Abstract
The Gen-Probe Aptima human immunodeficiency virus type 1 (HIV-1) RNA assay was adapted for the diagnosis of HIV infection in infants by using dried blood spots. The assay was 99% sensitive (128/129) and 100% specific (162/162). This may prove useful in resource-limited settings, since it precludes the need for a phlebotomist and maintenance of a cold chain.
Diagnosis of human immunodeficiency virus type 1 (HIV-1)-infected infants is essential for the evaluation of interventions for the prevention of this transmission and for identifying infants for initiation of therapy. Nucleic acid detection methods are necessary for infant diagnosis due to the presence of maternal antibodies in the babies for up to 18 months after birth. Dried blood spots (DBS) are an easy way to collect and ship specimens for diagnostic testing of HIV and preclude the necessity of a phlebotomist and maintenance of the cold chain (2, 7). However, the limited volume of specimen (50 μl) dried on filter paper decreases the sensitivity of any assay. Current diagnostic methods that have been used on DBS include DNA PCR (3, 14), the ultrasensitive p24 antigen assay (12), and viral load (VL) testing (1, 4, 8). The lower limits of detection in these RNA and DNA PCR detection methods were approximately 4,000 copies (cp)/ml (1, 4, 8, 14), likely due to the small sample size of DBS.
The Gen-Probe Aptima HIV-1 RNA qualitative assay is extremely sensitive for the detection of HIV in plasma (5). This assay has been evaluated recently as being a more sensitive screening tool for HIV-positive samples from a sexually transmitted disease clinic than typical antibody testing followed by pooled RNA testing (13). In addition, preliminary results from our laboratory suggest that plasma specimens with VLs of ≥20 cp/ml are detectable. Therefore, we evaluated this assay for use with DBS from infants and children as a rapid and less-expensive alternative to other assays for the diagnosis of HIV-1 infection in infants.
We formulated an elution buffer for the removal of blood containing HIV from DBS (1 mM EDTA, 1 mM EGTA, 3% lithium lauryl sulfate in phosphate-buffered saline). Optimization of elution resulted in the following protocol: two 6-mm circles were punched from each card and placed in a 2-ml screw-top tube. The hole punch was cleaned between cards by punching a clean Whatman 903 card twice. Elution buffer (525 μl) was added to each tube, and the specimens were rocked for 2 hours at room temperature. The specimens were spun down for 30 s at 10,000 rpm in a microcentrifuge, and the filter paper was removed from the tubes using disposable wooden applicator sticks. The Aptima HIV-1 RNA qualitative assay was performed according to the manufacturer's instructions, using 500 μl of the eluate.
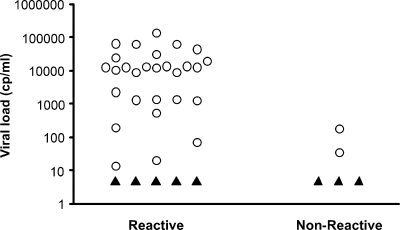
Initial experiments with DBS made from whole blood spiked with known quantities of HIV-1 indicated that our limit of detection was near 400 cp/ml, which seemed appropriate given the small sample volume represented by two punches from a single spot (approximately 10 μl plasma). However, once we began testing DBS collected from adults with low VLs, the detection limit for the Aptima assay fell to 20 to 200 cp/ml (Fig. 1). The lower detection limit with patient DBS, compared to the spiked level of DBS, may be explained by the contribution of cellular HIV RNA and possibly proviral DNA from infected cells in the blood, since all nucleic acid is included in the Aptima assay (a lower limit of 4 cp/reaction was achieved by Pasternak et al. [9] by using peripheral blood mononuclear cells). The real-time reverse transcription-PCR assay developed by Ou et al. (8) also involved isolation of total nucleic acid, which is likely why that assay was also very sensitive (a lower limit of approximately 8 cp/reaction or 4,000 cp/ml). Cellular RNA is not measured in plasma testing, so the level of RNA may be higher in whole blood due to the infected cells.
FIG. 1.
Aptima results (reactive or nonreactive) from 39 DBS collected from adults. VLs were determined using the Roche Monitor v1.5 RNA assay (using either the standard or ultrasensitive method) on plasma from the same blood collection that was used for the DBS. Open circles, determined VLs; filled triangles, VLs under the lower detection limit (50 cp/ml).
