Abstract
Class 1 and 2 integrons were detected in 45.8% (54/118) and 19.5% (23/118) of our tested Pseudomonas aeruginosa isolates, respectively. Three strains were positive for both the integrons. This is the first report of class 2 integrons in P. aeruginosa and also of isolates carrying class 1 and 2 integrons simultaneously.
Pseudomonas aeruginosa remains one of the most important pathogens in the nosocomial setting (14), and it not only is naturally resistant to many antimicrobial agents but also has the distinctive capacity via multiple mechanisms to become resistant to virtually all the antibiotics available commercially (9, 38). A genetic element, the integron, is potentially a major agent in the dissemination of multidrug resistance among gram-negative bacteria, especially in Pseudomonas (16). Gene cassettes, present in the variable region of integrons, are discrete mobile units comprising a gene, usually an antibiotic resistance gene, and a recombination site that is recognized by an integrase. The class 1 integron has been identified as a primary source of resistance genes within gram-negative and -positive bacteria (6, 20, 33, 36, 40, 41, 42), and the class 2 integron has been seen in Acinetobacter sp. isolates throughout the world (28). However, class 2 integrons in P. aeruginosa strains had not yet been investigated. In this study, 118 imipenem-resistant P. aeruginosa isolates were chosen for the investigation of class 1 and 2 integrons because of the relatively high integron-positive rate in imipenem-resistant isolates.
From 2001 to 2005, a total of 118 consecutive nonduplicated P. aeruginosa isolates which were intermediate or resistant (nonsusceptible) to imipenem (IMP; MIC > 8 μg/ml) were isolated from the First Affiliated Hospital of Jinan University, an 850-bed tertiary-level teaching hospital in Guangzhou, China. Identification of isolates to the species level and antimicrobial susceptibility testing were performed with the Vitek system (bioMerieux Vitek Systems Inc., Hazelwood, MO). The quality control strain used was P. aeruginosa ATCC 27853. Template DNA used for PCR was prepared as described previously (16). Detection and characterization of class 1 and 2 integrons were performed as described previously (35, 41). PCR products of the variable region were further characterized by restriction fragment length polymorphism (RFLP), and at least two different restriction endonucleases were chosen for each RFLP assay, and the DNA sequence for at least one of the variable region amplification products belonging to each of the individual RFLP patterns was determined as described previously (35). Seventy-four integron-positive P. aeruginosa isolates were subjected to genotyping analysis by randomly amplified polymorphic DNA PCR (RAPD-PCR) as described previously (35).
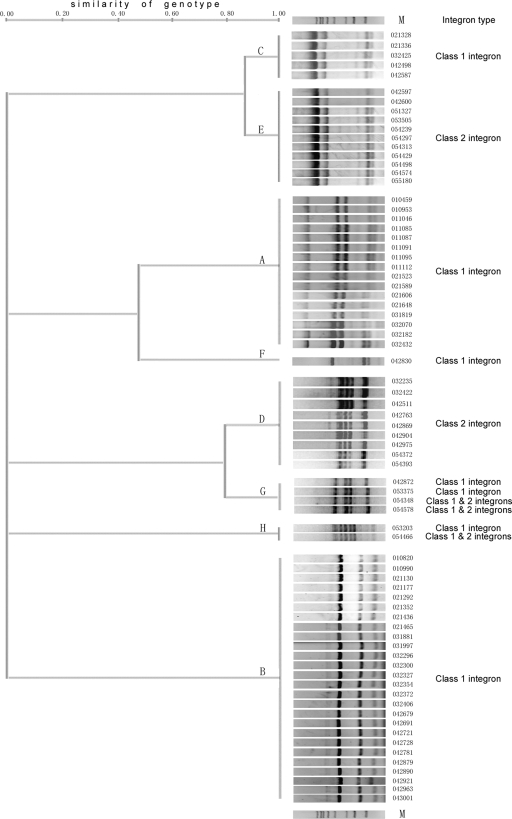
The multidrug resistance (defined as resistance to six or more antibiotics) rates of integron-positive and -negative strains were 93.2% and 18.2%, respectively (Table 1). Class 1 integron was detected in 54 isolates, and 51 strains carried the 3′ conserved region of qacEΔ1-sul1. Seven different sizes of variable region were found, with fragments with lengths ranging between 879 bp and 2,655 bp (Table 2). The array of the aacA4-catB3-dfrA1 noncoding gene cassette has been reported previously (16). The defective class 1 integron with a sul3 gene, which was identical with that seen in Salmonella enterica serovar Typhimurium (AY047357), had never been reported to be seen in isolates of P. aeruginosa. Class 2 integrons were found in 23 P. aeruginosa isolates, and all strains harbored the same array of three cassettes, dfrA1-sat1-aadA1, which was identical to that found in Tn7. Three strains had both class 1 and 2 integrase genes. No class 3 integrase gene was detected in any of the isolates examined. RAPD-PCR analysis divided 74 integron-positive P. aeruginosa strains into eight different groups with different RAPD patterns (genotypes A to H) (Fig. 1). Fifty-one class 1 integron-positive strains and 3 class 1 and 2 integron-positive strains were of types A, B, C, F, G, and H, and 20 class 2 integron-positive strains were of types D and E (Table 2).
