Abstract
Adeno-associated virus (AAV) replication and biology have been extensively studied using cell culture systems, but there is precious little known about AAV biology in natural hosts. As part of our ongoing interest in the in vivo biology of AAV, we previously described the existence of extrachromosomal proviral AAV genomes in human tissues. In the current work, we describe the molecular structure of infectious DNA clones derived directly from these tissues. Sequence-specific linear rolling-circle amplification was utilized to isolate clones of native circular AAV DNA. Several molecular clones containing unit-length viral genomes directed the production of infectious wild-type AAV upon DNA transfection in the presence of adenovirus help. DNA sequence analysis of the molecular clones revealed the ubiquitous presence of a double-D inverted terminal repeat (ITR) structure, which implied a mechanism by which the virus is able to maintain ITR sequence continuity and persist in the absence of host chromosome integration. These data suggest that the natural life cycle of AAV, unlike that of retroviruses, might not have genome integration as an obligatory component.
Since its initial description over 40 years ago (1, 2, 15), adeno-associated virus (AAV) has emerged as a unique member of the parvovirus family. AAV is replication incompetent and requires a helper virus (e.g., adenovirus) to produce new infectious virions (1, 6, 17, 25, 33, 34). In the absence of helper virus, the hallmark of AAV infection in cultured cells is site-specific integration of the viral genome into a specific locus (AAVS1) on human chromosome 19 (20, 21, 36). Virally encoded gene products, through the recognition and binding of similar viral and cellular sequences, mediate this unique site specificity. The integrated head-to-tail AAV DNA arrays contain rearranged viral inverted terminal repeats (ITRs) and flanking cellular sequences (22, 25, 43). Helper virus infection of cells harboring integrated AAV DNA results in rescue of the AAV genome, leading to the production of new infectious particles. This unique property of site-specific integration has been well documented in transformed cultured cells, but to date, there is only a single report of AAVS1 site-specific integration in two human testis tissue samples (24). We have not observed AAVS1 integration using a sensitive nested PCR assay in 54 AAV-positive human (38) and nonhuman (unpublished observations) primate nontesticular tissue samples.
While ubiquitous in nature, AAV has never been associated with any disease or pathological condition in humans (3-5, 13), and consequently, there have been relatively few studies of in vivo AAV biology. Most observations concerning AAV biology have utilized cultured cells, and these findings have been widely accepted as the model for natural infection in the primate host. The continued development of recombinant AAV vectors for human gene transfer has brought renewed interest in wild-type AAV infection in the natural host. A better understanding of the biology of natural infection will undoubtedly affect the design and use of recombinant AAV vectors in humans.
We recently characterized AAV genomes derived directly from human tissue samples (7). Most of the genomes were closely related to AAV serotype 2; a single isolate shared homology with both serotypes 2 and 3. Moreover, the majority of the predicted amino acid substitutions (relative to AAV2) were conserved among the individual samples, suggesting that a specific virus isolate was circulating in the local population during the time period of tissue sample procurement (winter 2002 to 2003). Interestingly, none of the AAV2 capsid sequences retained arginine residues at positions 585 and 588 (R585S and R588T substitutions), which have been shown to be critical for heparin sulfate proteoglycan receptor binding (18, 26). Consistent with this observation, we showed that two infectious AAV preparations generated using methods detailed herein failed to bind to heparin sulfate chromatography columns (7). These data suggest that this receptor is not required for natural infection in humans, at least for the viruses that we isolated for these studies (7). In contrast to the prevailing wisdom that AAV genomes integrate in humans, analyses of the molecular forms of the AAV DNA in these tissues revealed that the genome sequences in these tissues were extrachromosomal episomes (38). In fact, we did not identify any integration events at the AAVS1 locus, as judged by the failure to amplify AAV-AAVS1 junction fragments by using a sensitive nested-PCR-based assay.
In the present study, we set out to characterize extrachromosomal AAV genomes found in human tissues. In doing so, we developed a novel sequence-specific linear rolling-circle amplification method (SSLRCA) that amplified the extrachromosomal AAV genomes directly from tissue DNA. The resulting circular monomeric AAV genomes were shown to be biologically active when directly transfected into HeLa cells. Furthermore, the resulting clones gave rise to infectious virus, and several isolates were successfully passaged in cultured cells. Sequence analyses of infectious molecular clones revealed intact rep and cap open reading frames (ORFs), as well as a complete double-D ITR element. Conversely, analyses of noninfectious clones suggested that the deletion of internal ITR regions was responsible for the lack of infectivity. To our knowledge, these clones represent the first direct isolation of intact, full-length infectious DNA representing wild-type AAV.
