases of neonatal shigellosis caused by Shigella boydii have been described previously, prior to our finding, no persuasive evidence proved that this taxonomic unit is involved in the process of intra-amniotic inflammation or infection (25, 46). The uncultivable species Leptotrichia amnionii and Leptotrichia (Sneathia) sanguinegens were previously linked with postpartum and neonatal bacteremia (14, 21). At the time of this writing, we found only random reports of their involvement in the triggering of the inflammatory response associated with PTB and stillbirth (17, 47). Lastly, in the current as well as in our previous studies, we provided evidence that a Bergeyella sp., an uncultivated oral species, is a participant in the process of intra-amniotic infection (19). Further studies that employ molecular detection methods to identify and characterize these previously unrecognized species in our population remain to be performed.
A discrepancy between the PCR and the culture results has been reported before (26, 47). While it is expected that PCR will detect more bacteria due to its high sensitivity, it is rather surprising that several bacterial species were identified by culturing but not by the PCR methods. Such a discrepancy could be due to several reasons. The first is the misclassification of cultured species on the basis of standard microbiological criteria alone (4). This could explain cases 5, 10, 11, 15, and 16. In each of these five cases, a close relative was identified by PCR and clone analysis. Since 16S rRNA gene sequencing is the gold standard for bacterial species identification, it is likely that the same species was also isolated by culture but was named incorrectly. Second, it is theoretically plausible that the universal primers A17F and 1512R were not universal enough to detect all species. While this could be true with any universal primer, it does not appear to be the case in our study. With the exception of Klebsiella pneumoniae in case 4 and Eikenella corrodens in case 14, the species identified by culture, i.e., Prevotella in case 1, Streptococcus in cases 2 and 14, and Ureaplasma in cases 3 and 4, were also identified by PCR and clone analysis in other samples. Primers A17F and 1512R could identify our laboratory strains of Eikenella corrodens and Klebsiella pneumoniae (Fig. 2). Thus, it is likely that the species identified by culture existed in small quantities in a mixed infection, and by selecting 10 random clones for DNA sequencing analysis, we likely sequenced those most abundant or most easily detected by the A17F and 1512R primer set. A less likely alternative is that the DNA of these particular species had been degraded in the AF samples prior to PCR amplification.
Of notable interest is the apparent discrepancy observed in cases 10 and 11, with clinical cultures identifying Ureaplasma urealyticum but PCR results reporting Ureaplasma parvum. The taxonomy of human Ureaplasma spp. was changed in 2002 on the basis of phylogenetic analysis, and the two former Ureaplasma urealyticum biovars were classified as different species, i.e., Ureaplasma parvum (previously known as Ureaplasma urealyticum biovar 1) and Ureaplasma urealyticum (previously known as Ureaplasma urealyticum biovar 2) (27, 28). Ureaplasmas are common commensals of the urogenital tract and are recognized as important opportunistic pathogens during pregnancy linked to chorioamnionitis (38) and early-onset sepsis in premature infants (12). When they are distinguished by PCR techniques, the majority of human Ureaplasma isolates recovered in the lower genital tract and AF belonged to Ureaplasma parvum serovars, while Ureaplasma urealyticum (biovar 2) was isolated less often (29). This is in agreement with our present data and the conclusion of an earlier study of AF from pregnancies with adverse outcomes (35). Subtyping based on growth on differentiation medium and colony morphology, as performed by the clinical laboratory in this study, cannot distinguish between the Ureaplasma subtypes. Thus, our finding of Ureaplasma parvum in AF underscores the necessity of DNA-based bacterial identification in epidemiological studies to assess the biological significance of Ureaplasma infection in human pregnancy. Since no Ureaplasma was detected by PCR in cases 3, 4, and 16, it is unclear if the species identified by culture was really U. urealyticum or U. parvum.