TABLE 1.
Association between antibiotic susceptibility profile and integrons in 118 Pseudomonas aeruginosa isolates
| Antibiotica | % (no.) of isolates
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total (n = 118)
|
Integron-positive isolates (n = 74)
|
Integron-negative isolates (n = 44)
|
|||||||
| Resistant | Intermediate | Susceptible | Resistant | Intermediate | Susceptible | Resistant | Intermediate | Susceptible | |
| AMK | 33.1 (39) | 13.6 (16) | 53.3 (63) | 44.6 (33) | 18.9 (14) | 37.5 (27) | 13.6 (6) | 4.5 (2) | 81.9 (36) |
| ATM | 41.5 (49) | 14.4 (17) | 44.1 (52) | 55.4 (41) | 14.9 (11) | 29.7 (22) | 18.2 (8) | 13.6 (6) | 68.2 (30) |
| CAZ | 37.3 (44) | 7.6 (9) | 55.1 (65) | 50.0 (37) | 5.4 (4) | 44.6 (33) | 15.9 (7) | 11.4 (5) | 72.7 (32) |
| CIP | 70.3 (83) | 11.0 (13) | 18.6 (22) | 79.7 (59) | 12.2 (9) | 8.1 (6) | 54.5 (24) | 9.1 (4) | 36.4 (16) |
| CRO | 42.4 (50) | 6.8 (8) | 50.8 (60) | 50.0 (37) | 6.8 (5) | 43.2 (32) | 29.6 (13) | 6.8 (3) | 63.6 (28) |
| GEN | 63.6 (75) | 11.0 (13) | 25.4 (30) | 85.1 (63) | 14.9 (11) | 0 (0) | 27.3 (12) | 4.5 (2) | 68.2 (30) |
| LVX | 61.0 (72) | 19.5 (23) | 19.5 (23) | 70.3 (52) | 21.6 (16) | 8.1 (6) | 45.5 (20) | 15.9 (7) | 38.6 (17) |
| PIP | 55.1 (65) | 5.9 (7) | 39.0 (46) | 70.3 (52) | 8.1 (6) | 21.6 (16) | 29.6 (13) | 2.2 (1) | 68.2 (30) |
| SXT | 70.3 (83) | 16.9 (20) | 12.7 (15) | 79.7 (59) | 18.9 (14) | 1.4 (1) | 54.5 (24) | 13.6 (6) | 31.9 (14) |
| TCC | 45.8 (54) | 10.2 (12) | 44.1 (52) | 55.4 (41) | 16.2 (12) | 28.4 (21) | 29.6 (13) | 0 (0) | 70.5 (31) |
| TET | 78.0 (92) | 7.6 (9) | 14.4 (17) | 83.7 (62) | 9.5 (7) | 6.8 (5) | 68.2 (30) | 4.5 (2) | 27.3 (12) |
| TOB | 55.1 (65) | 21.2 (25) | 23.7 (28) | 75.7 (56) | 24.3 (18) | 0 (0) | 20.5 (9) | 15.9 (7) | 63.6 (28) |
AMK, amikacin; ATM, aztreonam; CAZ, ceftazidime; CRO, ceftriaxone; CIP, ciprofloxacin; GEN, gentamicin; LVX, levofloxacin; PIP, piperacillin; TET, tetracycline; TCC, ticarcillin-clavulanic acid; TOB, tobramycin; SXT, trimethoprim-sulfamethoxazole.
TABLE 2.
