Abstract
The recommended breakpoints for the cefoxitin disk diffusion test for Staphylococcus aureus were recently modified. In this large-sample study, cefoxitin sensitivity and specificity compared to those of oxacillin were 97.3% and 100%, respectively. This study validated the new cefoxitin breakpoints for the detection of mecA-mediated resistance in S. aureus.
Staphylococcus aureus is a serious current health care concern. Both community-associated methicillin-resistant S. aureus (MRSA) and health care-associated MRSA are growing threats to the immunocompromised, as well as to the general public. Accurate detection of methicillin resistance in S. aureus is of the utmost importance to ensure effective treatment for the affected patient and to prevent further transmission.
The mecA gene confers resistance to methicillin in S. aureus. The gene is located on the staphylococcal chromosome cassette mec and encodes penicillin binding protein 2a (PBP2a). PBP2a is located in the bacterial cell wall and has a low binding affinity for β-lactams (1, 2). This study evaluated the new Clinical Laboratory Standards Institute (CLSI) breakpoints for the cefoxitin disk test for determining mecA-mediated resistance in S. aureus (4).
CLSI recommends usage of cefoxitin instead of oxacillin when using the disk diffusion method to determine resistance against methicillin for S. aureus (4). Cefoxitin results are easier to interpret and are thus more sensitive for the detection of mecA-mediated resistance than oxacillin results (5, 6, 7, 9-11). The recommended resistance and susceptibility breakpoints for the 30-μg cefoxitin disk test used to detect mecA-mediated resistance in S. aureus were changed in January 2007 by CLSI from ≤19 mm and ≥20 mm to ≤21 mm and ≥22 mm, respectively. This study sampled a large number of recent S. aureus isolates to compare the performance of the cefoxitin disk test at the new breakpoints to that of the 1-μg oxacillin disk test.
Between August and September 2007, a total of 1,611 nonduplicate S. aureus isolates were collected by 53 Wisconsin clinical laboratories (up to 20 consecutive MRSA and methicillin-susceptible S. aureus isolates per laboratory) and submitted to the Wisconsin State Laboratory of Hygiene. Species identification was confirmed by colony morphology, coagulase slide test, subsequent tube test, and biochemicals. The isolation sites of the S. aureus isolates collected were as follows: skin and soft tissue (1,159 isolates; 71.9%), urine (136 isolates; 8.4%), respiratory tract (130 isolates; 8.1%), bloodstream (65 isolates; 4.0%), and other (121 isolates; 7.5%).
Susceptibility to antimicrobial agents for confirmed S. aureus isolates was evaluated by the CLSI disk diffusion method on Mueller-Hinton agar (3). All agar plates and antibiotic disks were obtained from Remel (Lenexa, KS). A direct colony suspension of each S. aureus isolate was prepared to a 0.5 McFarland standard and plated on Mueller-Hinton agar. The zones of inhibition were measured using the Biomic V3 system (Giles Scientific, Inc., Santa Barbara, CA) after 24 h of incubation at 35°C (4). The oxacillin disk was read using transmitted light as the CLSI document recommends. The ATCC 25923 S. aureus strain was included in each test sample set as a quality control. All results were within the acceptable limits for quality control strains according to the 2007 CLSI guidelines.
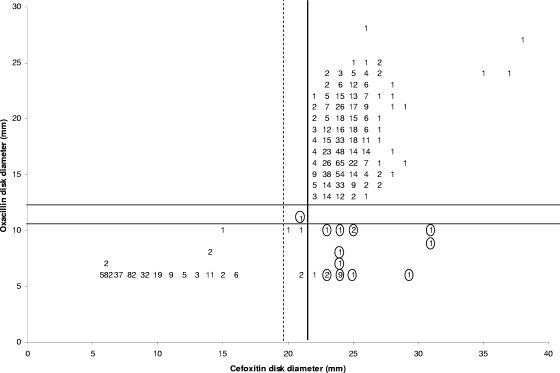
Figure 1 shows the scatterplot correlation of oxacillin (1 μg) disk diameter with the cefoxitin (30 μg) disk diameter. The correlation coefficient (R2) for the scattergram was determined to be 0.8389. The cefoxitin disk demonstrated a sensitivity and specificity of 97.3% and 100%, respectively, compared to the oxacillin disk.
FIG. 1.
Correlation of oxacillin (1 μg) and cefoxitin (30 μg) disk test zone diameters obtained from 1,611 Staphylococcus aureus isolates. A circled number indicates a PBP2a-negative isolate at that position. The dashed line represents the 2006 cefoxitin CLSI interpretive criteria for S. aureus; the solid vertical line represents the new cefoxitin CLSI interpretive criteria. The solid horizontal lines represent the CLSI interpretive criteria for oxacillin.
Discrepant results were repeated and, when necessary, resolved using the PBP2a latex agglutination kit (Denka Seiken Co. Ltd., Tokyo, Japan) in accordance with the manufacturer's instructions. The kit contains latex particles sensitized with a monoclonal antibody against PBP2a. Visible agglutination indicates a positive result and the presence of PBP2a, the mecA gene product. Following discrepancy analysis, the cefoxitin disk test sensitivity increased to 99.9%. Twenty-one of the 22 isolates that tested resistant to oxacillin and susceptible to cefoxitin were found to be negative by PBP2a latex agglutination. Absence of the PBP2a protein is indicative of non-mecA-mediated methicillin resistance. A 1.3% (21/1,611) rate of non-mecA-mediated resistance in S. aureus was measured in this study population. Alternatively, these isolates could represent false-positive resistance to the oxacillin disk test. Eighteen of the 21 PBP2a-negative isolates tested positive for beta-lactamase by nitrocefin testing. It may be possible that some of these isolates are hyper-beta-lactamase producers, thereby accounting for non-mecA-mediated methicillin resistance.