MATERIALS AND METHODS
Cells and viruses.
HeLa cells were purchased from the American Type Culture Collection (Rockville, MD) and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and penicillin and streptomycin. Wild-type adenovirus type 5 (Ad5) was grown, and its titers were determined, as described previously (9). Ad5 at a multiplicity of infection (MOI) of 20 was used in all assays involving helper virus.
Human tissues.
All tissue samples were acquired after approval from the Nationwide Children's Hospital Institutional Review Board, and where required, informed consent was obtained. Fresh human tonsil and adenoid specimens (n = 101) were collected from children aged 2 to 13 years at Nationwide Children's Hospital undergoing elective surgical excision. Additional human tissue samples (n = 74) representing normal liver, spleen, muscle, heart, and lung were obtained from the Cooperative Human Tissue Network at Nationwide Children's Hospital (7, 38). Samples were stored frozen at −80°C until DNA isolation.
Manipulation of nucleic acids.
For DNA isolation from human samples, freshly thawed tissue (0.2 to 0.5 g) was digested for 15 h in 3 ml of digestion buffer (10 mM Tris, pH 8.0; 100 mM NaCl; 0.5% sodium dodecyl sulfate [SDS]; 25 mM EDTA) supplemented with 2 mg/ml proteinase K at 55°C with constant agitation. DNA was subjected to three phenol-chloroform-isoamyl alcohol (25:24:1) extractions. A final chloroform extraction was performed, followed by DNA ethanol precipitation and resuspension in 10 mM Tris, pH 8.0. Low-molecular-weight DNA was isolated from cells using standard procedures as previously described (7). For Southern blot hybridization analysis, DNA was fractionated on 0.8% agarose gels and transferred to a nylon membrane. DNA hybridization conditions were 65°C for 16 h in a buffer containing 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1× Denhardt's reagent, and 200 μg/ml sonicated herring sperm DNA. Membranes were washed twice at 60°C in 2× SSC and 0.2% SDS for 30 min and then twice at 60°C in 0.2× SSC and 0.2% SDS for 30 min.
SSLRCA.
To detect the presence of double-stranded circular AAV genomes, 1 μg of genomic DNA was first digested in a 15-μl volume with a restriction enzyme that did not cut within the AAV genome (SpeI for human samples; EcoRV for Detroit 6 and plasmid spike samples). Linear DNA (both double and single stranded) was degraded by incubation with 10 U Plasmid-Safe ATP-dependent DNase (PS-DNase; Epicentre Technologies) in a 25-μl volume for 16 h at 37°C in 33 mM Tris (pH 7.8), 66 mM potassium acetate, 10 mM magnesium acetate, 0.5 mM dithiothreitol, and 1 mM ATP. PS-DNase was heat inactivated for 30 min at 70°C, and 2.5 μl (equivalent to 100 ng) was used as a template for SSLRCA using phage phi29 DNA polymerase (TempliPhi; GE Healthcare). Template DNA was mixed in a final volume of 15 μl in 10 mM Tris, pH 8.0, with 300 pmol each of two AAV cap primers (temp1, 5′-ATTGGCATTGCGATTCC-3′; temp2, 5′-TGGTGATGACTCTGTCG-3′) that contained a phosphothioation linkage between the last two 3′ nucleotides. This mixture was heated to 95°C for 3 min, cooled slowly to 4°C to allow primer annealing, and mixed with 15 μl of TempliPhi reaction buffer containing phi29 polymerase and incubated for 18 h at 30°C. Amplified products were heat inactivated at 65°C for 10 min and then digested with either XbaI or HindIII for Southern hybridization (these enzymes cut once in the AAV2 genome). For cloning, SSLRCA-amplified DNA was digested with a single-cut enzyme (XbaI or HindIII) and the unit-length DNA fragment cloned into plasmid pBlueScript KS− (Stratagene Corp.). DNA sequencing was performed by the Sequencing Core Laboratory. In addition, manual DNA sequencing of the AAV ITR region was accomplished using the SequiTherm Excel II DNA sequencing kit (Epicentre) following the manufacturer's protocol for isothermal sequencing with α-labeled 35S-dATP.
Analysis of SSLRCA-amplified ITR products.