Phenotypic and genotypic characteristics of 74 integron-positive Pseudomonas aeruginosa isolates
| Strain | Yr of isolation | Source | Age (yr), sexa | Genetic material in isolate withb:
|
RAPD pattern | Resistance profilec | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Class 1 integrons
|
Class 2 integrons
|
|||||||||
| intI1 | 3′ conserved sequence | Gene cassette | intI2 | Gene cassette | ||||||
| 010459 | 2001 | Blood | 67, M | + | + | dfrA12-orfF-aadA2 | − | − | A | ACiGLTToTs |
| 010820 | 2001 | Sputum | 79, F | + | + | aacA4-cmlA1 | − | − | B | AzCaCeCiGLPTTcToTs |
| 010953 | 2001 | Blood | 80, M | + | + | dfrA17-aadA5 | − | − | A | ACiGLTToTs |
| 010990 | 2001 | Sputum | 80, M | + | − | aacA4-cmlA1 | − | − | B | AzCaCeCiGLPTTcToTs |
| 011046 | 2001 | Sputum | 51, M | + | + | dfrA12-orfF-aadA2 | − | − | A | ACiGLTToTs |
| 011085 | 2001 | Sputum | 73, F | + | + | dfrA12-orfF-aadA2 | − | − | A | ACiGLTToTs |
| 011087 | 2001 | Sputum | 45, F | + | + | dfrA12-orfF-aadA2 | − | − | A | ACiGLTToTs |
| 011091 | 2001 | Blood | 45, M | + | + | dfrA12-orfF-aadA2 | − | − | A | ACiGLTToTs |
| 011095 | 2001 | Sputum | 54, M | + | + | dfrA12-orfF-aadA2 | − | − | A | ACiGLTToTs |
| 011112 | 2001 | Stool | 65, F | + | + | dfrA17-aadA5 | − | − | A | ACiGLTToTs |
| 021130 | 2002 | Stool | 54, M | + | + | aacA4-cmlA1 | − | − | B | AzCaCeCiGLPTTcToTs |
| 021177 | 2002 | Sputum | 30, M | + | + | aacA4-cmlA1 | − | − | B | AzCaCeCiGLPTTcToTs |
| 021292 | 2002 | Blood | 72, F | + | + | aacA4-cmlA1 | − | − | B | AzCaCeCiGLPTTcToTs |
| 021328 | 2002 | Sputum | 81, F | + | + | aacA4-cmlA1 | − | − | C | ACeGPToTs |
| 021336 | 2002 | Sputum | 69, M | + | + | aacA4-cmlA1 | − | − | C | ACeGPToTs |
| 021352 | 2002 | Sputum | 69, F | + | + | aacA4-cmlA1 | − | − | B | AzCaCeCiGLPTTcToTs |
| 021436 | 2002 | Pus | 45, M | + | + | dfrA12-orfF-aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 021465 | 2002 | Sputum | 30, F | + | + | dfrA12-orfF-aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 021523 | 2002 | Sputum | 65, F | + | − | dfrA12-orfF-aadA2 | − | − | A | ACiGLTToTs |
| 021589 | 2002 | Stool | 25, M | + | + | dfrA12-orfF-aadA2 | − | − | A | ACiGLTToTs |
| 021606 | 2002 | Sputum | 77, M | + | + | dfrA12-orfF-aadA2 | − | − | A | ACiGLTToTs |
| 021648 | 2002 | Blood | 42, M | + | + | dfrA12-orfF-aadA2 | − | − | A | ACiGLTToTs |
| 031819 | 2003 | Sputum | 75, M | + | + | dfrA12-orfF-aadA2 | − | − | A | ACiGLTToTs |
| 031881 | 2003 | Sputum | 83, F | + | + | dfrA12-orfF-aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 031997 | 2003 | Pus | 21, F | + | + | dfrA12-orfF-aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 032070 | 2003 | Sputum | 80, M | + | + | dfrA12-orfF-aadA2 | − | − | A | ACiGLTToTs |
| 032182 | 2003 | Stool | 26, F | + | + | dfrA12-orfF-aadA2 | − | − | A | ACiGLTToTs |
| 032235 | 2003 | Sputum | 73, F | − | − | − | + | dfrA1-sat1-aadA1 | D | AAzCaCeCiGLPTTcToTs |
| 032296 | 2003 | Blood | 72, M | + | + | dfrA12-orfF-aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 032300 | 2003 | Sputum | 78, F | + | + | aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 032327 | 2003 | Stool | 22, M | + | + | dfrA12-orfF-aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 032354 | 2003 | Sputum | 72, F | + | + | aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 032372 | 2003 | Blood | 76, M | + | + | aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 032406 | 2003 | Stool | 70, F | + | + | aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 032422 | 2003 | Sputum | 72, M | − | − | − | + | dfrA1-sat1-aadA1 | D | AAzCaCeCiGLPTTcToTs |
