Abstract
The prevalence of Streptococcus pneumoniae serotype 6C, a recently recognized serotype that cross-reacts serologically with serotype 6A, was investigated. Isolates of serotype 6A in various collections were recovered, and serotype 6C was differentiated from 6A by multiplex PCR of DNA extracts by using appropriate primers. Antimicrobial susceptibility was performed by Clinical and Laboratory Standards Institute broth microdilution, and selected isolates were typed by pulsed-field gel electrophoresis, repetitive sequence-based PCR typing, and rapid multilocus sequence typing (MLST) by electrospray ionization mass spectrometry of PCR products. A total of 60 serotype 6C isolates were found: 30 of 122 Cleveland isolates collected from 1979 to 2007, 19 of 39 pediatric isolates collected nationwide in 2005 and 2006, and 11 pediatric isolates from Massachusetts collected in 2006 and 2007. Only four isolates were recovered prior to introduction of the conjugate pneumococcal vaccine in 2000; the earliest isolate was recovered in 1989. The sources of the isolates included blood (n = 5), the lower respiratory tract (n = 27), the sinus (n = 5), the ear (n = 2), and the nasopharynx (n = 18); isolates were recovered from 49 children and 11 adults. Pediatric isolates were found in all six major U.S. geographic regions. Antimicrobial susceptibility showed that 22 isolates were nonsusceptible to penicillin, macrolides, and trimethoprim-sulfamethoxazole, 8 had other resistance patterns, and 30 were fully susceptible. The three typing methods used showed similar clusters of up to eight isolates per cluster. MLST showed five clusters related to serotype 6A, two clusters related to serotype 6B, one cluster related to serotype 3, and one cluster related to serotype 34. This study documents the occurrence, nationwide distribution, diversity, likely origins, and increasing incidence after 2001 of this recently recognized serotype. Serotype 6C warrants consideration for addition to future conjugate pneumococcal vaccines.
The polysaccharide capsule is the major virulence determinant of Streptococcus pneumoniae, preventing phagocytosis. A total of 25 serotypes have one antigenic capsular determinant, while 21 serogroups have a common and one or more unique determinants. The number of known capsular serotypes, which totaled 90 in 1995 (16), increased to 91 in 2007 with the description of serotype 6C (34, 35). This new serotype cross-reacts serologically with serotype 6A but is differentiated by a change in the wciN gene region of the capsular locus encoding for galactosyl transferase (32).
A variety of capsular serotypes have been used to develop vaccines, and two such vaccines are currently available. The first is a 23-valent purified capsular polysaccharide vaccine (Pneumovax; Merck & Co, Whitehouse Station, NJ) containing 23 serotypes (1, 2, 3, 4, 5, 6B, 7F, 8, 9N, 9V, 10A, 11A, 12F, 14, 15B, 17F, 18C, 19F, 19A, 20, 22F, 23F, and 33F). The second is a 7-valent vaccine containing saccharides purified from capsular polysaccharides conjugated to a protein vector (PCV7) to provide immunity in children under 2 years of age (Prevnar; Wyeth Pharmaceuticals, Inc., Philadelphia, PA) and contains serotypes 4, 6B, 9V, 14, 18C, 19F, and 23F.
In view of the inclusion of serotype 6B in PCV7 and the inability to differentiate serotypes 6A and 6C by serotyping, we recovered and reidentified isolates previously characterized as serotype 6A in various strain collections to determine the incidence of serotype 6C. Serotype 6C isolates were further characterized to determine their diversity and relationships to other serotypes.
MATERIALS AND METHODS
Strains.
Isolates of serotype 6A in two strain collections were recovered from frozen storage. The first collection consisted of 2,735 isolates recovered at University Hospitals Case Medical Center from 1979 through 2007 (invasive isolates from 1979 to 1996 and isolates from all sources from 1997 to 2007); details of these collections have been published (22, 23). The second collection comprised 393 isolates recovered from U.S. children during a nationwide surveillance study conducted in 2005 and 2006; these isolates were prospectively collected from 104 participating institutions geographically distributed across the United States, limited to one per patient, and collected from a variety of specimen sources (noninvasive, invasive, and carriage) (5). Random isolates of serotypes 6A (n = 10) and 6B (n = 5) were also selected from the nationwide surveillance collection for comparison. An additional 11 nasopharyngeal isolates provided by Stephen Pelton were also studied; these had been recovered from children in Massachusetts in 2006 and 2007 and were identified as atypical serotype 6A strains based on capsular swelling with factor 6a antiserum but not with pooled antiserum containing this factor (20).
Serotyping.
Serotyping was performed by capsular swelling reaction using commercial serogroup and serotype specific antisera (Statenserum Institute, Copenhagen, Denmark) according to the manufacturer's instructions (26).
Antimicrobial susceptibility.
MICs were determined by broth microdilution according to Clinical and Laboratory Standards Institute (CLSI) recommended procedures (30), using custom prepared frozen microdilution trays (TREK, Westlake, OH) as previously described (23). The following antimicrobial agents (and the testing ranges in μg/ml) were tested: penicillin (0.015 to 8), amoxicillin (0.015 to 16), amoxicillin-clavulanic acid (2:1 ratio) (0.015/0.008 to 16/8), cefuroxime (0.03 to 4), ceftriaxone (0.015 to 2), cefdinir (0.03 to 4), azithromycin (0.015 to 32), clindamycin (0.015 to 2), levofloxacin (0.25 to 32), cefpodoxime (0.03 to 4), and trimethoprim-sulfamethoxazole (SXT; 1:19 ratio, 0.06/1.14 to 4/76). MICs were interpreted according to current CLSI breakpoints; penicillin MICs were interpreted according to the new oral penicillin breakpoints (susceptible, ≤0.06 μg/ml; intermediate, 0.12 to 1 μg/ml; and resistant, ≥2 μg/ml) since these correspond to breakpoints previously in use (3).
Differentiation of serogroup 6 serotypes by PCR amplification of capsular gene targets.
