Abstract
Following an outbreak caused by staphylococcal cassette chromosome mec (SCCmec) type V methicillin (meticillin)-resistant Staphylococcus aureus (MRSA), a point-prevalence survey of the nasal carriage of staphylococci was conducted in a long-term-care facility in northern Finland in 2004. The focus was directed at methicillin-resistant coagulase-negative staphylococci (MR-CNS) and their SCCmec elements. A nasal swab was taken from 76 of the 80 residents 6 months after the onset of the outbreak. Staphylococcal isolates were identified by conventional methods and the GenoType Staphylococcus test, and their SCCmec elements were analyzed. Of the 76 individuals, 24 (32%) carried S. aureus and 67 (88%) CNS in their nostrils. Of the CNS carriers, 41 (61%) had at least one mecA-positive MR-CNS, and two individuals (3%) had both MRSA and methicillin-resistant Staphylococcus epidermidis (MRSE). Among the 61 MR-CNS isolates identified, 49 (80%) were MRSE. The distribution of the SCCmec types was diverse: 20 (33%) were of type IV, 11 (18%) of type V, 4 (6%) of type I or IA, 3 (4%) of type II, and 23 (38%) of new types (with six different combinations of ccr and other mec genes or only mecA). Both of the individuals with MRSA and MRSE shared SCCmec type V among their isolates. Nasal MR-CNS carriage was common among the residents of this long-term-care facility. A variety of SCCmec types, including many new types, were identified among the MR-CNS strains. The horizontal transfer of SCCmec elements is speculated based on the sharing of SCCmec type V between MRSA and MRSE.
Coagulase-negative staphylococci (CNS) belong to the normal microbial flora of the skin and mucous membranes of humans. The most frequently encountered CNS species in humans, in decreasing order of occurrence, are Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus saprophyticus, and Staphylococcus lugdunensis (8). CNS are an important cause of nosocomial infections, particularly causing foreign device-related infections and infections among immunocompromised patients. In a recent prospective laboratory-based surveillance in four Finnish acute-care hospitals, 76% of the blood culture CNS isolates were resistant to methicillin (meticillin) (23).
Methicillin resistance in staphylococci is caused by the expression of penicillin-binding protein PBP2a (PBP2′), which is encoded by the mecA gene. In S. aureus and CNS, mecA is located on a genetic element called the staphylococcal cassette chromosome (SCCmec) (15, 37). SCCmec is integrated into the chromosome of S. aureus at a unique site (attBscc) located near the S. aureus origin of replication. Up to now, six different SCCmec types (I to VI) have been recognized, each of which is different in size (21 to 67 kb) and characterized by a different set of ccr recombinase genes and mec gene complex (3, 12, 13, 22, 24, 32). In addition to the major types, a number of new SCCmec elements, including non-mecA-encoding cassettes, have been discovered (2, 3, 11, 16, 19, 20, 28). New types may be generated continuously (5, 9).
The SCCmec has been identified exclusively among staphylococci, but its origin remains unknown (9). It has been suggested that the ccr and mec genes from an unknown source were brought together in CNS (34, 38), and a deletion in the mec regulatory genes occurred before the cassette was transferred into S. aureus to create methicillin-resistant S. aureus (MRSA) (10, 30). The transfer of mecA from S. epidermidis to S. aureus has been suspected to occur in vivo (36). However, the mechanisms responsible for the possible horizontal transfer of mecA between staphylococcal species or between different gram-positive species are not known. Evaluations of the epidemiology of methicillin-resistant staphylococcal colonization and SCCmec typing are necessary to understand the apparent emergence of MRSA strains from CNS.
This point-prevalence study of the nasal carriage of CNS was conducted 6 months after an outbreak of MRSA in a long-term-care facility (LTF) in northern Finland in 2004. The MRSA outbreak was caused by a strain that had not been encountered previously in Finland, FIN-22, with SCCmec type V (17). In this study, we focused on the structure of the SCCmec elements of methicillin-resistant (MR)-CNS strains and a structural comparison of SCCmec elements of methicillin-resistant S. epidermidis (MRSE) and MRSA isolated from the same person at the same time, under the suspicion of horizontal SCCmec transfer in vivo.
(These data were presented in part at the 47th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL, 17 to 20 September 2007, as poster no. 2117.)