| 032425 | 2003 | Blood | 72, F | + | − | aadA2 | − | − | C | ACeGPToTs |
| 032432 | 2003 | Sputum | 47, M | + | + | dfrA12-orfF-aadA2 | − | − | A | ACiGLTToTs |
| 042498 | 2004 | Blood | 85, M | + | + | aadA2 | − | − | C | ACeGPToTs |
| 042511 | 2004 | Sputum | 67, F | − | − | − | + | dfrA1-sat1-aadA1 | D | AAzCaCeCiGLPTTcToTs |
| 042587 | 2004 | Blood | 24, M | + | + | aadA2 | − | − | C | ACeGPToTs |
| 042597 | 2004 | Sputum | 65, M | − | − | − | + | dfrA1-sat1-aadA1 | E | AAzCiGLPTTcToTs |
| 042600 | 2004 | Stool | 74, F | − | − | − | + | dfrA1-sat1-aadA1 | E | AAzCiGLPTTcToTs |
| 042679 | 2004 | Blood | 71, M | + | + | aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 042691 | 2004 | Pus | 36, M | + | + | aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 042721 | 2004 | Sputum | 83, F | + | + | dfrA12-orfF-aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 042728 | 2004 | Sputum | 48, M | + | + | aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 042763 | 2004 | Stool | 83, F | − | − | − | + | dfrA1-sat1-aadA1 | D | AAzCaCeCiGLPTTcToTs |
| 042781 | 2004 | Sputum | 61, M | + | + | dfrA12-orfF-aadA2; aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 042830 | 2004 | Sputum | 75, M | + | + | aacA4-catB3-dfrA1 (noncoding) | − | − | F | GPTTcTo |
| 042869 | 2004 | Stool | 66, F | − | − | − | + | dfrA1-sat1-aadA1 | D | AAzCaCeCiGLPTTcToTs |
| 042872 | 2004 | Sputum | 53, M | + | + | sul3 | − | − | G | AAzCaCeCiGLPTTcToTs |
| 042879 | 2004 | Blood | 71, F | + | + | dfrA12-orfF-aadA2; dfrA17-aadA5 | − | − | B | AzCaCeCiGLPTTcToTs |
| 042890 | 2004 | Sputum | 48, M | + | + | aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 042904 | 2004 | Pus | 28, F | − | − | − | + | dfrA1-sat1-aadA1 | D | AAzCaCeCiGLPTTcToTs |
| 042921 | 2004 | Sputum | 76, M | + | + | dfrA12-orfF-aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 042963 | 2004 | Blood | 86, F | + | + | dfrA12-orfF-aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 042975 | 2004 | Stool | 63, M | − | − | − | + | dfrA1-sat1-aadA1 | D | AAzCaCeCiGLPTTcToTs |
| 043001 | 2004 | Sputum | 71, F | + | − | aadA2 | − | − | B | AzCaCeCiGLPTTcToTs |
| 051327 | 2005 | Sputum | 49, M | − | − | − | + | dfrA1-sat1-aadA1 | E | AAzCiGLPTTcToTs |
| 053203 | 2005 | Blood | 62, F | + | + | blaVIM-4-pse1 | − | − | H | AAzCaCeCiGLPTTcToTs |
| 053375 | 2005 | Sputum | 33, F | + | + | sul3 | − | − | G | AAzCaCeCiGLPTTcToTs |
| 053505 | 2005 | Stool | 46, M | − | − | − | + | dfrA1-sat1-aadA1 | E | AAzCiGLPTTcToTs |
| 054239 | 2005 | Sputum | 56, F | − | − | − | + | dfrA1-sat1-aadA1 | E | AAzCiGLPTTcToTs |
| 054297 | 2005 | Blood | 38, F | − | − | − | + | dfrA1-sat1-aadA1 | E | AAzCiGLPTTcToTs |
| 054313 | 2005 | Sputum | 55, M | − | − | − | + | dfrA1-sat1-aadA1 | E | AAzCiGLPTTcToTs |
| 054348 | 2005 | Blood | 44, F | + | + | sul3 | + | dfrA1-sat1-aadA1 | G | AAzCaCeCiGLPTTcToTs |
| 054372 | 2005 | Sputum | 43, F | − | − | − | + | dfrA1-sat1-aadA1 | D | AAzCaCeCiGLPTTcToTs |
| 054393 | 2005 | Blood | 57, F | − | − | − | + | dfrA1-sat1-aadA1 | D | AAzCaCeCiGLPTTcToTs |
| 054429 | 2005 | Sputum | 49, M | − | − | − | + | dfrA1-sat1-aadA1 | E | AAzCiGLPTTcToTs |
| 054466 | 2005 | Blood | 60, F | + | + | blaVIM-4-pse1 | + | dfrA1-sat1-aadA1 | H | AAzCaCeCiGLPTTcToTs |
| 054498 | 2005 | Sputum | 51, M | − | − | − | + | dfrA1-sat1-aadA1 | E | AAzCiGLPTTcToTs |
| 054574 | 2005 | Blood | 29, F | − | − | − | + | dfrA1-sat1-aadA1 | E | AAzCiGLPTTcToTs |