To examine the fidelity of the SSLRCA assay, circular templates that contained defined ITR-ITR structures were generated and then subjected to the SSLRCA reaction. Products were examined by sequencing and Smith-Birnstiel end labeling of the linear amplified products (40). Briefly, a complete ITR-ITR end-joined template was generated by digesting a pSub201-based rAAV vector plasmid (rAAV/CMV/eGFP) with PvuII to release the intact rAAV expression cassette (including ITRs). This fragment was self-ligated and then treated with PS-DNase to remove all linear forms. A portion of this circular template (diluted 1:100) was used as the template in an SSLRCA reaction as described above. Both self-ligated and SSLRCA products were linearized by SalI enzyme digestion, treated with alkaline phosphatase, and end-labeled using polynucleotide kinase and [γ-32P]ATP. After labeling, the products were digested with NdeI to create linear, end-labeled fragments. Selected molecular clones isolated from tonsil sample T88 also underwent SSLRCA and end labeling in a similar manner as described above, but with the following minor modifications. T88 clones were released from the plasmid vector backbone by XbaI digestion prior to self-ligation and treatment with PS-DNase, and then they were linearized with SnaBI before treatment with alkaline phosphatase and polynucleotide kinase. DraIII was used to generate linear end-labeled fragments. To determine ITR-ITR junction heterogeneity occurring in vivo, SSLRCA products were treated identically as described above for T88-derived molecular clones. DNA fragments were fractionated on a 6% denaturing polyacrylamide gel and exposed directly to film. Specific band counts were determined using a Typhoon 9400 variable mode imager (GE Healthcare) with the ImageQuant 5.2 software.
Functional analysis of AAV molecular clones.
To determine if SSLRCA-generated AAV molecular clones could produce infectious virions, the pBlueScript vector backbone and cap ORF was recreated by digestion with XbaI and self-ligation to form head-to-tail circular monomeric genomes. The ligated products (1 μg) were directly transfected into 4 × 105 HeLa cells by using the FuGene 6 reagent (Roche Applied Science), followed by infection with Ad5. After maximum cytopathic effect development (48 h), the transfected cells (P0) were harvested and low-molecular-weight DNA prepared from half of the cells. The recovered DNA was subsequently treated with DpnI to digest input plasmid DNA. A clarified cell lysate was generated from the other half of P0 cells by three freeze-thaw cycles and a 60-min incubation at 56°C to heat-inactivate Ad5. The clarified lysate (1:10 dilution) was used to infect 4 × 105 HeLa cells (along with Ad5). After 48 h, the transduced cells (P1) were harvested and low-molecular-weight DNA isolated.
Western blot analysis.
To detect the presence of Rep and Cap proteins, a cell lysate was generated from transfected cells (P0) after 48 h and electrophoresed on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subsequently transferred to a polyvinylidene difluoride membrane (GE Healthcare). The membrane was blocked for 10 min at 25°C using TBST buffer (10 mM Tris [pH 8.0], 150 mM NaCl, 0.1% Tween 20) plus 5% nonfat dry milk. Primary antibodies (from American Research Products, Inc.) were used at a 1:100 dilution (Rep proteins, 1:1 mixture of anti-AAV Rep clone 303.9 and clone 226.7; Cap proteins, anti-AAV Cap clone B1) and hybridized in blocking solution for 1 h. The blot was washed three times for 10 min in TBST buffer and incubated for 30 min in TBST buffer plus 2.5% dry milk with a horseradish peroxidase-conjugated anti-mouse secondary antibody (Vector Laboratories) at a 1:10,000 dilution. The blot was washed as described above and developed using the ECL Plus detection system (GE Healthcare).
Real-time PCR.
Wild-type AAV genomes and AAV plasmids were quantified by real-time PCR using the ABI Prism 7000 sequence detection system in a standard 25-μl TaqMan PCR with the following primers and probe: ForCAPSS, 5′-AACGACAACCACTACTTTGGC-3′, 50 nM final concentration; RevCAPSS, 5′-AAGTGGCAGTGGAATCTGTTG-3′, 900 nM final concentration; and CAPSSProbe, 5′-[6-FAM]CTACAGCACCCCCTGGGGGTATTTTGA[TAMRA-6-FAM]-3′, 200 nM final concentration.
RESULTS
SSLRCA.