MATERIALS AND METHODS
Setting.
A 34-bed health care ward, situated in a municipality of 5,000 inhabitants in northern Finland, takes care of the elderly patients with multiple underlying diseases, but it also gives primary care. The associated 46-bed nursing home is only for the elderly. Each room has four patients. A total of 76 nasal swabs were collected from 76 out of the total of 80 patients on 26 February 2004. One swab per patient was taken from both nostrils. The median age of 76 patients was 80 years (range, 35 to 99 years), 36% were male (n = 27), 26% used antimicrobials (n = 20), and 5% used foreign devices (n = 4). The median length of nursing stay was 9 months (range, <1 to 90 months).
Bacterial cultures and identification of staphylococci.
The screening swabs (Probact transport swab; Schofield St-Heywood, United Kingdom) were cultivated on nonselective sheep blood agar (SBA; CM1008; Oxoid, United Kingdom) and on selective oxacillin resistance screening agar (ORSAB; CM1008; Oxoid, United Kingdom) plates. The SBA plates were incubated for 48 h and ORSAB plates were incubated for 96 h, and they were inspected daily. Based on colony morphology, staphylococcus-like colonies were picked and subcultured onto the SBA plate. The colonies were identified by conventional biochemical tests (1, 18). If the identification of staphylococcal species by using these tests was unclear, GenoType Staphylococcus (Hain Lifescience, Germany) was performed. For all CNS isolates, resistance to methicillin was determined by the oxacillin disk diffusion test (inhibition zone, ≤18 mm), and the oxacillin MIC (Etest; AB Biodisk, Solna, Sweden) was tested for every MR-CNS isolate (4).
SCCmec typing.
The SCCmec types were determined by two PCR methods. The first multiplex PCR method, modified slightly (11) from the original description (31) by Oliveira and de Lencastre, detects eight loci (A through H) within SCCmec and uses mecA as an internal control. Based on the first PCR, representative isolates (in each MR-CNS species) of the different SCCmec patterns, and four isolates from which only mecA was amplified, were analyzed for their ccr and mec components by using the multiplex PCR methods described by Kondo and coworkers (19). This assay identifies mecA and the ccr types (ccrAB1 to ccrAB4 and ccrC) as well as the mec classes A, B, and C. The following reference strains were used in the analysis: Iberian HPV107 (SCCmec type IA, ccrA1, class B), UK EMRSA-16 96/32010 (SCCmec type II, ccrA2, class A), Brazilian HSJ216 (SCCmec type IIIA, ccrA3, class A), Pediatric clone HDE288 (SCCmec type VI, ccrA4, class B) (32), and the Finnish MRSA FIN-7 (SCCmec type V, ccrC, class C) (18).
PFGE.
Pulsed-field gel electrophoresis (PFGE) was carried out as previously described for S. aureus (29). PFGE patterns were analyzed by BioNumerics (version 2.0; Applied Maths, Kortrijk, Belgium) and were further interpreted according to the criteria of Tenover et al. (35).
Ethical aspects.
We were at liberty to collect the samples from the residents with approval from the Ministry of Social Affairs and Health and the data protection authority. In addition, permission for sampling was asked from each patient.
RESULTS
Of the 76 patients, 73 (96%) were colonized with a staphylococcal species (Table 1): 67 (92%) were colonized by at least one CNS strain, 49 (73%) by CNS only, and 18 (27%) by CNS in combination with S. aureus. S. aureus alone was found in six persons (8%). Of 67 CNS carriers, 41 (61%) were colonized with at least one MR-CNS strain, and two of them carried MRSA as well. Twenty-six (39%) patients were colonized by methicillin-susceptible CNS strains, and one of them carried MRSA as well.
TABLE 1.
Number of persons with different combinations of MRSA and methicillin-susceptible S. aureus (MSSA) and CNS
| Strain | No. of MRSA isolates | No. of MSSA isolates | No. of CNS isolates | Total (n = 73) |
|---|---|---|---|---|
| MR-CNS | 2 | 11 | 28 | 41 |
| MS-CNSa | 1 | 4 | 21 | 26 |
| S. aureus | 2 | 4 | 6 |
MS, methicillin susceptible.