Cefoxitin incorrectly identified only one isolate as susceptible (zone diameter = 22 mm) that was oxacillin resistant and PBP2a positive. An additional isolate that tested intermediate to oxacillin and resistant to cefoxitin was found to be negative for PBP2a. Four of the isolates testing resistant to oxacillin from this study would have been misinterpreted as susceptible using the previous cefoxitin susceptibility breakpoint of ≥20 mm.
Cefoxitin will detect only MRSA with a mecA-mediated resistance mechanism (8). There is a CLSI comment in the M100-S17 document warning of this limitation of cefoxitin as a substitute for oxacillin. However, non-mecA-mediated methicillin resistance in S. aureus is a rare occurrence, as evidenced by this study. Even with this limitation, cefoxitin disk diffusion zones are much easier to read than those of oxacillin due to the frequent hazy oxacillin zones, which are commonly misinterpreted as evidence of oxacillin susceptibility. This rate of false susceptibility associated with the oxacillin disk diffusion test has been noted to be as high as 4.4% in some studies (7; Wisconsin State Laboratory of Hygiene, unpublished data), well above the CLSI-recommended acceptability limit of ≤1.5%. Oxacillin must also be read using transmitted light, unlike most other antimicrobials, including cefoxitin, to ensure correct interpretation.
This large-sample study validated the new CLSI breakpoints for the 30-μg cefoxitin disk test to detect mecA-mediated resistance in S. aureus. We found the cefoxitin disk diffusion test to be a superior test and a surrogate for the oxacillin disk diffusion test due to its ease of reading and higher sensitivity.
Acknowledgments
This work was supported in part by an appointment to the Emerging Infectious Disease Fellowship Program administered by the Association of Public Health Laboratories and funded by the Centers for Disease Control and Prevention (CDC) and by CDC Cooperative Agreement U38 HM000012.
The study contents are solely the responsibility of the authors and do not necessarily represent the official views of the CDC.
Footnotes
Published ahead of print on 19 November 2008.
REFERENCES
- 1.Appelbaum, P. 2007. Microbiology of antibiotic resistance in Staphylococcus aureus. Clin. Infect. Dis. 45S165-S170. [DOI] [PubMed] [Google Scholar]
- 2.Berger-Bachi, B., and S. Rohrer. 2002. Factors influencing methicillin resistance in staphylococci. Arch. Microbiol. 178165-171. [DOI] [PubMed] [Google Scholar]
- 3.CLSI. 2006. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-A9, ninth ed. CLSI, Wayne, PA.
- 4.CLSI. 2007. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. CLSI M100-S17. CLSI, Wayne, PA.
- 5.Felten, A., B. Grandry, P. H. Lagrange, and I. Casin. 2002. Evaluation of three techniques for detection of low-level methicillin-resistant Staphylococcus aureus (MRSA): a disk diffusion method with cefoxitin and moxalactam, the Vitek 2 system, and the MRSA-screen latex agglutination test. J. Clin. Microbiol. 402766-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mimica, M. J., E. N. Berezin, R. L. B. Carvalho, L. M. J. Mimica, M. A. P. Safadi, E. Schneider, and H. H. Caiaffa-Filho. 2007. Detection of methicillin resistance in Staphylococcus aureus isolated from pediatric patients: is the cefoxitin disk diffusion test accurate enough? Braz. J. Infect. Dis. 11415-417. [DOI] [PubMed] [Google Scholar]
- 7.Pottumarthy, S., T. R. Fritsche, and R. N. Jones. 2005. Evaluation of alternative disk diffusion methods for detecting mecA-mediated oxacillin resistance in an international collection of staphylococci: validation report from the SENTRY antimicrobial surveillance program. Diagn. Microbiol. Infect. Dis. 5157-62. [DOI] [PubMed] [Google Scholar]
- 8.Swenson, J. M., D. Lonsway, S. McAllister, A. Thompson, L. Jevitt, W. Zhu, and J. B. Patel. 2007. Detection of mecA-mediated resistance using reference and commercial testing methods in a collection of Staphylococcus aureus expressing borderline oxacillin MICs. Diagn. Microbiol. Infect. Dis. 5833-39. [DOI] [PubMed] [Google Scholar]
- 9.Swenson, J. M., F. C. Tenover, and the Cefoxitin Disk Study Group. 2005. Results of disk diffusion testing with cefoxitin correlate with presence of mecA in Staphylococcus spp. J. Clin. Microbiol. 433818-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velasco, D., M. del Mar Tomas, M. Cartelle, A. Becceiro, A. Perez, F. Molina, R. Moure, R. Villanueva, and G. Bou. 2005. Evaluation of different methods for detecting methicillin (oxacillin) resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 55379-382. [DOI] [PubMed] [Google Scholar]
- 11.Witte, W., B. Pasemann, and C. Cuny. 2007. Detection of low-level oxacillin resistance in mecA-positive Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 13408-412. [DOI] [PubMed] [Google Scholar]