In our previous work on characterization of AAV in human tissues, we found that the majority of AAV genomes were extrachromosomal and that none were integrated in the AAVS1 locus (38). As part of those efforts, we developed a novel method (SSLRCA) that allowed us to amplify extrachromosomal AAV genomes directly from tissue DNA. It occurred to us that these amplified products might represent intact, biologically active AAV genomes. To investigate this possibility, templates for SSLRCA were derived from total tissue DNA that was treated with a unique exonuclease (PS-DNase) which preferentially degraded linear DNA fragments, leaving circular, double-stranded DNA intact (Fig. 1). This treatment enriched the substrate DNA for circular templates and facilitated amplification using AAV-specific primers and phi29 DNA polymerase. Following overnight isothermic incubation, the resulting high-molecular-weight amplicons were digested into unit-length fragments and cloned into a standard plasmid cloning vector for sequence and functional analyses. Unit-length AAV genomes were generated by releasing inserts from the vector backbone and performing an intramolecular ligation. The resulting circular monomeric AAV genome was then directly transfected into HeLa cells to evaluate infectivity.
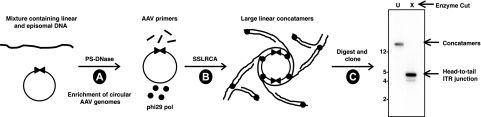
FIG. 1.
SSLRCA schematic for the isolation of infectious AAV molecular clones. Total cellular DNA is first digested with a restriction enzyme that does not cut within the AAV genome but restricts the host genome. This results in the creation of linear fragments that are substrates for exonuclease digestion (PS-DNase [step A]). PS-DNase does not degrade double-stranded, circular DNA molecules. To amplify remaining circular episomes, AAV cap gene-specific primers (complementary to each strand) are annealed to the circular episome, and phi29 DNA polymerase is used to initiate rolling-circle replication via strand displacement (step B). SSLRCA reaction products are high-molecular-weight linear AAV concatemers (U) that can be digested with a single-cut enzyme (X) to produce unit-length AAV genomic fragments, which can be cloned for functional analysis (step C).
To determine the limit of detection for circular AAV, decreasing amounts of a 15-kb plasmid containing AAV2 ITR and rep and cap genes were spiked into 100 ng of naïve tonsil DNA and subjected to SSLRCA. The amplification over a range of input copies was determined for four independent replicate experiments (Table 1). This experiment showed that we could reliably amplify as few as 25 plasmid copies (107-fold amplification).
TABLE 1.
SSLRCA assay sensitivity
| Plasmid input (no. of copies)a | Amplification (fold) for replicate:
|
Totalc | |||
|---|---|---|---|---|---|
| 1b | 2 | 3 | 4 | ||
| 100 | 3.1 × 104 | 3.3 × 105 | 3.7 × 107 | 1.6 × 106 | 4 of 4 |
| 75 | 4.6 × 105 | 2.2 × 106 | 2.7 × 107 | 4.1 × 107 | 4 of 4 |
| 50 | 9.5 × 105 | 1.5 × 106 | 7.4 × 107 | 1.5 × 106 | 4 of 4 |
| 25 | 0 | 2.1 × 106 | 1.1 × 107 | 4.6 × 107 | 3 of 4 |
| 10 | 0 | 1.0 × 107 | 0 | 0 | 1 of 4 |
| 7.5 | 0 | 2.1 × 107 | 0 | 0 | 1 of 4 |
| 5 | 0 | 0 | 0 | 0 | 0 of 4 |
| 1 | 0 | 0 | 0 | 0 | 0 of 4 |
| 0 | 0 | 0 | 0 | 0 | 0 of 4 |
Copies of a circular 15-kb plasmid, containing AAV2 rep/cap genes, were spiked into 100 ng of total cellular DNA isolated from a naïve human tonsil sample and used as a template for SSLRCA.
Amplification was determined by dividing the number of AAV cap copies present after SSLRCA by the number of cap copies present before the SSLRCA assay was performed.
Number of total replicates scored as positive for the detection of circular template.
As an important control, SSLRCA substrate specificity was examined using Detroit 6 cells (19), which are a well-characterized transformed cell line that contains integrated AAV2 genomes (head-to-tail tandem arrays) in the AAVS1 locus on chromosome 19 (q19.1). As noted above, AAV genomes integrated within the Detroit 6 DNA should not amplify using the SSLRCA method. Accordingly, 1,000 and 100,000 integrated AAV2 copies from Detroit 6 total DNA were spiked into 100 ng of naïve tonsil DNA, and SSLRCA was performed. The AAV2 genome copy number was determined by cap gene quantitative PCR before and after SSLRCA, and the average amplification was 1.3- ± 1.0-fold over 10 independent replicate assays (data not shown). These data demonstrate the strict requirement for circular templates to achieve robust SSLRCA-dependent amplification.