In all, we tested 291 DBS from infants and children born to HIV-1-infected mothers (Table 1). The samples had been collected as part of the prevention of mother-to-child transmission studies in South Africa (subtype C) (10-12); Malawi (subtype C); Tanzania (HIVNET 024; subtypes A, C, and D) (6), Vietnam (CRF 01_AE); Dominican Republic, Haiti, and Trinidad (subtype B); and North Carolina (subtype B). The spots had been made from 50 μl whole blood per spot on Whatman 903 cards with blood drawn by heel stick. The samples were dried at room temperature, sealed individually in plastic bags with desiccant pouches, and stored at 4°C (for Vietnam and North Carolina infants) or room temperature prior to testing. The spots were stored in our laboratory for between <1 year (North Carolina) and 3 to 5 years (HIVNET 024) prior to testing for this study. HIV status for the samples had originally been determined by NucliSens HIV-1 QT manual VL assay (bioMérieux; DBS from Dominican Republic, Haiti, Trinidad, Malawi, and Tanzania), DNA PCR assay (Roche Amplicor v1.5; DBS from South Africa), or an in-house HIV assay (DBS from Vietnam). All samples had been collected with appropriate Institutional Review Board approval for each country, and the UNC IRB also approved the study.
TABLE 1.
Sources, subtypes, and Aptima results for infant DBS
| Site of collection | Presumed subtype(s) | Initial testing method | No. of Aptima-R samples/no. of HIV-positive samples (%)a | No. of Aptima-NR samples/no. of HIV-negative samples (%)a |
|---|---|---|---|---|
| North Carolina | B | DNA PCR | 2/2 | 74/74 |
| Haiti | B | DNA PCR | 2/2 | 0/0 |
| Trinidad | B | DNA PCR | 4/4 | 0/0 |
| Dominican Republic | B | DNA PCR/NucliSens | 18/18 | 8/8 |
| South Africab | C | DNA PCR | 47/47 | 32/32 |
| Malawi | C | NucliSens | 26/26 | 10/10 |
| Tanzania (HIVNET 024)c | A, C, D | Roche Monitor v1.5 (standard) | 25/26d | 28/28 |
| Vietnam | CRF01_AE | DNA PCR | 4/4 | 10/10 |
| Total | 128/129 (99.2) | 162/162 (100) |
The Aptima assay was very sensitive for infant diagnosis using DBS. One hundred twenty-nine of the 291 samples had previously tested positive, and 128 of the 129 were reactive on the Aptima assay (99.2% sensitivity) (Table 1). The one false-negative infant specimen had been stored for 4 years at room temperature and had the lowest VL of the HIVNET 024 samples tested (10,954 cp/ml in May 2003). We have shown in a separate study that these DBS lost an average of 1 log10 in VL over this length of time (J. A. E. Nelson, A. M. Loftis, D. Kamwendo, W. W. Fawzi, T. E. Taha, R. L. Goldenberg, and S. A. Fiscus, submitted for publication). The 162 samples that had previously tested negative were all nonreactive on the Aptima assay (100% specificity) (Table 1).
The results of the present study indicate that DBS can be used in the Gen-Probe Aptima HIV-1 RNA qualitative assay for infant diagnosis without the loss of sensitivity or specificity achieved by currently used methods. In our laboratory, we have found that the Aptima assay is less expensive than the NucliSens HIV-1 QT manual assay (bioMérieux, Inc.) for DBS, with increased sensitivity and less labor involved, making it a good choice for infant diagnosis from DBS. In addition, this very sensitive assay would be ideal for the diagnosis of acute HIV infection via DBS.
Acknowledgments
Overall support for the International Maternal Pediatric Adolescent AIDS Clinical Trials Group (IMPAACT) was provided by the National Institute of Allergy and Infectious Diseases (U01 AI068632), the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the National Institute of Mental Health, and the NICHD International and Domestic Pediatric and Maternal HIV Clinical Trials Network (N01-DK-9-001/HHSN2672008000001C). This work was also supported by the UNC Center for AIDS Research (P30 AI50410).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
DBS for this study were provided by Melissa Miller, Gayle Sherman, Charles van der Horst, Annette Sohn, and Consuela Beck-Sague. Technical assistance was provided by Takesha McMillion and Mark Turner.
Footnotes
Published ahead of print on 12 November 2008.