Serotype 6C was differentiated from serotypes 6A and 6B by multiplex PCR of DNA extracts using two forward primers, 5101 (5′-ATTTGGTGTACTTCCTCC), which attaches to nucleotides 6949 to 6966 in the gene wciN, and 5106 (5′-TACCATGCAGGGTGGAATGT), which attaches to nucleotides 5897 to 5916 in gene wchA, and a common reverse primer, 3101 (5′-CCATCCTTCGAGTATTGC), which attaches to nucleotides 7888 to 7905 in gene wciO (27, 34). Primer pair 5101-3101 produces products of 958 or 1267 kb with serotypes 6A and 6B, while no product is produced with serotype 6C. Primer pair 5106-3101 produces wciN PCR products of 2.0 and 1.8 kb with serotypes 6A and 6C, respectively, whereas no product is produced with serotype 6B. Control strains of serotypes 6A, 6B, and 6C were included in each run; the 6A and 6B control strains were selected from the nationwide surveillance collection, whereas the 6C control was Massachusetts isolate L4097 that had been identified as serotype 6C by In Ho Park and Moon Nahm at the University of Alabama, Birmingham. DNA was extracted from isolate suspensions by heating 10 colonies suspended in 100 μl of water at 95°C for 10 min. PCRs were performed in a total volume of 25 μl consisting of 1 μl of DNA lysate, 12.5 μl of HotStar Taq Plus Mastermix, 2.5 μl of CoralLoad loading dye, 6 μl of RNase-free water (Qiagen, Valencia, CA), 1 pmol of each forward primer (0.5 μl each), 2 pmol (1 μl) of the reverse primer (Integrated DNA Technologies, Coralville, IA), and 1 μl of 25 mM MgCl2 (Promega, Madison, WI). Amplification of PCR products was performed by a denaturation step at 95°C for 5 min and 35 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 2 min, followed by a final extension at 72°C for 10 min in a thermal cycler (Bio-Rad, Hercules, CA). PCR products were detected on 1% agarose gels incorporating 50 μg of ethidium bromide, run for 1 h at 200 V and visualized under UV light. A 1-kb DNA ladder (Promega) was included on each gel.
Typing by PFGE.
Isolates were subcultured onto Trypticase soy sheep blood agar plates and incubated at 35°C in 5% CO2 for no more than 16 h. Growth from plates was suspended in 1 M NaCl-10 mM Tris-HCl (pH 7.6) to achieve the turbidity of a 1.0 McFarland standard. Extraction of genomic DNA in 1% CleanCut agarose plugs (Bio-Rad) was performed by standard methods and included 2 mg of lysozyme/ml for bacterial lysis. DNA was digested with 2 U of the restriction endonuclease SmaI at 25°C. Products were separated by electrophoresis in a GenePath (Bio-Rad) apparatus with pulse times optimized for S. pneumoniae. PulseMarker 30 to 1,000 kb (Sigma, St. Louis, MO) was included in each gel as a molecular weight standard. Pulsed-field gel electrophoresis (PFGE) fingerprint profiles were interpreted according to standard guidelines as indistinguishable (identical fragments), closely related (2 to 3 differences), possibly related (4 to 6 differences), or different (≥7 differences) (37).
Rep-PCR typing.
Genomic DNA was extracted from several colonies of an overnight subculture grown on blood agar plates and purified using an UltraClean microbial DNA isolation kit (MO BIO Laboratories, Carlsbad, CA). PCR amplification of 25 ng of purified DNA was performed with reagents provided in the DiversiLab DNA Enterococcus fingerprinting kit (bioMerieux, Hazelwood, MO) according to the manufacturer's instructions (the Enterococcus kit is recommended for S. pneumoniae) (14). PCR cycling conditions included an initial denaturation at 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 50 C for 30 s, and 70°C for 90 s, with a final extension at 70°C for 3 min. Products were separated in a microfluidics DNA chip (bioMerieux) in an Agilent 2100 BioAnalyzer (Agilent Technologies, Palo Alto, CA) according to the manufacturer's instructions. Repetitive sequence-based PCR (Rep-PCR) fingerprints were compared by using the Pearson correlation coefficient with DiversiLab 3.1 software (bioMerieux) to assess both band position and intensity. Interpretation of fingerprint patterns was based on best fit of the current data to known serotype groupings. Isolates considered “indistinguishable” had a similarity index of ≥97% and no obvious band differences. Strains characterized as “similar” had ≥97% similarity and one band difference. Isolates characterized as “different” had <97% similarity or two or more bands differing.
MLST with detection and analysis of PCR products by ESI-MS.
Multilocus sequence typing (MLST) analysis of S. pneumoniae was carried out based on the gene targets described by Enright and Spratt (10). Briefly, internal fragments of the aroe, gdh, gki, recP, spi, xpt, and ddl genes were amplified by multiplex PCR from chromosomal DNA. The primer pairs used in the present study (Table 1) are internal to the loci described by Enright and Spratt and adapted for detection using PCR-electrospray ionization mass spectrometry (ESI-MS), wherein the PCR products are detected and analyzed by a technique that uses ESI to produce ions from macromolecules that are then separated by MS (12, 28). PCR was performed in 40-μl reaction mixtures consisting of 10× PCR buffer, deoxynucleoside triphosphates, primers, genomic sample, and Taq polymerase (2.4 U per reaction mixture). The reactions were performed in 96-well plates (Bio-Rad) with an Eppendorf thermal cycler (Westbury, NY). The PCR buffer consisted of 4 U of AmpliTaq Gold (Applied Biosystems, Foster City, CA), 1× buffer II (Applied Biosystems), 2.0 mmol/liter MgCl2, 0.4 mol of betaine/liter, and 800 μmol of deoxynucleoside triphosphate mix/liter. The following PCR conditions were used to amplification: 95°C for 10 min, followed by 50 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 30 s. After PCR amplification, wells were rigorously desalted by use of a protocol based on the weak anion-exchange method, and ESI-MS was then performed using an Ibis T5000 Biosensor System (Ibis Biosciences, Carlsbad, CA) (8, 18). Base compositions are derived by the use of an algorithm constrained by Watson and Crick base pairing and acceptable mass error limits. For MLST analysis on the T5000 platform, where the amplicon size is typically less than 150 bp and mass errors are routinely less than 20 ppm, unique base compositions are obtained. Base composition signatures from multiple loci are used to generate the complete signature profile for each input sample. An automated algorithm computes the sequence types (ST) consistent with observations over all of the PCRs run on the input sample. The ST identified are then compared to ST of the strains in the MLST database to determine relationships to previously characterized strains (http://spneumoniae.mlst.net/) (10, 11). The allelic profiles of the isolates are then compared to each other, as well as with other isolates in the pneumococcal MLST database, using software available at the MLST website (http://www.mlst.net). Phylogenetic analysis of ST are performed using the program eBURST, which uses a model of bacterial evolution to produce a cluster of closely related genotypes that are all descended from the founding genotype, which is referred to as a “clonal complex” (http://eburst.mlst.net) (11).