Direct amplification of AAV genomes from human tissue.
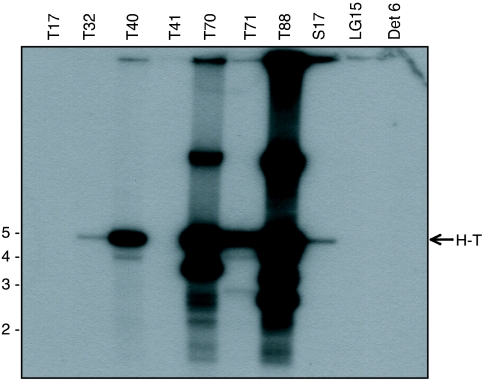
As noted above, we previously identified nine human DNA samples that contained variable amounts of AAV DNA ranging from 80 to 33,000 copies/μg of total cellular DNA (7). These nine DNA samples were subjected to SSLRCA, and six yielded AAV2-specific amplification. Digestion of the amplified products with a restriction enzyme that was predicted to cut once inside the AAV2 genome (HindIII) yielded the expected 4.7-kb unit-length fragment (Fig. 2). In addition, both high- and low-molecular-weight forms were present in samples T70 and T88. The high-molecular-weight forms might represent unresolved large concatemeric DNA that was undigested due to other genome species that do not contain the restriction site(s). The low-molecular-weight forms might correspond to genome species that contain an additional restriction site(s) or could be derived from in vivo deleted viral genomes. The amount of amplified AAV2 product correlated with the input AAV copy number previously determined from the various samples (Table 1). The three AAV samples that failed to amplify (LG15, T17, and T41) harbored copy numbers that were at or below the limit of detection for the assay (see Table 2). The low level of amplification for T32 and T41 was also consistent with the low AAV2 copy numbers in these tissues.
FIG. 2.
SSLRCA method detects episomal AAV in human tissue DNA. Human total cellular DNA (100 ng) was subjected to SSLRCA, and the resulting product was digested with HindIII (cuts once within the AAV genome) and subjected to Southern blot hybridization using a 1-kb PCR-amplified AAV2 rep-cap DNA fragment as the probe. A 4.7-kb unit-length head-to-tail (H-T) DNA fragment was detected in six of the AAV-positive samples, along with high- and low-molecular-weight DNA in samples T88 and T70. No evidence of SSLRCA-mediated amplification of integrated AAV genomes was seen using 100 ng of Detroit 6 (Det 6) DNA (corresponding to 83,000 integrated copies).
TABLE 2.
Summary of molecular clones isolated by SSLRCA
| Tissue sample | No. of AAV copiesa | SSLRCA resultb | No. of clones analyzedc | % Infectious clonesd |
|---|---|---|---|---|
| T17 | 56 | − | 0 | NA |
| T32 | 21 | + | 1/1 | 100 (1/1) |
| T40 | 655 | + | 6/60 | 17 (1/6) |
| T41 | 59 | − | 0 | NA |
| T70 | 780 | + | 15/80 | 93 (14/15) |
| T71 | 760 | + | 6/26 | 67 (4/6) |
| T88 | 3,300 | + | 14/80 | 71 (10/14) |
| S17 | 20 | + | 3/3 | 0 (0/3) |
| LG15 | 8 | − | 0 | NA |
| Detroit 6 | 83,000 | − | 0 | NA |
AAV copy number in 100 ng of total cellular DNA based on cap quantitative PCR.
SSLRCA-positive result based on Southern blot detection of AAV genomes.
The fraction represents the number of molecular clones that were analyzed for infectivity (passage) over the number of total clones isolated by bacterial colony hybridization.
Since only a subset of clones was screened for passage, the percentage reflects our ability to obtain replicating monomeric and dimeric AAV genomic species following passage (P1). Fraction in parentheses represents the number of infectious clones over the number of clones analyzed for infectivity. NA, not applicable.
Isolation of infectious AAV molecular clones.
