Abstract
Specimen-to-specimen carryover during ThinPrep slide preparation was evaluated by comparing human papillomavirus genotypes detected prior and subsequent to the ThinPrep processing of 121 PreservCyt samples. Overall, 52 samples generated concordant genotypes and 38 had additional and 21 had fewer genotypes postprocessing. PreservCyt samples should be aliquoted for PCR testing prior to ThinPrep processing.
Liquid-based cytology is becoming increasingly utilized across laboratories as an alternative to conventional cytopreparation methodology (Pap smear). Compared to the Pap smear, liquid-based cytology has been associated with improved specimen adequacy and consistency and is a more effective means of detecting atypical cells, cervical cancer, and precursor lesions (2). In addition, the collection of cells in a liquid-based medium enables adjunctive testing of the same specimen for human papillomavirus (HPV), thereby providing the capacity for improved cytological assessment (1, 5). This, in turn, may provide the opportunity for improved triage and management of patients with cervical abnormalities, as well as for determining HPV type-specific persistent infection, which is a significant risk factor for the development of high-grade cervical disease.
One system for preparing a cell monolayer for Pap smear analysis utilizes cervical cells collected in PreservCyt solution (Cytyc Corporation, Boxborough, MA) and the ThinPrep 2000 System (TP2000). PreservCyt solution can be first used to obtain a cell monolayer for cytological analysis and the residual material for molecule-based assays for the detection of nucleic acids from a variety of infectious disease agents, including HPV (4). Increasingly, highly sensitive target amplification assays such as PCR are being used for testing of samples first processed for cytology. However, there are concerns regarding the possibility of vial-to-vial carryover of HPV during sample processing for cytology. Therefore, if sensitive target amplification methods are used for the triage of samples that have already been processed for cytology by the ThinPrep System, there is the possibility of false-positive results.
In this study, 121 consecutively presenting participants enrolled in a cross-sectional study of HPV prevalence in anal ThinPrep specimens collected for anal cytology in two ongoing cohort studies, the Health in Men and positive Health studies of human immunodeficiency virus (HIV)-negative and HIV-positive men who have sex with men (MSM) in Sydney, Australia, were utilized. The study was approved by the human research ethics committee of the University of New South Wales.
Anal samples were collected by a trained nurse using a sterile swab moistened with saline. The swab was inserted 3 to 5 cm into the anus, rotated for 1 min, and then slowly removed and rinsed into ThinPrep vials containing 20 ml PreservCyt fixative solution (Cytyc). One milliliter of each collected PreservCyt sample was aliquoted with an individual sterile pipette and sent to the molecular microbiology laboratory of the Royal Women's Hospital, Melbourne, Australia, and the remainder was sent to the routine cytology laboratory for preparation of ThinPrep slides. The slides were prepared with a TP2000 (Cytyc) instrument and a dedicated filter for each sample, as recommended by the manufacturer. The residual PreservCyt sample was then forwarded to the Royal Women's Hospital for postprocessing sample evaluation.
Testing to detect and determine HPV genotype profiles was performed with the Roche LINEAR ARRAY (LA) HPV genotyping test (Roche Molecular Systems, Alameda, CA) on aliquots collected prior and subsequent to cytology processing.
Cellular and viral DNA was extracted from PreservCyt specimens with the automated MagNA Pure LC (MP) isolation and purification system (Roche Molecular Systems) by a modified protocol (8). Briefly, 1-ml aliquots of PreservCyt specimens were centrifuged at 13,000 × g for 20 min prior to supernatant discarding and resuspension of cell pellets in 200 μl of sterile phosphate-buffered saline, followed by extracted by MP with the DNA-I isolation kit into a final volume of 100 μl.
The LA HPV genotyping test included PCR amplification of target DNA, followed by hybridization with a reverse line blot system for the simultaneous detection of up to 37 anogenital HPV genotypes (i.e., genotypes 6, 11, 16, 18, 26, 31, 33, 35, 39, 40, 42, 45, 51, 52, 53, 54, 55, 56, 58, 59, 61, 62, 64, 66, 67, 68, 69, 70, 71, 72, 73, 81, 82, 83, 84, IS39, and CP6108) (11). The LA HPV genotyping test amplifies a 450-bp region within the HPV L1 gene by using a pool of biotinylated PGMY primers. It also incorporates the amplification of a region within the β-globin gene as an internal control. PCR was performed with a reaction volume of 100 μl and 50 μl of MP-extracted DNA (equivalent to 500 μl of the original specimen) (8). Denatured PCR amplicons were hybridized and detected by the recommended LA HPV genotyping protocol, with the modification of using an orbital microplate shaker and a dry-air incubator at 53°C in place of the shaking water bath (7, 10).