From the 67 patients with CNS, 127 isolates were obtained. The number of isolates per individual varied from one to five. Of the 127 CNS isolates, 61 (48%) were shown to be methicillin resistant. These included 49 (80%) of 82 having S. epidermidis isolates, 10 (66%) of 15 having S. capitis isolates, 1 (14%) of 7 having S. haemolyticus isolates, and 1 (50%) of 2 having S. hominis isolates.
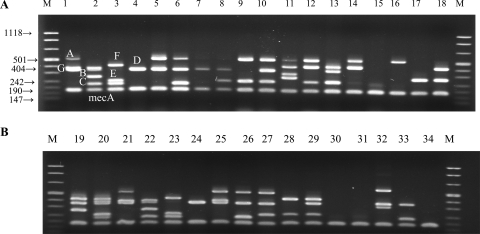
Among the 49 MRSE isolates, three MRSE strains could be classified as SCCmec type I (Fig. 1, lane 5) and two as SCCmec type IV (Fig. 1, lane 7); in one MRSE isolate, only mecA was amplified (Fig. 1, lane 15). The remaining 43 MRSE isolates could not be interpreted as belonging to any of the currently described SCCmec types (31). Within the other MR-CNS species, one methicillin-resistant S. capitis isolate could be classified as SCCmec type IA (Fig. 1, lane 25), while seven methicillin-resistant S. capitis isolates and one methicillin-resistant S. haemolyticus isolate (Fig. 1, lane 32) could not be recognized as any previously known SCCmec type. In two methicillin-resistant S. capitis isolates and one methicillin-resistant S. hominis isolate, only mecA was amplified (Table 2 and Fig. 1, lanes 30, 31, and 34).
FIG. 1.
SCCmec multiplex patterns of MR-CNS strains (31). M, molecular size markers (in kilodaltons). Lanes 1 to 4 and 21 to 24, control MRSA of type IA (locus A, upstream of the pls gene; locus G, left of the junction between IS431 and pUB110; and locus D, the dcs region), type II (loci G, D, and B, kdp operon; locus C, mecI gene), type IIIA (locus F, between Tn554 and orfX; locus E, between integrated pI258 and Tn554; and locus C) and SCCmec type IV (locus D) and internal control mecA. The sizes of amplicons, by locus, are the following: A, 495 bp; B, 284 bp; C, 209 bp; D, 342 bp; E, 243 bp; F, 414 bp; G, 381 bp; and mecA, 160 bp. Lanes 5 to 20, MRSE isolates; lanes 25 to 31 and 33, methicillin-resistant S. capitis; lane 32, methicillin-resistant S. haemolyticus; and lane 34, methicillin-resistant S. hominis.
TABLE 2.
Distribution of the SCCmec types or elements within the CNS isolates
| Staphylococcal isolate(s) | SCCmec type | Locus (loci) amplified by the Oliveira strategy (31) | Locus (loci) amplified by the Kondo strategy (19)
|
No. of isolates | |
|---|---|---|---|---|---|
| ccr type | mec class | ||||
| S. epidermidis (n = 49) | I | mecA, D, A | 1 | B | 3 |
| II | mecA, C, D, A | 2 | A | 2 | |
| IV | mecA, D | 2 | B | 2 | |
| IV | mecA, E, D | 2 | B | 2 | |
| IV | mecA, E, A | 2 | B | 1 | |
| IV | mecA, E, D, A | 2 | B | 13 | |
| V | mecA, B, H, F, A | ccrC | C | 1 | |
| V | mecA, E, F, A | ccrC | C | 3 | |
| V | mecA, E (very faint), F, A | ccrC | C | 5 | |
| New (IV + ccrC) | mecA, E, D, F | 2, ccrC | B | 1 | |
| New (V + ccrA4) | mecA, F, A | 4, ccrC | C | 1 | |
| NTa | mecA | None | None | 1 | |
| NT | mecA, A | None | None | 1 | |
| NT | mecA, B | None | None | 1 | |
| New | mecA, B, F | ccrC | A | 8 | |
| New | mecA, B, D, F | 4, ccrC | A | 3 | |
| New | mecA, C, E, D, G, F | 3, 4, ccrC | A and B | 1 | |
| S. capitis (n = 10) | IA | mecA, D, G, A | 1 | B | 1 |
| II | mecA, C, D, A | 2 | A | 1 | |
| IV | mecA, E, D, A | 2 | B | 1 | |
| V | mecA, E, F | ccrC | C | 2 | |
| New (IV + ccrC) | mecA, E, D, F | 2, ccrC | B | 2 | |
| New | mecA | 1 | A | 2 | |
| New | mecA, C, D | 3, 4 | A and B | 1 | |
| S. haemolyticus (n = 1) | IV | mecA, D, H, A (>501 bp) | 2 | B | 1 |
| S. hominis (n = 1) | New | mecA | 1 | A | 1 |
NT, nontypeable.