TABLE 1.
Primer pairs used in this study for MLSTa
| Primer pair name | Gene target | MLST sequence coordinatesb | MLST gene coordinatesc | Primer
|
|
|---|---|---|---|---|---|
| Orientationd | Sequence (5′-3′)a | ||||
| BCT3260 | spi | c364415-363943 | 43-184 | F | TGATCCGACCCTAGCGGATGG |
| R | TATGGTGTCGCCAGGCATTCC | ||||
| BCT3262 | ddl | c1492971-1492531 | 126-265 | F | TGCCTTCCGATATGACAGCCG |
| R | TGTAGTCATAAAAGGCAACGTCCTTGAC | ||||
| BCT3263 | ddl | c1492971-1492531 | 231-369 | F | TGAAGTTGTCAAGGACGTTGCCTT |
| R | TGCACGGAAGGCTGTTTCTGC | ||||
| BCT3257 | gdh | 1123551-1124010 | 116-256 | F | TGTGCCTTCTTTGTAGACAGCGATC |
| R | TGGAGCAAGTGGTTCTCCAAAGATAGA | ||||
| BCT3258 | gki | 600427-600909 | 160-300 | F | TTCGCAGAAGGCAAATTGCTTCA |
| R | TGTTGCTGAAGCGACTGTCTCAA | ||||
| BCT3259 | recP | c1817785-1817336 | 16-154 | F | TTAACCGCGACCGCTTTATTCTTTCA |
| R | TGACCTGGTGTTTTTGAACCCCATT | ||||
| BCT3261 | xpt | 1635367-1635850 | 201-317 | F | TGAACATCACCATGAACGAAGGCATC |
| R | TCAAAACCTTGTCCTCTGGTGAGAGG | ||||
| BCT3367 | aroE | c1232155-1231720 | 295-431 | F | TCGGATGGCGTCAGTCAGATTTC |
| R | TGCAGCTCAGAAGCATATTCTAAAGCA | ||||
Two primer pairs are used for the amplification of different regions of ddl.
All coordinates are based on the S. pneumoniae R6 complete genome sequence (GenBank no. NC 003098). MLST sequence coordinates are based on published MLST loci; “c” refers to complementary strand sequence based on the orientation of the reference genome.
Target regions within the MLST loci used in this study.
F, forward; R, reverse.
RESULTS
Isolates previously identified by capsular swelling as serotype 6A, consisting of Cleveland strains collected from 1979 to 2007 (122 of 2,735 [4.5%]), pediatric isolates collected nationwide in 2005 and 2006 (39 of 393 [9.9%]), and 11 Massachusetts strains were tested by multiplex PCR to differentiate serotype 6C from serotype 6A. A total of 30 (24.6%) of the 122 Cleveland isolates, 19 (48.7%) of the 39 nationwide pediatric isolates, and all 11 Massachusetts isolates were shown to be serotype 6C; the remaining isolates were serotype 6A. The serotype 6C isolates originated in blood (5), the lower respiratory tract (27), the paranasal sinus (5), the middle ear (2), the nasopharynx (18), and from miscellaneous or unknown sources (3). The serotype 6A isolates originated in blood (39), the lower respiratory tract (32), cerebrospinal fluid (2), the paranasal sinus (2), the middle ear (2), and miscellaneous or unknown sources (35).
Cleveland strains collected from 1979 to 2007.
Of the 30 serotype 6C isolates recovered, two were from invasive sources collected from 1979 to 1996, and 28 were from all sources from 1997 to 2007 (2 from invasive and 26 from noninvasive sites). The earliest serotype 6C isolate was recovered from blood in 1989, with additional single isolates from this specimen source from 1994, 2003, and 2007. All age groups were represented, with isolates from five patients <2 years old, eight patients 2 to 5 years old, six patients 6 to 18 years old, eight patients 19 to 65 years old, and three patients >65 years old. During the period when isolates from all sources were collected (from 1997 to 2007), the annual incidence increased from zero per year prior to 2002 to two to six per year thereafter, with most isolates recovered from lower respiratory sources (one in 1998, one in 2002, two in 2003, two in 2004, four in 2005, five in 2006, and four in 2007). In contrast, the annual incidence of serotype 6A decreased from 8 to 13 per year in 1998 to 2002 to 4 to 7 per year thereafter.
Pediatric isolates collected nationwide in 2005 and 2006.
The 19 serotype 6C isolates originated from all six major U.S. geographic regions, with fewer isolates from northeast (n = 2), northwest (n = 2), and southwest (n = 1) regions than from north central (n = 6), south central (n = 4), and southeast regions (n = 4). Isolates originated from eight patients <2 years old, four patients 2 to 5 years old, and seven patients 6 to 18 years old. Isolates were recovered from blood (1), the middle ear (2), the lower respiratory tract (8), the nasopharynx (4), and the paranasal sinus (4).
Antimicrobial susceptibility.
Susceptibility results are shown in Table 2. A total of 30 isolates (50%) were susceptible to all agents tested (9 of 11 Massachusetts isolates, 16 of 30 Cleveland isolates, and 5 of 19 regional isolates). Of the 28 penicillin nonsusceptible isolates, 27 were penicillin intermediate. The 28 nonsusceptible strains showed five patterns, with 22 isolates having the most frequent pattern (penicillin nonsusceptible; azithromycin and SXT resistant); these isolates originated from Cleveland (n = 10); the north central, northwestern, south central, and southeastern regions (n = 10, with 2 to 3 per region); and Massachusetts (n = 1). Four isolates were penicillin intermediate and SXT resistant, one was penicillin intermediate and azithromycin resistant, two were SXT resistant, and one was penicillin intermediate.
TABLE 2.