PTB remains a significant public health problem, accounting for 70% of the cases of perinatal mortality and nearly half of the cases of long-term neurological morbidity (43). There is abundant literature indicating that a significant number of spontaneous PTBs are associated with intrauterine microbial infection and inflammation (16, 49). However, despite significant research efforts and the development of powerful molecular tools, much more needs to be discovered. It is clear that we continue to have a poor understanding of the pathophysiology that links intrauterine infection to PTB. The immune mechanisms are not fully elucidated, and the time course of infection and its subsequent inflammation are not well defined. Regrettably, even the source of the intra-amniotic bacteria is not well established. Although the primary source is thought to be the lower genital tract, other potential sources (i.e., the oral cavity) need to be considered (3, 19). By demonstrating the involvement of several taxa (F. nucleatum and Bergeyella spp.), we again provide evidence that, in addition to the vaginal species, bacteria from oral sources may also play a significant role in intra-amniotic infection.
In this study we could not demonstrate causation, nor could we investigate the role of viruses or other pathogens in triggering PTB. Much more work is needed in that regard. Taking advantage of the cutting-edge DNA-based technologies for the detection, identification, and characterization of infectious agents, we aspire to provide a comprehensive mapping of the bacteria in AF. Powerful discovery tools such as multi-isotope imaging mass spectrometry or nucleotide sequence analysis allowed the rapid identification of pathogens and even a comprehensive description of their metabolic activity in the environment (32, 54). Although over 50 new infectious agents have been discovered during the last 40 years (e.g., Ebola virus, the severe acute respiratory syndrome-associated coronavirus, Bartonella henselae, Escherichia coli O157, and Clostridium difficile), most of the infectious agents are already known pathogens (15). Still, their reemergence as important infectious pathogens is due to the development of novel molecular methods for detection and identification. The species described in our study were all previously known. However, for some of them, their association with the process of intra-amniotic infection and inflammation is a novel finding. Even though prior trials aimed to prove the effectiveness of antibiotic treatment in preventing PTB failed to prove their benefit, we believe that future studies are warranted (42). One of the reasons which may explain the failure of antibiotics to prevent PTB is the lack of recognition of the true pathogen and knowledge of its antibiotic sensitivity. On the basis of the concept that molecular diagnostic tools should allow the rapid and accurate identification of the pathogens involved, we propose that in the future targeted antibiotic and anti-inflammatory treatment will be necessary to prevent PTB and its consequences for the neonate.
Prior studies demonstrated that approximately 30% of bacterial cultures turn out to be negative, even though proteomic analysis or other laboratory tests suggest intra-amniotic inflammation (6). This should not come as a surprise when the fact that patients may receive antibiotics prior to the amniocentesis or prior to their transfer for medical care in a tertiary-care medical center is taken into account (1, 6). Thus, cultivable and uncultivated bacteria may not be found if the clinicians are relying on culture conditions alone. Since molecular bacterial footprints may persist even after antimicrobial treatment, our proposed 16S rRNA gene-based culture-independent technology enables the detection of bacterial DNA which can be amplified regardless of antibiotic treatment. Further studies are needed to investigate these aspects.
This study shows that the process of intra-amniotic infection is complex and suggests that the involvement of uncultivated or difficult-to-cultivate species has been underestimated. Taken together, our findings confirm that the development of powerful DNA and proteomic technologies under the spectrum of multidimensional biology may provide in the future the necessary breakthrough to yield information once considered inaccessible (8, 31).
Acknowledgments
We are indebted to the nurses, fellows, and residents in the Department of Obstetrics and Gynecology and Reproductive Sciences, Yale New Haven Hospital; all patients who participated in the study; Vineet Bhandari for scoring early-onset neonatal sepsis; and Loretta Anthony for assisting with the technical aspects of cultivating Mycoplasma and Ureaplasma.
This work was supported in part from National Institutes of Health grants RO1DE14924, R21DE17165, and KO2DE16102 (to Y.W.H.) and RO1HD47321 (to I.A.B.). C.S.B. is supported by grant RO3HD50249 and the Yale WRHR Career Development Center (grant K12 HD1027766).