By analyzing the ccr and mec components (19), the MRSE isolates could be categorized as follows: 3 (6%) harbored SCCmec type I, 2 (4%) type II, 18 (37%) type IV, and 9 (12%) type V. Three isolates were of a nontypeable SCCmec type, as neither ccr genes nor mec genes could be amplified. The remaining 14 isolates had ccr and mec complex gene combinations for which no names have been assigned previously (Table 2). Among the 10 methicillin-resistant S. capitis isolates, three harbored SCCmec types IA, II, and IV, and two harbored type V. The remaining five isolates harbored a new SCCmec type (Table 2). The single methicillin-resistant S. haemolyticus isolate harbored SCCmec type IV, and the methicillin-resistant S. hominis isolate harbored a new SCCmec type (Table 2).
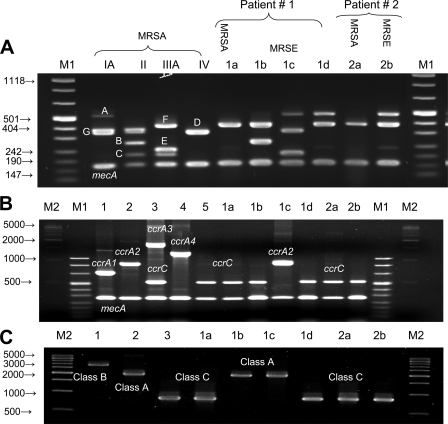
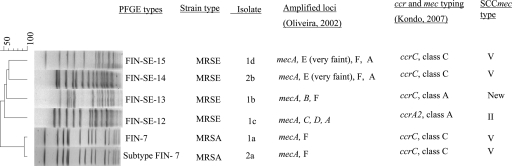
Two patients were colonized by both MRSE and MRSA. The first one carried an MRSA strain and three MRSE strains. The MRSA strain and one of the MRSE strains harbored SCCmec type V (ccrC and class C), while the two MRSE strains were of different SCCmec types, type II (ccrA2 and class A) and a new SCCmec type (ccrC and mec, class A). The second carrier had MRSA and MRSE, both of which harbored SCCmec type V (Fig. 2B and C and 3). The MRSE multiplex SCCmec patterns varied among the isolates (Fig. 2A). Genotyping by PFGE revealed that the two MRSA strains were representatives of a Finnish epidemic strain (FIN-7 and FIN-7 subtype), and the four MRSE strains had unique PFGE profiles (Fig. 3).
FIG. 2.
Three multiplex PCRs for the SCCmec type assignment of the staphylococcal isolates from the two patients colonized with MRSA and MRSE. Lanes M1 and M2, molecular size markers. Lanes 1 to 5, control MRSA for SCCmec type IA (ccrA1, class B), SCCmec type II (ccrA2, class A), SCCmec type IIIA (ccrA3, class A), SCCmec type VI (ccrA4, class B), and SCCmec type V (ccrC and class C). (A) Multiplex PCR patterns (31). Lanes 1 to 4, controls; lanes 1a and 2a, MRSA isolates; lanes 1b, 1c, 1d, and 2b, MRSE isolates. (B) Multiplex PCR for the typing of ccr genes. Lanes 1 to 5, control MRSA isolates for ccr types. (C) Multiplex PCR for typing of mec genes (19). Lanes 1 to 3, control MRSA isolates for mec classes. Lanes 1a and 2a, MRSA (ccrC, class C); lane 1b, MRSE (ccrC, class A); lane 1c, MRSE (ccrA2, class A); lanes 1d and 2b, MRSE (ccrC, class C); lane 2a, MRSA (ccrC) and MRSE (ccrC).