Antimicrobial susceptibilities of the 60 isolates of S. pneumoniae serotype 6C
| Antimicrobial agent | Susceptible breakpoint (μg/ml) | MIC (μg/ml)
|
% Susceptible | ||
|---|---|---|---|---|---|
| Range | MIC50 | MIC90 | |||
| Penicillin | ≤0.06 | ≤0.015-2 | 0.12 | 1 | 50 |
| Amoxicillin | ≤2 | ≤0.015-1 | 0.12 | 1 | 100 |
| Ceftriaxone | ≤1 | ≤0.015-0.5 | 0.12 | 0.25 | 100 |
| Cefuroxime | ≤1 | ≤0.03-4 | 0.5 | 2 | 55 |
| Cefdinir | ≤0.5 | ≤0.03-4 | 0.5 | 2 | 60 |
| Cefpodoxime | ≤0.5 | ≤0.03-2 | 0.12 | 1 | 57 |
| Azithromycin | ≤0.5 | ≤0.03-32 | 0.12 | 8 | 58 |
| Clindamycin | ≤0.25 | ≤0.015-0.06 | 0.03 | 0.06 | 100 |
| Levofloxacin | ≤2 | 1-2 | 1 | 1 | 100 |
| SXTa | ≤0.5 | ≤0.06-≥4 | 1 | >4 | 50 |
MICs are expressed as the trimethoprim component.
PFGE.
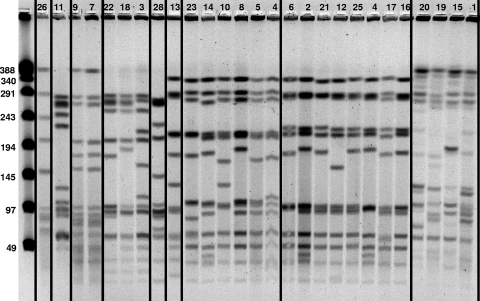
Twenty-five representative isolates were selected for testing by PFGE based on time period, antimicrobial susceptibility patterns, and regional source. Eight indistinguishable, closely related or possibly related PFGE patterns were identified (Fig. 1). Three patterns were unique (i.e., contained single isolates), while other patterns contained two to eight isolates. Within each pattern, only one pattern included strains with identical fragments (strains 7 and 9). The other patterns showed considerable variation within each group of isolates.
FIG. 1.
PFGE of 26 representative strains in lanes 2 to 27, with unrelated strains separated by vertical lines. Strains 1 to 7 are regional isolates, strains 8 to 12 are Massachusetts isolates, and strains 13, 14, and 16 to 26 are Cleveland isolates of serotype 6C. Strain 15 is a serotype 6A control, and strain 28 is a serotype 6B control. A DNA ladder with known fragment sizes is shown in lane 1, with sizes of bands (in kilobases) shown.
Rep-PCR.
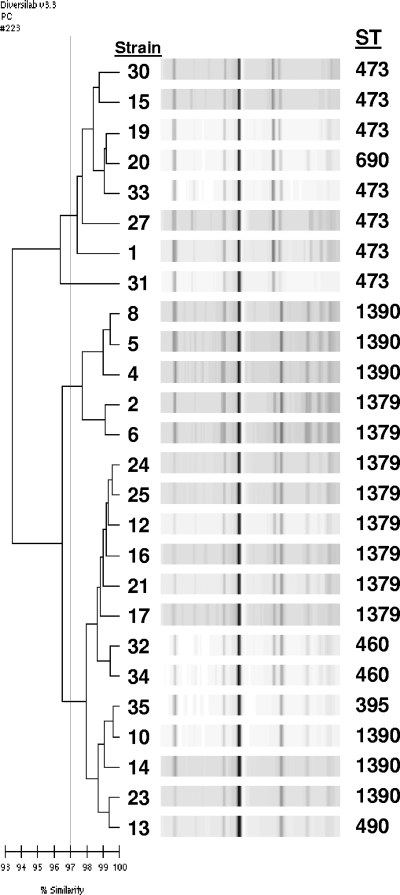
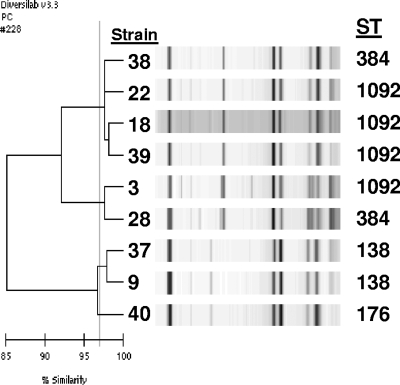
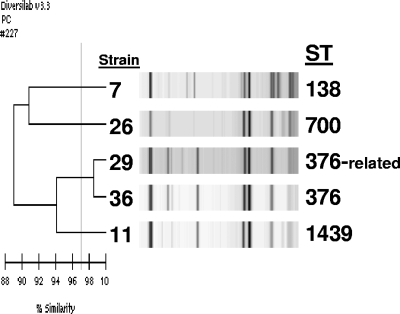
The same 25 serotype 6C isolates tested by PFGE were tested by this method, as well as nine randomly selected serotype 6A and five serotype 6B isolates. Among the 25 serotype 6C isolates, nine groups were identified as “indistinguishable” with a similarity index of ≥97% and no obvious band differences or as “similar” with ≥97% similarity and one band difference. Groups contained 1 to 10 isolates per group, with susceptibility patterns tending to cluster with groups. However, geographic clustering was not noted. The three largest groups contained 10, 6, and 4 isolates, with two of these groups closely related to several serotype 6A isolates (Fig. 2). Three smaller groups were closely related to several serotype 6B isolates (Fig. 3). The remaining serotype 6C isolates were “different” with <97% similarity to each other and to all other isolates studied; however, one of these isolates had 94% similarity to two serotype 6A isolates (Fig. 4).
FIG. 2.
Rep-PCR analysis showing three groups of serotype 6C isolates closely related to serotype 6A isolates. Strains 1, 2, 4 to 6, 8, 10, 12 to 14, 16, 17, 19 to 21, and 23 to 25 are serotype 6C isolates, and strains 15, 27, and 30 to 35 are serotype 6A isolates.
FIG. 3.
Rep-PCR analysis showing three groups of serotype 6C isolates closely related to serotype 6B isolates. Strains 3, 9, 18, and 22 are serotype 6C isolates, and strains 28 and 37 to 40 are serotype 6B isolates.