REFERENCES
- 1.Brambilla, D., C. Jennings, G. Aldrovandi, J. Bremer, A. M. Comeau, S. A. Cassol, R. Dickover, J. B. Jackson, J. Pitt, J. L. Sullivan, A. Butcher, L. Grosso, P. Reichelderfer, and S. A. Fiscus. 2003. Multicenter evaluation of use of dried blood and plasma spot specimens in quantitative assays for human immunodeficiency virus RNA: measurement, precision, and RNA stability. J. Clin. Microbiol. 411888-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cassol, S., T. Salas, M. J. Gill, M. Montpetit, J. Rudnik, C. T. Sy, and M. V. O'Shaughnessy. 1992. Stability of dried blood spot specimens for detection of human immunodeficiency virus DNA by polymerase chain reaction. J. Clin. Microbiol. 303039-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Creek, T., A. Tanuri, M. Smith, K. Seipone, M. Smit, K. Legwaila, C. Motswere, M. Maruping, T. Nkoane, R. Ntumy, E. Bile, M. Mine, L. Lu, G. Tebele, L. Mazhani, M. K. Davis, T. H. Roels, P. H. Kilmarx, and N. Shaffer. 2008. Early diagnosis of human immunodeficiency virus in infants using polymerase chain reaction on dried blood spots in Botswana's national program for prevention of mother-to-child transmission. Pediatr. Infect. Dis. J. 2722-26. [DOI] [PubMed] [Google Scholar]
- 4.Fiscus, S. A., D. Brambilla, L. Grosso, J. Schock, and M. Cronin. 1998. Quantitation of human immunodeficiency virus type 1 RNA in plasma by using blood dried on filter paper. J. Clin. Microbiol. 36258-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giachetti, C., J. M. Linnen, D. P. Kolk, J. Dockter, K. Gillotte-Taylor, M. Park, M. Ho-Sing-Loy, M. K. McCormick, L. T. Mimms, and S. H. McDonough. 2002. Highly sensitive multiplex assay for detection of human immunodeficiency virus type 1 and hepatitis C virus RNA. J. Clin. Microbiol. 402408-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldenberg, R. L., A. Mwatha, J. S. Read, S. Adeniyi-Jones, M. Sinkala, G. Msmanga, F. Martinson, I. Hoffman, W. Fawzi, M. Valentine, L. Emel, E. Brown, V. Mudenda, and T. E. Taha. 2006. The HPTN 024 Study: the efficacy of antibiotics to prevent chorioamnionitis and preterm birth. Am. J. Obstet. Gynecol. 194650-661. [DOI] [PubMed] [Google Scholar]
- 7.Mei, J. V., W. H. Hannon, T. L. Dobbs, C. J. Bell, C. Spruill, and M. Gwinn. 1998. Radioimmunoassay for monitoring zidovudine in dried blood spot specimens. Clin. Chem. 44281-286. [PubMed] [Google Scholar]
- 8.Ou, C. Y., H. Yang, S. Balinandi, S. Sawadogo, V. Shanmugam, P. M. Tih, C. Adje-Toure, S. Tancho, L. K. Ya, M. Bulterys, R. Downing, and J. N. Nkengasong. 2007. Identification of HIV-1 infected infants and young children using real-time RT PCR and dried blood spots from Uganda and Cameroon. J. Virol. Methods 144109-114. [DOI] [PubMed] [Google Scholar]
- 9.Pasternak, A. O., K. W. Adema, M. Bakker, S. Jurriaans, B. Berkhout, M. Cornelissen, and V. V. Lukashov. 2008. Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. J. Clin. Microbiol. 462206-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patton, J. C., E. Akkers, A. H. Coovadia, T. M. Meyers, W. S. Stevens, and G. G. Sherman. 2007. Evaluation of dried whole blood spots obtained by heel or finger stick as an alternative to venous blood for diagnosis of human immunodeficiency virus type 1 infection in vertically exposed infants in the routine diagnostic laboratory. Clin. Vaccine Immunol. 14201-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patton, J. C., A. H. Coovadia, T. M. Meyers, and G. G. Sherman. 2008. Evaluation of the ultrasensitive human immunodeficiency virus type 1 (HIV-1) p24 antigen assay performed on dried blood spots for diagnosis of HIV-1 infection in infants. Clin. Vaccine Immunol. 15388-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patton, J. C., G. G. Sherman, A. H. Coovadia, W. S. Stevens, and T. M. Meyers. 2006. Ultrasensitive human immunodeficiency virus type 1 p24 antigen assay modified for use on dried whole-blood spots as a reliable, affordable test for infant diagnosis. Clin. Vaccine Immunol. 13152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren, A., B. Louie, L. Rauch, L. Castro, S. Liska, J. D. Klausner, and M. W. Pandori. 2008. Screening and confirmation of human immunodeficiency virus type 1 infection solely by detection of RNA. J. Med. Microbiol. 571228-1233. [DOI] [PubMed] [Google Scholar]
- 14.Zhang, Q., L. Wang, Y. Jiang, L. Fang, P. Pan, S. Gong, J. Yao, Y. W. Tang, S. H. Vermund, and Y. Jia. 2008. Early infant human immunodeficiency virus type 1 detection suitable for resource-limited settings with multiple circulating subtypes by use of nested three-monoplex DNA PCR and dried blood spots. J. Clin. Microbiol. 46721-726. [DOI] [PMC free article] [PubMed] [Google Scholar]