The funding source had no involvement in the design of the study, the interpretation of the data, the writing of the report, or the decision to submit the paper for publication.
Footnotes
Published ahead of print on 29 October 2008.
REFERENCES
- 1.ACOG Committee on Practice Bulletins—Obstetrics. 2007. ACOG Practice Bulletin No. 80: premature rupture of membranes. Clinical management guidelines for obstetrician-gynecologists. Obstet. Gynecol. 1091007-1019. [DOI] [PubMed] [Google Scholar]
- 2.Barron, E. J., J. H. Jorgensen, M. L. Landry, and M. A. Pfaller. 2007. Bacteriology, p. 974. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. L. Landry, and M. A. Pfaller (ed.), Manual of clinical microbiology, 9th ed. ASM Press, Washington, DC.
- 3.Bearfield, C., E. S. Davenport, V. Sivapathasundaram, and R. P. Allaker. 2002. Possible association between amniotic fluid micro-organism infection and microflora in the mouth. Br. J. Obstet. Gynaecol. 109527-533. [DOI] [PubMed] [Google Scholar]
- 4.Bosshard, P. P., R. Zbinden, S. Abels, B. Böddinghaus, M. Altwegg, and E. C. Böttger. 2006. 16S rRNA gene sequencing versus the API 20 NE system and the VITEK 2 ID-GNB card for identification of nonfermenting gram-negative bacteria in the clinical laboratory. J. Clin. Microbiol. 441359-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowen, J. M., L. Chamley, J. A. Keelan, and M. D. Mitchell. 2002. Cytokines of the placenta and extra-placental membranes: roles and regulation during human pregnancy and parturition. Placenta 23257-273. [DOI] [PubMed] [Google Scholar]
- 6.Buhimschi, C. S., V. Bhandari, B. D. Hamar, M. O. Bahtiyar, G. Zhao, A. K. Sfakianaki, C. M. Pettker, L. Magloire, E. Funai, E. R. Norwitz, M. Paidas, J. A. Copel, C. P. Weiner, C. J. Lockwood, and I. A. Buhimschi. 2007. Proteomic profiling of the amniotic fluid to detect inflammation, infection, and neonatal sepsis. PLoS Med. 4e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buhimschi, C. S., I. A. Buhimschi, S. Abdel-Razeq, V. A. Rosenberg, S. F. Thung, G. Zhao, E. Wang, and V. Bhandari. 2007. Proteomic biomarkers of intra-amniotic inflammation: relationship with funisitis and early-onset sepsis in the premature neonate. Pediatr. Res. 61318-324. [DOI] [PubMed] [Google Scholar]
- 8.Buhimschi, C. S., V. A. Rosenberg, A. T. Dulay, S. Thung, A. K. Sfakianaki, M. O. Bahtiyar, and I. A. Buhimschi. 2008. Multidimensional system biology: genetic markers and proteomic biomarkers of adverse pregnancy outcome in preterm birth. Am. J. Perinatol. 25175-187. [DOI] [PubMed] [Google Scholar]
- 9.Buhimschi, I. A., R. Christner, and C. S. Buhimschi. 2005. Proteomic biomarker analysis of amniotic fluid for identification of intra-amniotic inflammation. Br. J. Obstet. Gynaecol. 112173-181. [DOI] [PubMed] [Google Scholar]
- 10.Buhimschi, I. A., E. Zambrano, C. M. Pettker, M. O. Bahtiyar, M. Paidas, V. A. Rosenberg, S. Thung, C. M. Salafia, and C. S. Buhimschi. 2008. Using proteomic analysis of the human amniotic fluid to identify histologic chorioamnionitis. Obstet. Gynecol. 111403-412. [DOI] [PubMed] [Google Scholar]