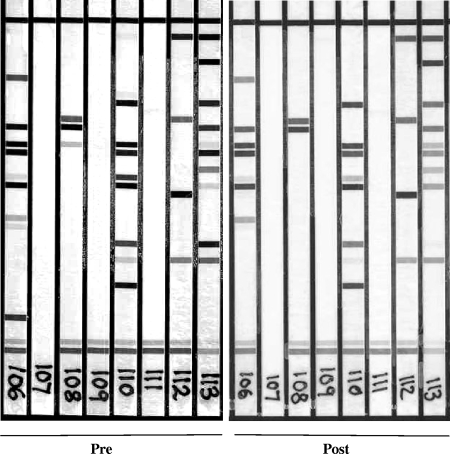
The LA HPV genotyping test strips were manually interpreted by using the HPV reference guide provided and classified high-risk (HR) positive if at least 1 of 13 HR-HPV genotypes (i.e., 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, or 68) was detected. HPV genotype differences were identified by comparing the bands detected on the LA from pre- and postsampling reactions, as shown in a representative group of eight samples (Fig. 1).
FIG. 1.
Representative cohort of samples showing the differences seen with the banding pattern observed pre- and post-ThinPrep processing.
Of the 242 pre- and post-TP2000 processed samples, 5 were negative for β-globin. Three of these five samples tested negative for β-globin prior, but not subsequent, to TP2000 processing, with another sample negative for β-globin both pre- and postprocessing. The three samples with discordant β-globin results were also negative for any HPV genotype prior to ThinPrep processing; however, all became β-globin positive and HPV positive subsequent to processing.
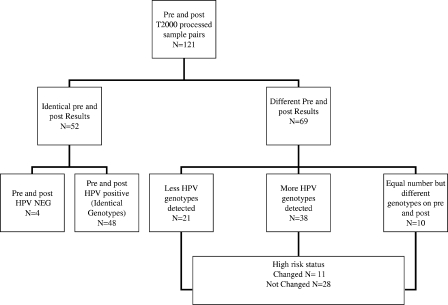
Of 121 sample pairs, 52 (43%) had concordant results, with 4 sample pairs negative for HPV both pre- and postprocessing and 48 samples having the same HPV genotype profile pre- and postprocessing (Fig. 2). Sixty-nine (57%) sample pairs had different results pre- and post-ThinPrep processing, with the majority (38/69 [55%]) identified as having an increased number of HPV genotypes following processing. No preference for particular HPV genotypes was seen. Subsequent to processing, 21 sample pairs had fewer HPV genotypes, while 10 pairs were identified with the same number of genotypes, though with a different HPV genotype composition. Changes in HR-HPV types were detected among 39 sample pairs, with nine patient results changing from HR-HPV negative to positive and two samples changing from HR-HPV positive to negative.
FIG. 2.
Overall HPV genotyping results from pre- and post-TP2000 processed samples.
In recent years, utilization of liquid-based cytology as an alternative method to conventional Pap smear preparation has become more broadly applied. This method has been shown to produce a single cell layer which not only allows for better viewing of cell morphology but can also accommodate the utilization of the residual cells within the vial for adjunct testing such as detection of HPV.
Most laboratories utilizing a TP2000 processor to prepare slides would process multiple samples during each working shift. Although the filters used are single use, the filter cap is not changed and the technician preparing the slides does not routinely change gloves between samples. As a result, there is a possibility of transfer of residual material from a previously processed sample to the next samples. With highly sensitive amplification methods being utilized commonly in laboratories, such cross-contamination may cause false positivity and may therefore have implications for treatment regimens or patient follow-up.
MSM are at a greatly increased risk of HPV-related anogenital disease, including anal dysplasia, anal cancer, and anogenital warts. In the largest study to date, the U.S. EXPLORE Study, the prevalence of anal HPV infection in HIV-negative MSM was 57% by PCR. In addition, within this present study population, the prevalence of HR genotypes was 26%, with 45% of the participants infected with multiple HPV types (data not shown). Therefore, anal samples from MSM populations provide an ideal cohort, with a high prevalence of HPV genotypes for the evaluation of cross-contamination during the preparation of slides for cytology.
Our data demonstrate that such cross-contamination can occur during slide preparation on the TP2000 processor; additional or different HPV genotypes were detected in the post- versus pre-ThinPrep samples in as many as 40% of the samples. Among these, 11 samples would have had a result of HR HPV presence changed, which incidentally may have altered the management of those patients had HPV triage been utilized. Assay reproducibility was not assessed in this study; however, previous studies have demonstrated excellent reproducibility for the detection of HPV genotypes by LA HPV genotyping (6-9) and would not attribute the observed changes to assay variability.