FIG. 4.
Rep-PCR analysis showing three serotype 6C isolates not closely related to each other or to the other study isolates. Strains 7, 11, and 26 are serotype 6C isolates, and strains 29 and 36 are serotype 6A isolates.
MLST.
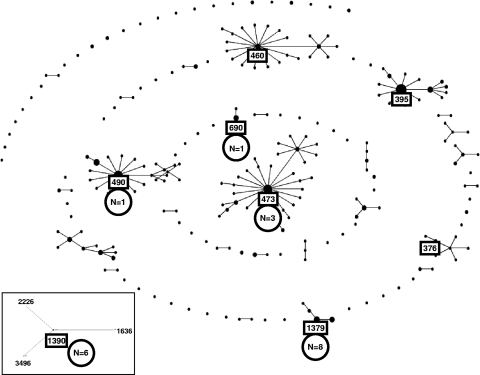
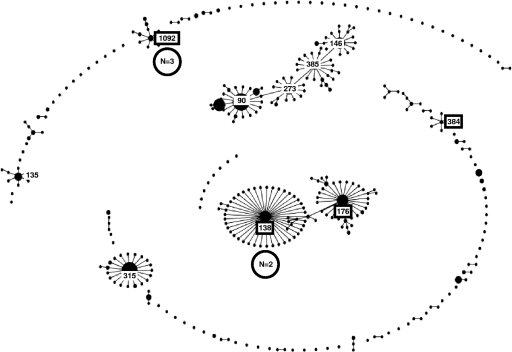
The same 40 isolates typed by Rep-PCR were typed by this method. Among the 25 serotype 6C isolates, nine clusters were identified, with up to 8 isolates per cluster. ST of these clusters were compared to ST patterns present in the MLST database for S. pneumoniae by eBURST analysis; the major clonal complexes associated with serotypes 6A and 6B are shown in Fig. 5 and 6. These comparisons showed that five clusters of 19 serotype 6C isolates were related to clonal complexes associated with serotype 6A, with ST or predicted founder ST being ST473 (n = 3), ST1379 (n = 8), ST490 (n = 1), ST690 (n = 1), and ST1390 (n = 6) (Fig. 5). The ST or predicted founder ST detected for the nine serotype 6A isolates were ST473 (n = 4), ST460 (n = 2), ST395 (n = 1), and ST376 (n = 2). Two clusters of five serotype 6C strains were related to clonal complexes associated with serotype 6B, with the ST or predicted founder ST being ST1092 (n = 3) and ST138 (n = 2) (Fig. 6). The ST or predicted founder ST detected for the five serotype 6B isolates were ST90 (n = 2), ST1092 (n = 1), ST138 (n = 1), and ST176 (n = 1). The last two clusters of serotype 6C strains were single isolates of ST700, an ST associated only with serotype 3 isolates from four countries in Africa, and ST1439, the earliest isolate detected (isolated in 1989), an ST associated only with serotype 34.
FIG. 5.
Clustering by eBURST analysis of the 318 ST of serotype 6A strains included in the MLST database, showing the major clonal complexes associated with this serotype based on a minimum similarity of four identical loci. The ST1390 clonal complex is unrelated to other serogroup 6 clonal complexes and shares three identical loci with strains of serogroups 15, 18, and 21. Clonal complexes shown in boxes are ST or predicted founder ST of the serotype 6A strains detected in the present study. Numbers in circles (N) indicate the number of serotype 6C isolates associated with each clonal complex.
FIG. 6.
Clustering by eBURST analysis of the 379 ST of serotype 6B strains included in the MLST database, based on a minimum similarity of four identical loci, showing the major clonal complexes associated with this serotype. Clonal complexes shown in boxes are ST or predicted founder ST of the serotype 6B strains detected in the present study. Numbers in circles (N) indicate the number of serotype 6C isolates associated with each clonal complex.
Correlation between typing methods.
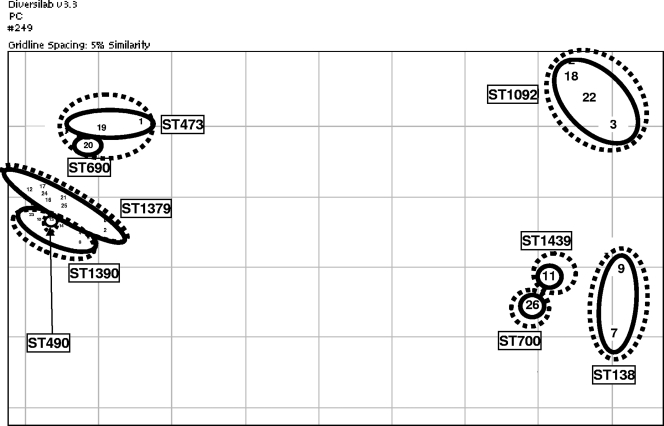
The relationships between serotype, PFGE, Rep-PCR, and MLST findings are shown in Fig. 7. Clustering of serotype 6C isolates identified by the three typing methods were generally similar, although there was some overlap between methods. In addition, six of the nine serotype 6A and all five of the serotype 6B isolates clustered with various serotype 6C clonal complexes, indicating that these are likely to be the founding genotypes of the serotype 6C isolates within these clusters. All seven serotype 6C strains studies that were nonsusceptible to penicillin, macrolides, and SXT clustered together and belonged to ST1379, while three of the four serotype 6C strains nonsusceptible to penicillin and SXT clustered together and belonged to ST1092.
FIG. 7.
Correlation between the three typing methods for the 26 serotype 6C isolates studied based on scatterplot similarity of Rep-PCR results analyzed with the Pearson correlation coefficient method. Strains with the same ST or predicted founder ST are enclosed by solid-line circles or ovals, with corresponding ST indicated. Strains with related PFGE patterns are enclosed by dashed-line circles or ovals. Strains 1 to 7 are regional isolates, strains 8 to 12 are Massachusetts isolates, and strains 13, 14, and 16 to 26 are Cleveland isolates, with isolate 26 being the oldest. ST473, ST490, ST690, ST1379, and ST1390 are ST associated with serotype 6A in the MLST database, while ST138 and ST1092 are associated with serotype 6B, ST700 is associated with serotype 3, and ST1439 is associated with serotype 34.