- 11.Buhimschi, I. A., G. Zhao, V. A. Rosenberg, S. Abdel-Razeq, S. Thung, and C. S. Buhimschi. 2008. Multidimensional proteomics analysis of amniotic fluid to provide insight into the mechanisms of idiopathic preterm birth. PLoS One 3e2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cassell, G. H., K. B. Waites, H. L. Watson, D. T. Crouse, and R. Harasawa. 1993. Ureaplasma urealyticum intrauterine infection: role in prematurity and disease in newborns. Clin. Microbiol. Rev. 1669-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho, J. C., and J. M. Tiedje. 2001. Bacterial species determination from DNA-DNA hybridization by using genome fragments and DNA microarrays. Appl. Environ. Microbiol. 673677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Martino, S. J., I. Mahoudeau, J. P. Brettes, Y. Piemont, H. Monteil, and B. Jaulhac. 2004. Peripartum bacteremias due to Leptotrichia amnionii and Sneathia sanguinegens, rare causes of fever during and after delivery. J. Clin. Microbiol. 425940-5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong, J., J. P. Olano, J. W. McBride, and D. H. Walker. 2008. Emerging pathogens: challenges and successes of molecular diagnostics. J. Mol. Diagn. 10185-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elovitz, M. A. 2006. Anti-inflammatory interventions in pregnancy: now and the future. Semin. Fetal Neonatal Med. 11327-332. [DOI] [PubMed] [Google Scholar]
- 17.Gardella, C., D. E. Riley, J. Hitti, K. Agnew, J. N. Krieger, and D. Eschenbach. 2004. Identification and sequencing of bacterial rDNAs in culture-negative amniotic fluid from women in premature labor. Am. J. Perinatol. 21319-323. [DOI] [PubMed] [Google Scholar]
- 18.Goldenberg, R. L., J. C. Hauth, and W. W. Andrews. 2000. Intrauterine infection and preterm delivery. N. Engl. J. Med. 3421500-1507. [DOI] [PubMed] [Google Scholar]
- 19.Han, Y. W., A. Ikegami, N. F. Bissada, M. Herbst, R. W. Redline, and G. G. Ashmead. 2006. Transmission of an uncultivated Bergeyella strain from the oral cavity to amniotic fluid in a case of preterm birth. J. Clin. Microbiol. 441475-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han, Y. W., R. W. Redline, M. Li, L. Yin, G. B. Hill, and T. S. McCormick. 2004. Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: implication of oral bacteria in preterm birth. Infect. Immun. 722272-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanff, P. A., J. A. Rosol-Donoghue, C. A. Spiegel, K. H. Wilson, and L. H. Moore. 1995. Leptotrichia sanguinegens sp. nov., a new agent of postpartum and neonatal bacteremia. Clin. Infect. Dis. 20S237-S239. [DOI] [PubMed] [Google Scholar]
- 22.Hill, G. B. 1993. Investigating the source of amniotic fluid isolates of fusobacteria. Clin. Infect. Dis. 16S423-S424. [DOI] [PubMed] [Google Scholar]
- 23.Hill, G. B. 1998. Preterm birth: associations with genital and possibly oral microflora. Ann. Periodontol. 3222-332. [DOI] [PubMed] [Google Scholar]
- 24.Hitti, J., D. E. Riley, M. A. Krohn, S. L. Hillier, K. J. Agnew, J. N. Krieger, and D. A. Eschenbach. 1997. Broad-spectrum bacterial rDNA polymerase chain reaction assay for detecting amniotic fluid infection among women in premature labor. Clin. Infect. Dis. 241228-1232. [DOI] [PubMed] [Google Scholar]