In addition, this study also showed that in 21 of 121 samples, fewer HPV genotypes were detected, which suggests that cells infected with such HPV genotypes (i.e., no difference between low-risk and HR types were seen) were depleted during preparation of the ThinPrep slide and hence not present in the aliquot taken for HPV genotyping. The potential for samples with a low viral load to generate false-negative outcomes has been demonstrated when tested utilizing hybrid capture. In a recent study, it was shown that samples with small lesions could lead to a lower number of abnormal cells collected (3). Comparing the difference between detection of additional types (31.4%; n = 38) and loss of types (17.4%; n = 21) with the two-tailed Fisher exact test showed a statistically significant difference (P = 0.016) between the detection of additional types and the loss of types.
This study demonstrates that HPV specimen depletion and carryover can occur during slide preparation on the TP2000 processor, resulting in the generation of false-negative and/or -positive results, particularly if the residual material is tested with a target amplification test. In order to avoid such false results, we recommend that cytology laboratories remove a 1-ml aliquot in a mode that does not contaminate the sample and prior to preparing the ThinPrep slide. This will allow for the aliquot to be tested for HPV should the need arise. Alternatively, steps could be implemented in order to reduce the rate of false positivity, such as changing gloves and decontaminating the filter cap between preparations. A separate study is needed to evaluate the effectiveness of such methods in reducing specimen carryover.
Footnotes
Published ahead of print on 12 November 2008.
REFERENCES
- 1.Atypical Squamous Cells of Undetermined Significance/Low-Grade Squamous Intraepithelial Lesions Triage Study (ALTS) Group. 2000. Human papillomavirus testing for triage of women with cytologic evidence of low-grade squamous intraepithelial lesions: baseline data from a randomized trial. J. Natl. Cancer Inst. 92397-402. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein, S. J., L. Sanchez-Ramos, and B. Ndubisi. 2001. Liquid-based cervical cytologic smear study and conventional Papanicolaou smears: a metaanalysis of prospective studies comparing cytologic diagnosis and sample adequacy. Am. J. Obstet. Gynecol. 185308-317. [DOI] [PubMed] [Google Scholar]
- 3.Eltoum, I. A., D. C. Chhieng, D. R. Crowe, J. Robertson, G. Jin, and T. R. Broker. 2007. Significance and possible causes of false-negative results of reflex human papillomavirus infection testing. Cancer 11154-159. [DOI] [PubMed] [Google Scholar]
- 4.Lentrichia, B. B., S. S. Hecht, D. Lapen, and M. K. Corkill. 1998. Potential for routine concurrent determination of chlamydia and cervical abnormalities by single fluid-based sampling. Prim. Care Update Ob. Gyns. 5149-150. [DOI] [PubMed] [Google Scholar]
- 5.Solomon, D., M. Schiffman, and R. Tarone. 2001. Comparison of three management strategies for patients with atypical squamous cells of undetermined significance: baseline results from a randomized trial. J. Natl. Cancer Inst. 93293-299. [DOI] [PubMed] [Google Scholar]
- 6.Steinau, M., D. C. Swan, and E. R. Unger. 2008. Type-specific reproducibility of the Roche linear array HPV genotyping test. J. Clin. Virol. 42412-414. [DOI] [PubMed] [Google Scholar]
- 7.Stevens, M. P., S. M. Garland, and S. N. Tabrizi. 2006. Human papillomavirus genotyping using a modified linear array detection protocol. J. Virol. Methods 135124-126. [DOI] [PubMed] [Google Scholar]
- 8.Stevens, M. P., E. Rudland, S. M. Garland, and S. N. Tabrizi. 2006. Assessment of MagNA Pure LC extraction system for detection of human papillomavirus (HPV) DNA in PreservCyt samples by the Roche AMPLICOR and LINEAR ARRAY HPV tests. J. Clin. Microbiol. 442428-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevens, M. P., S. M. Garland, and S. N. Tabrizi. 2008. Validation of an automated detection platform for use with the Roche linear array human papillomavirus genotyping test. J. Clin. Microbiol. 463813-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabrizi, S. N., M. Stevens, S. Chen, E. Rudland, J. R. Kornegay, and S. M. Garland. 2005. Evaluation of a modified reverse line blot assay for detection and typing of human papillomavirus. Am. J. Clin. Pathol. 123896-899. [DOI] [PubMed] [Google Scholar]
- 11.van Hamont, D., M. A. van Ham, J. M. Bakkers, L. F. Massuger, and W. J. Melchers. 2006. Evaluation of the SPF10-INNO LiPA human papillomavirus (HPV) genotyping test and the Roche linear array HPV genotyping test. J. Clin. Microbiol. 443122-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]