DISCUSSION
Pneumococci have been demonstrated both to lose capsules entirely and to change the serotypes of their capsules by genetic recombination at the capsular locus. This capability can lead to decreased efficacy of capsular polysaccharide-based vaccines. Since 2003, 37 such “vaccine escape” strains (with reference to serotypes contained in PCV7) have been detected, and a novel vaccine escape recombinant strain that switched serotype from serotype 4 to serotype 19A and simultaneously acquired penicillin nonsusceptibility in a single genetic event has recently been described (2).
Although serotype 6C was only described in 2007, examination of banked S. pneumoniae isolates has shown the presence of this serotype for many years and in many locations. The seminal study of serotype 6C indicated its presence as early as 1980 (34). In our institution the earliest isolate was from 1989, while a study in Uruguay found the new serotype as early as 1995 (29). The prevalence of serotype 6C isolates prior to conjugate vaccine use was low among healthy Dutch children, as well as in South African children with invasive disease (15, 17). A surveillance study in Birmingham, Alabama, covering the years 2002 to 2007, found that serotype 6C accounted for 15% of the vaccine-related serotypes isolated, a proportion equal to that of serotype 6A (4). A Centers for Disease Control study examining previously identified serotype 6A isolates in their Active Bacterial Core surveillance from 1999 to 2006 found an increase in the proportion, over time, of serotype 6C versus serotype 6A, with 16.7% of isolates previously identified as serotype 6A being serotype 6C in 1999, increasing to 61% by 2006 (6). They also found that rates of invasive disease caused by 6C increased significantly, from 0.25 cases per 100,000 population in 1999 to 0.56 cases per 100,000. During the same period, the rates of serotype 6A decreased significantly, from 1.3 to 0.36 cases per 100,000. Breakdown by age of patients showed that serotype 6C isolates increased significantly, from 3% of isolates in 1999 to 75% in 2006 in children <2 years, and from 28 to 68% in adults ≥65 years. MLST of serotype 6A and 6C isolates revealed nine distinct clusters previously associated with serotype 6A, with the majority of 6C isolates sharing three clusters with serotype 6A isolates. The authors of that study concluded that invasive disease due to serotype 6C in the United States may be increasing, while that due to serotype 6A is decreasing. Data from our institution displayed similar trends (22). These studies raise the question of the cross-immunogenicity of serotype 6B polysaccharide included in PCV7 to serotypes 6A and 6C. Several groups have presented findings, based on antibody responses and invasive disease surveillance, suggesting that serotype 6B conjugate vaccine has proved adequately immunogenic against serotype 6A but not against serotype 6C (4, 6, 24, 33). Similarly, inclusion of serotype 19F in PCV7 has not provided cross-immunogenicity to serotype 19A (1, 2, 19, 25, 31, 36). Use of other antigenic targets has been suggested, such as pneumococcal surface protein A (PspA), which comprises five clades in two families, and findings suggest that use of just two of the clades provides broad antibody cross-reactivity to all five clades (7). The diversity of antigenic targets in S. pneumoniae and the ability of this species to transform genetically will continue to challenge control of pneumococcal disease with use of vaccines.
Analysis of ST profiles of serogroup 6 isolates present in the MLST database revealed that there are several major related clonal complexes of serotype 6A, with ST473, ST490, ST1379, and ST395 the major predicted founders, sharing four or more loci (Fig. 5). One serotype 6A clonal complex, with ST1390 the predicted founder, is unrelated to other serogroup 6 strains, and has three common loci with strains of serogroups 15, 18, and 21. The major predicted founders of serotype 6B are ST138, ST176, ST90, ST273, ST385, ST146, ST315, and ST1092 (Fig. 6). A recent study of the genetic relatedness of 43 serotype 6A isolates and 32 serotype 6C isolates by eBURST MLST analysis found nine distinct genetic profiles (called eBURST serogroups) that had previously only been found in serotype 6A (6). The majority of the serotype 6C isolates shared three eBURST serogroups with serotype 6A isolates, as was the case with our isolates. The MLST database currently lists nine strains of serotype 6C, belonging to seven unrelated ST, with predicted founder ST of serotypes 6A, 6B, and 23F.
Our findings confirm many of the relationships discussed above and document additional relationships. Of the nine clusters of serotype 6C isolates detected, five were closely related to serotype 6A and two were closely related to serotype 6B strains, indicating likely capsular switching from serotypes 6A and 6B to serotype 6C. Two other clusters were, surprisingly, related to serotypes 3 and 34, indicating possible capsular switching from these serotypes. Our findings also demonstrate an increasing incidence of serotype 6C over time in Cleveland, as well as widespread distribution of isolates throughout the United States. These isolates were genetically diverse, indicating that they had likely developed over a considerable time period. Furthermore, our studies provide information on the applicability to S. pneumoniae of two typing methods, Rep-PCR and MLST with ESI-MS. The Barcodes Rep-PCR method has only been reported to have been used to type S. pneumoniae on one occasion, with PFGE used for comparison (14). In that study the investigators concluded that Rep-PCR was able to delineate S. pneumoniae with a discriminatory power equal to that of PFGE and suggested that it could be used as a stand-alone method for this species, with the only qualification that subtype groupings were more difficult to define by Rep-PCR. MLST with ESI-MS has been successfully used to type isolates of Streptococcus pyogenes, Acinetobacter baumannii, and Campylobacter species (8, 9, 13, 21, 38).
Our study highlights the presence of serotype 6C throughout the USA and the increasing incidence of this serotype after the introduction of PCV7. Although the clonality of serotype 6C strains was equally well characterized by PFGE, Rep-PCR, and MLST, MLST was the only method that could be used to detect the likely origin of strains. MLST was performed by using a novel detection method, ESI-MS, which avoids the need to perform nucleotide sequencing of amplification products.
Acknowledgments
The Veterans Affairs Merit Review Program, VISN 10 Geriatrics Research, Education, and Clinical Care (GRECC), and the National Institutes of Health (RO1 AI072219) supported the work of R.A.B.
We acknowledge the use of the pneumococcal MLST database, which is located at Imperial College London and is funded by the Wellcome Trust.
Footnotes
Published ahead of print on 29 October 2008.