- 25.Huskins, W. C., J. K. Griffiths, A. S. Faruque, and M. L. Bennish. 1994. Shigellosis in neonates and young infants. J. Pediatr. 12514-22. [DOI] [PubMed] [Google Scholar]
- 26.Jalava, J., M. L. Mäntymaa, U. Ekblad, P. Toivanen, M. Skurnik, O. Lassila, and A. Alanen. 1996. Bacterial 16S rDNA polymerase chain reaction in the detection of intraamniotic infection. Br. J. Obstet. Gynaecol. 103664-669. [DOI] [PubMed] [Google Scholar]
- 27.Kong, F., and G. L. Gilbert. 2004. Postgenomic taxonomy of human ureaplasmas—a case study based on multiple gene sequences. Int. J. Syst. Evol. Microbiol. 541815-1821. [DOI] [PubMed] [Google Scholar]
- 28.Kong, F., G. James, Z. Ma, S. Gordon, B. Wang, and G. L. Gilbert. 1999. Phylogenetic analysis of Ureaplasma urealyticum—support for the establishment of a new species, Ureaplasma parvum. Int. J. Syst. Bacteriol. 491879-1889. [DOI] [PubMed] [Google Scholar]
- 29.Kong, F., Z. Ma, G. James, S. Gordon, and G. L. Gilbert. 2000. Species identification and subtyping of Ureaplasma parvum and Ureaplasma urealyticum using PCR-based assays. J. Clin. Microbiol. 381175-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar, P. S., A. L. Griffen, J. A. Barton, B. J. Paster, M. L. Moeschberger, and E. J. Leys. 2003. New bacterial species associated with chronic periodontitis. J. Dent. Res. 82338-344. [DOI] [PubMed] [Google Scholar]
- 31.Kuypers, M. M. 2007. Microbiology. Sizing up the uncultivated majority. Science 3171510-1511. [DOI] [PubMed] [Google Scholar]
- 32.Lechene, C. P., Y. Luyten, G. McMahon, and D. L. Distel. 2007. Quantitative imaging of nitrogen fixation by individual bacteria within animal cells. Science 3171563-1566. [DOI] [PubMed] [Google Scholar]
- 33.Liu, H., R. Redline, and Y. W. Han. 2007. Fusobacterium nucleatum induces fetal death in mice by stimulating TLR4-mediated inflammatory response. J. Immunol. 1792501-2508. [DOI] [PubMed] [Google Scholar]
- 34.Madianos, P. N., S. Lieff, A. P. Murtha, K. A., Boggess, R. L. Auten Jr., J. D. Beck, and S. Offenbacher. 2001. Maternal periodontitis and prematurity. Part II. Maternal infection and fetal exposure. Ann. Periodontol. 6175-182. [DOI] [PubMed] [Google Scholar]
- 35.Martínez, M. A., A. Ovalle, A. Santa-Cruz, B. Barrera, R. Vidal, and R. Aguirre. 2001. Occurrence and antimicrobial susceptibility of Ureaplasma parvum (Ureaplasma urealyticum biovar 1) and Ureaplasma urealyticum (Ureaplasma urealyticum biovar 2) from patients with adverse pregnancy outcomes and normal pregnant women. Scand. J. Infect. Dis. 33604-610. [DOI] [PubMed] [Google Scholar]
- 36.Naef, R. W., J. R. Allbert III, E. L. Ross, B. M. Weber, R. W. Martin, and J. C. Morrison. 1998. Premature rupture of membranes at 34 to 37 weeks' gestation: aggressive versus conservative management. Am. J. Obstet. Gynecol. 178126-130. [DOI] [PubMed] [Google Scholar]
- 37.Naeye, R. L. 1992. Disorders of the placenta and decidua, p. 118-247. In R. L. Naeye (ed.), Disorders of the placenta, fetus and neonate: diagnosis and clinical significance. Mosby, St. Louis, MO.