| 054578 | 2005 | Sputum | 69, F | + | + | sul3 | + | dfrA1-sat1-aadA1 | G | AAzCaCeCiGLPTTcToTs |
| 055180 | 2005 | Sputum | 66, F | − | − | − | + | dfrA1-sat1-aadA1 | E | AAzCiGLPTTcToTs |
M, male; F, female.
+, present; −, absent.
A, amikacin; Az, aztreonam; Ca, ceftazidime; Ce, ceftriaxone; Ci, ciprofloxacin; G, gentamicin; L, levofloxacin; P, piperacillin; T, tetracycline; Tc, ticarcillin-clavulanic acid; To, tobramycin; Ts, trimethoprim-sulfamethoxazole.
FIG. 1.
RAPD-PCR patterns of 74 integron-positive Pseudomonas aeruginosa isolates.
Integrons have been identified as a primary source of resistance genes and were suspected to serve as reservoirs of antimicrobial resistance genes within microbial populations (34), and integron-mediated resistance to antibiotics in clinical isolates of P. aeruginosa has been reported (11, 16, 18, 24, 26). However, all of these studies were concerned with class 1 integrons, with no exception. Class 2 integrons were most frequently associated with members of the family Enterobacteriaceae, such as Escherichia coli and Salmonella enterica, and also are commonly found in Acinetobacter baumannii and Burkholderia cepacia (1, 3, 4, 19, 25, 27, 37). However, class 2 integrons in P. aeruginosa had never been reported. In this study, we detected 51 class 1 integron-positive strains, 20 class 2 integron-positive strains, and 3 class 1 and 2 integron-positive strains from total of 118 strains. This is the first report, to our knowledge, of class 2 integrons with dfrA1-sat1-aadA1 in P. aeruginosa. Furthermore, it is also the first time clinical P. aeruginosa isolates carrying class 1 and 2 integrons simultaneously have been identified.
Class 1 integrons were commonly found in the tested P. aeruginosa isolates (45.8%, 54/118), but the class 1 integron-positive rates had been decreasing during the 5-year study period, with rates of 66.6% (10/15) in 2001, 60.0% (12/20) in 2002, 52.0% (13/25) in 2003, 40.0% (14/35) in 2004, and 21.7% (5/23) in 2005. Class 2 integron appeared in 2003, with the class 2 integron-positive rates rising for the next three years, with rates of 8.0% (2/25) in 2003, 20.0% (7/35) in 2004, and 60.8% (14/23) in 2005, indicating that class 2 integron had been prevalent in recent years. The rate of integron-positive isolates had changed in a small scale, with rates of 66.6% in 2001, 60% in 2002 to 2004, and 69.5% in 2005, while the proportion of class 1 integrons had decreased more than 45% and the occurrence of class 2 integron began in 2003. The class 2 integron-positive rate increased to >60% in 2005, suggesting that class 2 integrons were increasing and suggesting and the possibility of this class replacing class 1 integron in recent years. The evolutionary success of an integron was determined by two important factors: the resistance cassettes it carries and the host range of the plasmid on which it occurs (13). The two most frequently detected resistance genes in 74 integron-positive isolates were of the aadA and dfrA families, with rates of 79.7% (59/74) and 64.9% (48/74), respectively. Since the two cassettes, dfrA1 and aadA1, have been observed in all class 2 integron-positive isolates, it is reasonable to presume the transferring of cassettes among different integrons (13). So whether class 2 integrons have more fitness and better survival ability than class 1 integrons under selective pressure and whether some cassettes appear to have been transferred among integron classes require further investigation.