REFERENCES
- 1.Beall, B., M. C. McEllistrem, R. E. Gertz, Jr., S. Wedel, D. J. Boxrud, A. L. Gonzalez, M. J. Medina, R. Pai, T. A. Thompson, L. H. Harrison, L. McGee, and C. G. Whitney. 2006. Pre- and postvaccination clonal compositions of invasive pneumococcal serotypes for isolates collected in the United States in 1999, 2001, and 2002. J. Clin. Microbiol. 44999-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brueggemann, A. B., R. Pai, D. W. Crook, and B. Beall. 2007. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog. 31628-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing; 18th informational supplement M100-S18. Clinical and Laboratory Standards Institute, Wayne, PA.
- 4.Coats, M., M. Nahm, D. Briles, and M. Crain. 2008. Clinical importance of Streptococcus pneumoniae capsular serotypes related to those in the heptavalent pneumococcal conjugate vaccine (PCV7), abstr. P3-142. Sixth International Symposium on Pneumococci and Pneumococcal Diseases, Reykjavik, Iceland.
- 5.Critchley, I. A., M. R. Jacobs, S. D. Brown, M. M. Traczewski, G. S. Tillotson, and N. Janjic. 2008. Prevalence of serotype 19A Streptococcus pneumoniae among isolates from U.S. children in 2005-2006 and activity of faropenem. Antimicrob. Agents Chemother. 522639-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.da Gloria Carvalho, M., C. Henderson, A. Trujillo, H. H. Joshi, I. H. Park, S. Hollingshead, C. Whitney, M. Nahm, B. Beall, and A. Team. 2008. Emergence of genetically diverse invasive pneumococcal serotype 6C, abstr. S01-O1. Sixth International Symposium on Pneumococci and Pneumococcal Diseases, Reykjavik, Iceland.
- 7.Darrieux, M., A. T. Moreno, D. M. Ferreira, F. C. Pimenta, A. L. de Andrade, A. P. Lopes, L. C. Leite, and E. N. Miyaji. 2008. Recognition of pneumococcal isolates by antisera raised against PspA fragments from different clades. J. Med. Microbiol. 57273-278. [DOI] [PubMed] [Google Scholar]
- 8.Ecker, D. J., R. Sampath, L. B. Blyn, M. W. Eshoo, C. Ivy, J. A. Ecker, B. Libby, V. Samant, K. A. Sannes-Lowery, R. E. Melton, K. Russell, N. Freed, C. Barrozo, J. Wu, K. Rudnick, A. Desai, E. Moradi, D. J. Knize, D. W. Robbins, J. C. Hannis, P. M. Harrell, C. Massire, T. A. Hall, Y. Jiang, R. Ranken, J. J. Drader, N. White, J. A. McNeil, S. T. Crooke, and S. A. Hofstadler. 2005. Rapid identification and strain typing of respiratory pathogens for epidemic surveillance. Proc. Natl. Acad. Sci. USA 1028012-8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ecker, J. A., C. Massire, T. A. Hall, R. Ranken, T. T. Pennella, C. Agasino Ivy, L. B. Blyn, S. A. Hofstadler, T. P. Endy, P. T. Scott, L. Lindler, T. Hamilton, C. Gaddy, K. Snow, M. Pe, J. Fishbain, D. Craft, G. Deye, S. Riddell, E. Milstrey, B. Petruccelli, S. Brisse, V. Harpin, A. Schink, D. J. Ecker, R. Sampath, and M. W. Eshoo. 2006. Identification of Acinetobacter species and genotyping of Acinetobacter baumannii by multilocus PCR and mass spectrometry. J. Clin. Microbiol. 442921-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144(Pt. 11)3049-3060. [DOI] [PubMed] [Google Scholar]
- 11.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 1861518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenn, J. B., M. Mann, C. K. Meng, S. F. Wong, and C. M. Whitehouse. 1989. Electrospray ionization for mass spectrometry of large biomolecules. Science 24664-71. [DOI] [PubMed] [Google Scholar]
- 13.Hannis, J. C., S. M. Manalili, T. A. Hall, R. Ranken, N. White, R. Sampath, L. B. Blyn, D. J. Ecker, R. E. Mandrell, C. K. Fagerquist, A. H. Bates, W. G. Miller, and S. A. Hofstadler. 2008. High-resolution genotyping of Campylobacter species by use of PCR and high-throughput mass spectrometry. J. Clin. Microbiol. 461220-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrington, S. M., F. Stock, A. L. Kominski, J. D. Campbell, J. C. Hormazabal, S. Livio, L. Rao, K. L. Kotloff, S. O. Sow, and P. R. Murray. 2007. Genotypic analysis of invasive Streptococcus pneumoniae from Mali, Africa, by semiautomated repetitive-element PCR and pulsed-field gel electrophoresis. J. Clin. Microbiol. 45707-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hattingh, O. 2008. Detection of serotype 6C among South African serotype 6A pneumococci causing invasive disease, 2005-2006, abstr. P2-002. Sixth International Symposium on Pneumococci and Pneumococcal Diseases, Reykjavik, Iceland.
- 16.Henrichsen, J. 1995. Six newly recognized types of Streptococcus pneumoniae. J. Clin. Microbiol. 332759-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermans, P. W., M. Blommaart, I. H. Park, M. H. Nahm, and D. Bogaert. 2008. Low prevalence of recently discovered pneumococcal serotype 6C isolates among healthy Dutch children in the pre-vaccination era. Vaccine 26449-450. [DOI] [PubMed] [Google Scholar]
- 18.Hofstadler, S., R. Sampath, L. Blyn, M. Eshoo, T. Hall, Y. Jiang, J. J. Drader, J. Hannis, K. Sannes-Lowery, L. Cummins, B. Libby, D. Walcott, A. Schink, C. Massire, R. Ranken, J. Gutierrez, S. Manalili, C. Ivy, R. Melton, H. Levene, G. Barrett-Wilt, F. Li, V. Zapp, N. White, V. Samant, J. McNeil, D. J. Knize, D. Robbins, K. Rudnick, A. Desai, E. Moradi, and D. Ecker. 2005. TIGER: the universal biosensor. Int. J. Mass Spectrom. 24223-41. [Google Scholar]
- 19.Hsu, K. K., J. E. Kellenberg, S. I. Pelton, D. S. Friedman, M. R. Moore, and H. T. Jordan. 2007. Emergence of antimicrobial-resistant serotype 19A Streptococcus pneumoniae-Massachusetts, 2001-2006. MMWR Morb. Mortal. Wkly. Rep. 561077-1080. [PubMed] [Google Scholar]
- 20.Huang, S., V. Hinrichsen, A. Stevenson, S. Rifas-Shiman, K. Kleinman, G. Lee, S. Pelton, W. Hanage, and J. Finkelstein. 2008. Continued impact of pneumococcal conjugate vaccine on serotypes, antibiotic resistance, and risk factors for carriage in young children, abstr. P3-078. Sixth International Symposium on Pneumococci and Pneumococcal Diseases, Reykjavik, Iceland.