- 38.Pettker, C. M., I. A. Buhimschi, L. K. Magloire, A. K. Sfakianaki, B. D. Hamar, and C. S. Buhimschi. 2007. Value of placental microbial evaluation in diagnosing intra-amniotic infection. Obstet. Gynecol. 109739-749. [DOI] [PubMed] [Google Scholar]
- 39.Riesenfeld, C. S., P. D. Schloss, and J. Handelsman. 2004. Metagenomics: genomic analysis of microbial communities. Annu. Rev. Genet. 38525-552. [DOI] [PubMed] [Google Scholar]
- 40.Rodwell, R. L., A. L. Leslie, and D. I. Tudehope. 1988. Early diagnosis of neonatal sepsis using a hematologic scoring system. J. Pediatr. 112761-767. [DOI] [PubMed] [Google Scholar]
- 41.Rodwell, R. L., K. M. Taylor, D. I. Tudehope, and P. H. Gray. 1993. Hematologic scoring system in early diagnosis of sepsis in neutropenic newborns. Pediatr. Infect. Dis. J. 12372-376. [DOI] [PubMed] [Google Scholar]
- 42.Romero, R., B. Sibai, S. Caritis, R. Paul, R. Depp, M. Rosen, M. Klebanoff, V. Sabo, J. Evans, and E. Thom. 1993. Antibiotic treatment of preterm labor with intact membranes: a multicenter, randomized, double-blinded, placebo-controlled trial. Am. J. Obstet. Gynecol. 169764-774. [DOI] [PubMed] [Google Scholar]
- 43.Saigal, S., and L. W. Doyle. 2008. An overview of mortality and sequelae of preterm birth from infancy to adulthood. Lancet 371261-269. [DOI] [PubMed] [Google Scholar]
- 44.Salafia, C. M., C. A. Vogel, A. M. Vintzileos, K. F. Bantham, J. Pezzullo, and L. Silberman. 1991. Placental pathologic findings in preterm birth. Am. J. Obstet. Gynecol. 165934-938. [DOI] [PubMed] [Google Scholar]
- 45.Salafia, C. M., C. Weigl, and L. Silberman. 1989. The prevalence and distribution of acute placental inflammation in uncomplicated term pregnancies. Obstet. Gynecol. 73383-389. [PubMed] [Google Scholar]
- 46.Sawardekar, K. P. 2005. Shigellosis caused by Shigella boydii in a preterm neonate, masquerading as necrotizing enterocolitis. Pediatr. Infect. Dis. J. 24184-185. [DOI] [PubMed] [Google Scholar]
- 47.Shukla, S. K., P. R. Meier, P. D. Mitchell, D. N. Frank, and K. D. Reed. 2002. Leptotrichia amnionii sp. nov., a novel bacterium isolated from the amniotic fluid of a woman after intrauterine fetal demise. J. Clin. Microbiol. 403346-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stackebrandt, E., C. Sproer, F. A. Rainey, J. Burghardt, O. Päuker, and H. Hippe. 1997. Phylogenetic analysis of the genus Desulfotomaculum: evidence for the misclassification of Desulfotomaculum guttoideum and description of Desulfotomaculum orientis as Desulfosporosinus orientis gen. nov., comb. nov. Int. J. Syst. Bacteriol. 471134-1139. [DOI] [PubMed] [Google Scholar]
- 49.Steer, P. J. 2006. The epidemiology of preterm labour—why have advances not equated to reduced incidence? Br. J. Obstet. Gynaecol. 113(Suppl. 3)1-3. [DOI] [PubMed] [Google Scholar]
- 50.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 241596-1599. [DOI] [PubMed] [Google Scholar]
- 51.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 244876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watts, D. H., M. A. Krohn, S. L. Hillier, and D. A. Eschenbach. 1992. The association of occult amniotic fluid infection with gestational age and neonatal outcome among women in preterm labor. Obstet. Gynecol. 79351-357. [DOI] [PubMed] [Google Scholar]
- 53.Weng, L., E. M. Rubin, and J. Bristow. 2006. Application of sequence-based methods in human microbial ecology. Genome Res. 16316-322. [DOI] [PubMed] [Google Scholar]
- 54.Yuen, K. Y., P. K. Chan, M. Peiris, D. N. Tsang, T. L. Que, K. F. Shortridge, P. T. Cheung, W. K. To, E. T. Ho, R. Sung, and A. F. Cheng. 1998. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351467-471. [DOI] [PubMed] [Google Scholar]