FIG. 3.
PFGE dendrogram of the methicillin-resistant staphylococcal isolates from two patients colonized with MRSA and MRSE. The distribution of the SCCmec types/elements for each strain is included. The scale bar at the top of the dendrogram represents similarity.
DISCUSSION
The nasal carriage of MR-CNS was found to be common among the residents of the studied LTF. The MRSE isolate was the most prevalent CNS species. A diversity of SCCmec types, with many new combinations of elements as well as nontypeable types, were recognized among the MR-CNS strains. The horizontal transfer of SCCmec elements is speculated based on the sharing of SCCmec type V between MRSA and MRSE in two patients.
The prevalence of CNS carriage among patients participating in this study was high, at 92%. The proportion of the nasal carriage of MR-CNS among patients in an LTF in this study was somewhat higher (48%) than that in a similar study in the United States (40%) (21). However, there is a very limited number of reports on MR-CNS nasal carriage among patients in long-term-care settings. Consistently with one such previous report (33), the most common MR-CNS species in our study was MRSE.
The SCCmec typing of MR-CNS isolates revealed that 62% of the isolates harbored previously recognized SCCmec types (I, IA, II, and IV). For the remaining 37%, ccr and mec complexes could not be amplified at all, or a variety of new combinations was detected. One-third of the MR-CNS isolates had SCCmec type IV, and SCCmec type IV was most prevalent among the S. epidermidis strains (37%). Among the 20 strains, 18 harbored a modified SCCmec type IV. While the originally described type IV contains only locus D (dcs region), we identified several additional loci amplified from type IV strains in different combinations (Fig. 1, lanes 8 to 10 and 32, and Table 2). The combinations were not species specific. Modified patterns also were found among other SCCmec types; SCCmec type V was found among 11 MR-CNS strains, which represented four different multiplex SCCmec patterns (Fig. 1 and Table 2). Three MR-CNS strains harboring SCCmec type II did not have locus B (kdp operon). The remaining 23 strains harbored a new SCCmec type; these strains carried either (i) known SCCmec types with additional elements (i.e., type IV and ccrC or type V and ccrA4) or (ii) combinations of ccr and mec that could not be interpreted as belonging to any of the presently described SCCmec types (Table 2). Previous studies also have shown variations in SCCmec cassettes. These include (i) strains containing both SCCmec type IV and ccrC, (ii) strains carrying multiple ccr genes (3, 7, 14, 26), (iii) strains carrying ccr genes without a mec complex, (iv) strains carrying a mec complex without ccr genes, and (v) a mecA-positive MRSA strain with neither ccr genes nor a mec complex (3, 25, 26). In our study, only mecA from three MR-CNS strains could be detected. The failure to amplify ccr and mec may indicate that the target sequences for primers have changed.
Defining SCCmec types in MR-CNS strains based solely on amplifying sequences between and flanking the ccr genes and the mec complex raises some concerns (26). These areas do not contain specific loci for a specific SCCmec type. For instance, locus A was previously thought to be part of SCCmec type I and IA only, but according to this study, it also is present in types II, IV, and V. Moreover, the SCCmec types IV and V contained a variety of loci. Locus B previously has been defined to be specific for SCCmec type II, but we recognized locus B as being present in type V and in three new types. Therefore, the detection of these intervening sequences provides valuable additional information on the discrimination of SCCmec types.
We have previously reported on the MRSA nasal carriage of this study population (17). In total, five different MRSA strains were identified, and all of them had SCCmec type V. In the present study, we analyzed in detail the two patients who carried both MRSE and MRSA strains simultaneously, and all of these isolates shared SCCmec type V (ccrC, class C). However, differences in the J-region sequences were identified between MRSA and MRSE strains (additional loci E and A in MRSE) (Fig. 2A and 3). Although we are not able to rule out the possibility that the similar SCCmec cassettes were acquired through different routes, this observation supports the possibility of SCCmec transfer. If such a transfer has happened, it was not complete. Further studies revealing the mechanisms of SCCmec transfer are needed. The hypothesis for the transfer of SCCmec between S. epidermidis and S. aureus has been previously reported (6, 7, 27, 37).
Acknowledgments
This work was supported by grants from the Paulo Foundation and the Maud Kuistila Memorial Foundation.