- 21.Hujer, K. M., A. M. Hujer, E. A. Hulten, S. Bajaksouzian, J. M. Adams, C. J. Donskey, D. J. Ecker, C. Massire, M. W. Eshoo, R. Sampath, J. M. Thomson, P. N. Rather, D. W. Craft, J. T. Fishbain, A. J. Ewell, M. R. Jacobs, D. L. Paterson, and R. A. Bonomo. 2006. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 504114-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs, M. R., C. E. Good, S. Bajaksouzian, and A. Windau. 2008. Emergence of serotypes 19A, 6C, and 22F and serogroup 15 Streptococcus pneumoniae in Cleveland in relation to introduction of the protein conjugated pneumococcal vaccine. Clin. Infect. Dis. 471388-1395. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs, M. R., C. E. Good, B. Beall, S. Bajaksouzian, A. R. Windau, and C. G. Whitney. 2008. Changes in serotypes and antimicrobial susceptibility of invasive Streptococcus pneumoniae strains in Cleveland: a quarter century of experience. J. Clin. Microbiol. 46982-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, K. H., H. Lee, R. Burton, and M. Nahm. 2008. Immune responses to vaccine-related serotypes, 6A, 6C, and 19A in infants immunized with 7-valent pneumococcal conjugate vaccine, abstr. P3-057. Sixth International Symposium on Pneumococci and Pneumococcal Diseases, Reykjavik, Iceland.
- 25.Kyaw, M. H., R. Lynfield, W. Schaffner, A. S. Craig, J. Hadler, A. Reingold, A. R. Thomas, L. H. Harrison, N. M. Bennett, M. M. Farley, R. R. Facklam, J. H. Jorgensen, J. Besser, E. R. Zell, A. Schuchat, and C. G. Whitney. 2006. Effect of introduction of the pneumococcal conjugate vaccine on drug-resistant Streptococcus pneumoniae. N. Engl. J. Med. 3541455-1463. [DOI] [PubMed] [Google Scholar]
- 26.Lund, E., and J. Henrichsen. 1978. Laboratory diagnosis, serology and epidemiology of Streptococcus pneumoniae. Methods Microbiol. 12244-255. [Google Scholar]
- 27.Mavroidi, A., D. Godoy, D. M. Aanensen, D. A. Robinson, S. K. Hollingshead, and B. G. Spratt. 2004. Evolutionary genetics of the capsular locus of serogroup 6 pneumococci. J. Bacteriol. 1868181-8192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meng, C. K., and J. B. Fenn. 1990. Analyzing organic molecules with electrospray mass spectrometry. Am. Biotechnol. Lab. 854-60. [PubMed] [Google Scholar]
- 29.Nahm, M. H., I. H. Park, T. Camou, and M. Hortal. 2008. Serotype 6C has been present in Uruguay, abstr. P1-063. Sixth International Symposium on Pneumococci and Pneumococcal Diseases, Reykjavik, Iceland.
- 30.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A6. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 31.Pai, R., M. R. Moore, T. Pilishvili, R. E. Gertz, C. G. Whitney, and B. Beall. 2005. Postvaccine genetic structure of Streptococcus pneumoniae serotype 19A from children in the United States. J. Infect. Dis. 1921988-1995. [DOI] [PubMed] [Google Scholar]
- 32.Park, I. H., S. Hollingshead, D. G. Pritchard, R. Cartee, J. Lin, S. Park, A. Brandao, M. C. Brandileone, and M. Nahm. 2008. Discovery of a new pneumococcal serotype 6C with distinct properties: implications for vaccine efficacy, abstr. P2-008. Sixth International Symposium on Pneumococci and Pneumococcal Diseases, Reykjavik, Iceland.
- 33.Park, I. H., M. R. Moore, J. T. Treanor, S. Pelton, T. Pilishvili, B. Beall, M. Shelly, G. Gallagher, B. Mahon, and M. H. Nahm. 2008. Reduction in serotype 6A invasive pneumococcal disease after accounting for effect of serotype 6C, abstr. P1-006. Sixth International Symposium on Pneumococci and Pneumococcal Diseases, Reykjavik, Iceland.
- 34.Park, I. H., S. Park, S. K. Hollingshead, and M. H. Nahm. 2007. Genetic basis for the new pneumococcal serotype, 6C. Infect. Immun. 754482-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park, I. H., D. G. Pritchard, R. Cartee, A. Brandao, M. C. Brandileone, and M. H. Nahm. 2007. Discovery of a new capsular serotype (6C) within serogroup 6 of Streptococcus pneumoniae. J. Clin. Microbiol. 451225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelton, S. I., H. Huot, J. A. Finkelstein, C. J. Bishop, K. K. Hsu, J. Kellenberg, S. S. Huang, R. Goldstein, and W. P. Hanage. 2007. Emergence of 19A as virulent and multidrug-resistant pneumococcus in Massachusetts following universal immunization of infants with pneumococcal conjugate vaccine. Pediatr. Infect. Dis. J. 26468-472. [DOI] [PubMed] [Google Scholar]
- 37.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 332233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wortmann, G., A. Weintrob, M. Barber, P. Scott, S. T. Zoll, M. W. Eshoo, R. Sampath, D. J. Ecker, and C. Massire. 2008. Genotypic evolution of Acinetobacter baumannii strains in an outbreak associated with war trauma. Infect. Control Hosp. Epidemiol. 29553-555. [DOI] [PubMed] [Google Scholar]