In conclusion, this study showed the occurrence and characteristics of class 1 and 2 integrons in clinical P. aeruginosa. Nevertheless, further studies need to be conducted to investigate the cause of the appearance and prevalence of class 2 integrons in P. aeruginosa in recent years. The findings will help to develop control strategies for infections in hospitals.
Nucleotide sequence accession number.
The nucleotide sequence accession number of the defective class 1 integron with sul3 gene in GenBank is AB281182.
Acknowledgments
This work was supported by Science Foundation of Ministry of Education of China (grant 706046), National Natural Science Foundation of China (grant 20436020) and State Scholarship Fund of China Scholarship Council (grant 2008615044).
Footnotes
Published ahead of print on 19 November 2008.
REFERENCES
- 1.Ahmed, A. M., H. Nakano, and T. Shimamoto. 2005. Molecular characterization of integrons in non-typhoid Salmonella serovars isolated in Japan: description of an unusual class 2 integron. J. Antimicrob. Chemother. 55371-374. [DOI] [PubMed] [Google Scholar]
- 2.Reference deleted.
- 3.Barlow, R. S., J. M. Pemberton, P. M. Desmarchelier, and K. S. Gobius. 2004. Isolation and characterization of integron-containing bacteria without antibiotic selection. Antimicrob. Agents Chemother. 48838-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biskri, L., and D. Mazel. 2003. Erythromycin esterase gene ere(A) is located in a functional gene cassette in an unusual class 2 integron. Antimicrob. Agents Chemother. 473326-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reference deleted.
- 6.Clark, N. C., Ø. Olsvik, J. M. Swenson, C. A. Spiegel, and F. C. Tenover. 1999. Detection of a streptomycin/spectinomycin adenylyltransferase gene (aadA) in Enterococcus faecalis. Antimicrob. Agents Chemother. 43157-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reference deleted.
- 8.Reference deleted.
- 9.Falagas, M. E., and S. K. Kasiakou. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin. Infect. Dis. 401333-1341. [DOI] [PubMed] [Google Scholar]
- 10.Reference deleted.
- 11.Gutiérrez, O., C. Juan, E. Cercenado, F. Navarro, E. Bouza, P. Coll, J. L. Perez, and A. Oliver. 2007. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Spanish hospitals. Antimicrob. Agents Chemother. 514329-4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reference deleted.
- 13.Hansson, K., L. Sundstrom, A. Pelletier, and P. H. Roy. 2002. IntI2 integron integrase in Tn7. J. Bacteriol. 1841712-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarvis, W. R., and W. J. Martone. 1992. Predominant pathogens in hospital infections. J. Antimicrob. Chemother. 2919-24. [DOI] [PubMed] [Google Scholar]
- 15.Reference deleted.
- 16.Li, X., L. Shi, W. Yang, L. Li, and S. Yamasaki. 2006. New array of aacA4-catB3-dfrA1 gene cassettes and a noncoding cassette from a class-1-integron-positive clinical strain of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 502278-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reference deleted.
- 18.Martinez-Freijo, P., A. C. Fluit, F. J. Schmitz, V. S. Grek, J. Vorhoef, and M. E. Jones. 1998. Class I integrons in gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J. Antimicrob. Chemother. 42689-696. [DOI] [PubMed] [Google Scholar]
- 19.Miko, A., K. Pries, A. Schroeter, and R. Helmuth. 2003. Multiple-drug resistance in d-tartrate-positive Salmonella enterica serovar Paratyphi B isolates from poultry is mediated by class 2 integrons inserted into the bacterial chromosome. Antimicrob. Agents Chemother. 473640-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nandi, S., J. J. Maurer, and C. Hofacre. 2004. Gram-positive bacteria are a major reservoir of class 1 antibiotic resistance integrons in poultry litter. Proc. Natl. Acad. Sci. USA 1017118-7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reference deleted.
- 22.Reference deleted.