Footnotes
Published ahead of print on 29 October 2008.
REFERENCES
- 1.Bannerman, T. L., and S. J. Peacock. 2007. Staphylococcus, Micrococcus and other catalase positive cocci, p. 390-404. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and M. L. Landry, Manual of clinical microbiology, 9th ed., ASM Press, Washington, DC.
- 2.Boyle-Vavra, S., B. Ereshefsky, C. C. Wang, and R. S. Daum. 2005. Successful multiresistant community-associated methicillin-resistant Staphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel Staphylococcal chromosome cassette mec (SCCmec) type VT or SCCmec type IV. J. Clin. Microbiol. 434719-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chongtrakool, P., T. Ito, X. X. Ma, Y. Kondo, S. Trakulsomboon, C. Tiensasitorn, M. Jamklang, T. Chavalit, J. H. Song, and K. Hiramatsu. 2006. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob. Agents Chemother. 501001-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement. CLSI document M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Grundmann, H., M. Aires-de-Sousa, J. Boyce, and E. Tiemersma. 2006. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368874-885. [DOI] [PubMed] [Google Scholar]
- 6.Hanssen, A. M., G. Kjeldsen, and J. U. Sollid. 2004. Local variants of Staphylococcal cassette chromosome mec in sporadic methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci: evidence of horizontal gene transfer? Antimicrob. Agents Chemother. 48285-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanssen, A. M., and J. U. Sollid. 2007. Multiple staphylococcal cassette chromosomes and allelic variants of cassette chromosome recombinases in Staphylococcus aureus and coagulase-negative staphylococci from Norway. Antimicrob. Agents Chemother. 511671-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heikens, E., A. Fleer, A. Paauw, A. Florijn, and A. C. Fluit. 2005. Comparison of genotypic and phenotypic methods for species-level identification of clinical isolates of coagulase-negative staphylococci. J. Clin. Microbiol. 432286-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiramatsu, K., L. Cui, M. Kuroda, and T. Ito. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9486-493. [DOI] [PubMed] [Google Scholar]
- 10.Hürlimann-Dalel, R. L., C. Ryffel, F. H. Kayser, and B. Berger-Bachi. 1992. Survey of the methicillin resistance-associated genes mecA, mecR1-mecI, and femA-femB in clinical isolates of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 362617-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibrahem, S., S. Salmenlinna, O. Lyytikäinen, M. Vaara, and J. Vuopio-Varkila. 2008. Molecular characterization of methicillin-resistant Staphylococcus epidermidis strains isolated from blood stream infections in Finland. Clin. Microbiol. Infect. 141020-1027. [DOI] [PubMed] [Google Scholar]
- 12.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 451323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 482637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kardén-Lilja, M., S. Ibrahem, J. Vuopio-Varkila, S. Salmenlinna, O. Lyytikäinen, L. Siira, and A. Virolainen. 2007. Panton-Valentine leukocidin genes and staphylococcal chromosomal cassette mec types amongst Finnish community-acquired methicillin-resistant Staphylococcus aureus strains, 1997-1999. Eur. J. Clin. Microbiol. Infect. Dis. 26729-733. [DOI] [PubMed] [Google Scholar]
- 15.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 441549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katayama, Y., F. Takeuchi, T. Ito, X. X. Ma, Y. Ui-Mizutani, I. Kobayashi, and K. Hiramatsu. 2003. Identification in methicillin-susceptible Staphylococcus hominis of an active primordial mobile genetic element for the staphylococcal cassette chromosome mec of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 1852711-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerttula, A. M., O. Lyytikäinen, J. Vuopio-Varkila, S. Ibrahem, N. Agthe, M. Broas, H. J. ägerroos, and A. Virolainen. 2005. Molecular epidemiology of an outbreak caused by methicillin-resistant Staphylococcus aureus in a health care ward and associated nursing home. J. Clin. Microbiol. 436161-6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kloos, W. E., and K. H. Schleifer. 1975. Simplified scheme for routine identification of human Staphylococcus species. J. Clin. Microbiol. 182-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kondo, Y., T. Ito, X. X. Ma, S. Watanabe, B. N. Kreiswirth, J. Etienne, and K. Hiramatsu. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuroda, M., A. Yamashita, H. Hirakawa, M. Kumano, K. Morikawa, M. Higashide, A. Maruyama, Y. Inose, K. Matoba, H. Toh, S. Kuhara, M. Hattori, and T. Ohta. 2005. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc. Natl. Acad. Sci. USA 10213272-13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, L. Y., C. Thomas, T. Chi, G. Stone, S. Greg, and T. Lauri. 2000. Nasal colonization by methicillin resistant coagulase-negative Staphylococcus in community skilled nursing facility patients. Am. J. Infect. Control 28269-272. [DOI] [PubMed] [Google Scholar]
- 22.Luong, T. T., S. Ouyang, K. Bush, and C. Y. Lee. 2002. Type 1 capsule genes of Staphylococcus aureus are carried in a staphylococcal cassette chromosome genetic element. J. Bacteriol. 1843623-3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lyytikäinen, O., J. Lumio, H. Sarkkinen, E. Kolho, A. Kostiala, and P. Ruutu. 2002. Nosocomial bloodstream infections in Finnish hospitals during 1999-2000. Clin. Infect. Dis. 3514-19. [DOI] [PubMed] [Google Scholar]
- 24.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 461147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miragaia, M., I. Couto, and H. de Lencastre. 2005. Genetic diversity among methicillin-resistant Staphylococcus epidermidis (MRSE). Microb. Drug Resist. 1183-93. [DOI] [PubMed] [Google Scholar]
- 26.Miragaia, M., J. C. Thomas, I. Couto, M. C. Enright, and H. de Lencastre. 2007. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J. Bacteriol. 1892540-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mombach Pinheiro Machado, A. B., K. C. Reiter, R. M. Paiva, and A. L. Barth. 2007. Distribution of staphylococcal cassette chromosome mec (SCCmec) types I, II, III, and IV in coagulase-negative staphylococci from patients attending a tertiary hospital in southern Brazil. J. Med. Microbiol. 561328-1333. [DOI] [PubMed] [Google Scholar]
- 28.Mongkolrattanothai, K., S. Boyle, T. V. Murphy, and R. S. Daum. 2004. Novel non-mecA-containing staphylococcal chromosomal cassette composite island containing pbp4 and tagF genes in a commensal staphylococcal species: a possible reservoir for antibiotic resistance islands in Staphylococcus aureus. Antimicrob. Agents Chemother. 481823-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 411574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musser, J. M., and V. Kapur. 1992. Clonal analysis of methicillin-resistant Staphylococcus aureus strains from intercontinental sources: association of the mec gene with divergent phylogenetic lineages implies dissemination by horizontal transfer and recombination. J. Clin. Microbiol. 302058-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 462155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliveira, D. C., C. Milheirico, and H. de Lencastre. 2006. Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob. Agents Chemother. 503457-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silva, F. R., E. M. Mattos, M. V. Coimbra, B. T. Ferreira-Carvalho, and A. M. Figueiredo. 2001. Isolation and molecular characterization of methicillin-resistant coagulase-negative staphylococci from nasal flora of healthy humans at three community institutions in Rio de Janeiro City. Epidemiol. Infect. 12757-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki, E., K. Kuwahara-Arai, J. F. Richardson, and K. Hiramatsu. 1993. Distribution of mec regulator genes in methicillin-resistant Staphylococcus clinical strains. Antimicrob. Agents Chemother. 371219-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wielders, C. L., M. R. Vriens, S. Brisse, L. A. de Graaf-Miltenburg, A. Troelstra, A. Fleer, F. J. Schmitz, J. Verhoef, and A. C. Fluit. 2001. In-vivo transfer of mecA DNA to Staphylococcus aureus. Lancet 3571674-1675. [DOI] [PubMed] [Google Scholar]
- 37.Wisplinghoff, H., A. E. Rosato, M. C. Enright, M. Noto, W. Craig, and G. L. Archer. 2003. Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrob. Agents Chemother. 473574-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, S., C. Piscitelli, H. de Lencastre, and A. Tomasz. 1996. Tracking the evolutionary origin of the methicillin resistance gene: cloning and sequencing of a homologue of mecA from a methicillin susceptible strain of Staphylococcus sciuri. Microb. Drug Resist 2435-441. [DOI] [PubMed] [Google Scholar]