- 23.Reference deleted.
- 24.Pitout, J. D. D., B. L. Chow, D. B. Gregson, K. B. Laupland, S. Elsayed, and D. L. Church. 2007. Molecular epidemiology of metallo-β-lactamase-producing Pseudomonas aeruginosa in the Calgary health region: emergence of VIM-2-producing isolates. J. Clin. Microbiol. 45294-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ploy, M., F. Denis, P. Courvalin, and T. Lambert. 2000. Molecular characterization of integrons in Acinetobacter baumannii: description of a hybrid class 2 integron. Antimicrob. Agents Chemother. 442684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poirel, L., M. Magalhaes, M. Lopes, and P. Nordmann. 2004. Molecular analysis of metallo-β-lactamase gene blaSPM-1-surrounding sequences from disseminated Pseudomonas aeruginosa isolates in Recife, Brazil. Antimicrob. Agents Chemother. 481406-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramírez, M. S., L. J. Vargas, V. Cagnoni, M. Tokumoto, and D. Centron. 2005. Class 2 integron with a novel cassette array in a Burkholderia cenocepacia isolate. Antimicrob. Agents Chemother. 494418-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramírez, M. S., C. Quiroga, and D. Centron. 2005. Novel rearrangement of a class 2 integron in two non-epidemiologically related isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 495179-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reference deleted.
- 30.Reference deleted.
- 31.Reference deleted.
- 32.Reference deleted.
- 33.Shi, L., M. Zheng, Z. Xiao, M. Asakura, J. Su, L. Li, and S. Yamasaki. 2006. Unnoticed spread of class 1 integrons in gram-positive strains isolated in Guangzhou, China. Microbiol. Immunol. 50463-467. [DOI] [PubMed] [Google Scholar]
- 34.Stokes, H. W., and R. M. Hall. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 31669-1683. [DOI] [PubMed] [Google Scholar]
- 35.Su, J., L. Shi, L. Yang, Z. Xiao, X. Li, and S. Yamasaki. 2006. Analysis of integrons in clinical isolates of Escherichia coli in China during the last six years. FEMS Microbiol. Lett. 25475-80. [DOI] [PubMed] [Google Scholar]
- 36.Tauch, A., S. Gotker, A. Puhler, J. Kalinowski, and G. Thierbach. 2002. The 27.8-kb R-plasmid pTET3 from Corynebacterium glutamicum encodes the aminoglycoside adenyltransferase gene cassette aadA9 and the regulated tetracycline efflux system Tet 33 flanked by active copies of the widespread insertion sequence IS6100. Plasmid 48117-129. [DOI] [PubMed] [Google Scholar]
- 37.van Essen-Zandbergen, A., H. Smith, K. Veldman, and D. Mevius. 2007. Occurrence and characteristics of class 1, 2 and 3 integrons in Escherichia coli, Salmonella and Campylobacter spp. in The Netherlands. J. Antimicrob. Chemother. 59746-750. [DOI] [PubMed] [Google Scholar]
- 38.Wang, C. Y., J. S. Jerng, K. Y. Chen, L. N. Lee, C. J. Yu, P. R. Hsueh, and P. C. Yang. 2006. Pandrug-resistant Pseudomonas aeruginosa among hospitalized patients: clinical features, risk-factors and outcomes. Clin. Microbiol. Infect. 1263-68. [DOI] [PubMed] [Google Scholar]
- 39.Reference deleted.
- 40.Xu, Z., L. Shi, C. Zhang, L. Zhang, X. Li, Y. Cao, L. Li, and S. Yamasaki. 2007. Nosocomial infection caused by class 1 integron-carrying Staphylococcus aureus in a hospital in South China. Clin. Microbiol. Infect. 13980-984. [DOI] [PubMed] [Google Scholar]
- 41.Xu, Z., L. Shi, M. J. Alam, L. Li, and S. Yamasaki. 2008. Integron-bearing methicillin-resistant coagulase-negative staphylococci in South China, 2001-2004. FEMS Microbiol. Lett. 278223-230. [DOI] [PubMed] [Google Scholar]
- 42.Xu, Z., L. Li, M. J. Alam, S. Yamasaki, and L. Shi. 2008. First confirmation of integron-bearing methicillin-resistant Staphylococcus aureus. Curr. Microbiol. 57264-268. [DOI] [PubMed] [Google Scholar]