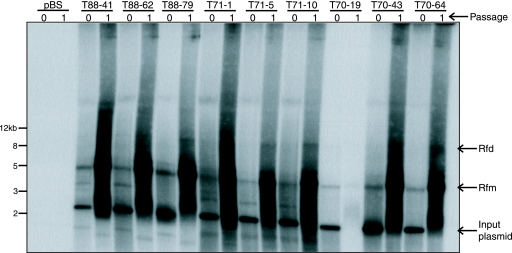
Having amplified AAV genomes from human tissue samples, we next asked whether SSLRCA could be used to isolate intact full-length molecular clones representing infectious wild-type AAV. The unit-length amplified AAV2 DNA (Fig. 1 and 2) was inserted into a plasmid vector by using the unique HindIII restriction enzyme site located in the AAV2 cap gene. To determine whether the resulting clones would give rise to infectious virus, the AAV DNA was released from the cloning vector backbone and self-ligated to restore the cap ORF and regenerate a circular monomeric AAV genome. The ligation reaction was directly transfected into Ad5-infected HeLa cells (MOI = 20). After adenovirus-induced cytopathic effect was observed, a cell lysate was prepared, clarified, and passed onto naïve HeLa cells (again, in the presence of adenovirus). Hirt DNA was isolated from both the transfected cells (P0) and the infected cells (P1) and analyzed by Southern blot hybridization. Analyses of nine clones isolated from three individual tonsil-adenoid tissue samples are shown in Fig. 3. DNA replication in P0 was observed for all clones, as judged by the presence of characteristic AAV monomeric (Rfm) and dimeric (Rfd) replicating DNA forms. Also present in the P0 Hirt DNA was a distinct 2.5-kb hybridizing fragment that was derived from the transfected DNA following DpnI enzyme digestion. Importantly, all the molecular clones gave rise to infectious virus, as judged by replicating AAV DNA in P1 cells. In summary, we were able to isolate infectious molecular clones from five of the six SSLRCA-positive tissue samples (Table 2). For sample S17, it remains possible that we simply did not isolate enough clones to identify an infectious clone.
FIG. 3.
Identification of infectious AAV molecular clones. Nine individual AAV molecular clones isolated from three tonsil tissue samples (T70, T71, and T88) were analyzed for their ability to replicate their respective genomes and produce infectious particles. Shown is an autoradiograph of a Southern blot hybridization (a 1-kb AAV2 rep-cap DNA fragment as the probe) of Hirt DNA isolated from cells at pass 0 (P0) and pass 1 (P1). Residual input plasmid in P0 Hirt DNA was digested with DpnI to reduce the input plasmid molecular weight (seen as a 2.5-kb DNA fragment). Replicating monomeric (Rfm) and dimeric (Rfd) AAV DNA forms were observed in P0 and P1 cells, indicative of replication competency (P0) and formation of infectious virus (P1). Upon prolonged exposure, the autoradiograph showed both Rfm and Rfd forms of AAV DNA in sample T70-19.
Characterization of AAV produced from infectious molecular clones.
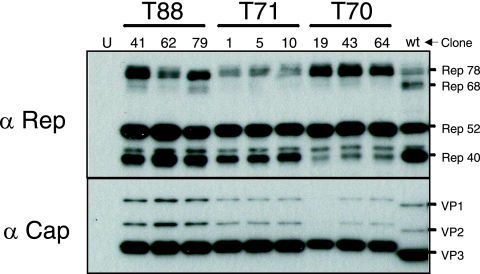
To further characterize AAV produced by our clones, we serially passaged infectious virions derived from clones T70-43 and T88-41 and showed that particles were DNase I resistant and purified at the predicted density in iodixanol gradients (data not shown). The cap gene sequence for both of these isolates predicted replacement of charged arginine residues at positions 585 and 588 with serine and threonine, respectively. As predicted, purified virions representing both clones did not bind to a heparin sulfate matrix (7). We also analyzed AAV Rep and Cap protein expression profiles of these and selected other molecular clones (Fig. 4). As expected, all of the infectious molecular clones produced the four Rep and three Cap proteins. Interestingly, the Cap proteins (VP1, VP2, and VP3) derived from prototype AAV2 migrated faster than did the Cap proteins derived from the infectious molecular clones. This difference in migration was not predicted based on the calculated molecular weights. We (data not shown) and others (30) have observed this phenomenon with Cap proteins from multiple serotypes. It is possible that the differences in electrophoretic mobility might be due to particle charge, residual secondary structure, or possibly posttranslational modifications, such as capsid phosphorylation, as recently reported by Zhong et al. (44).
FIG. 4.
Rep and Cap expression of T88 molecular clones. Individual molecular clones were analyzed for AAV Rep (Rep78, 68, 52, 40) and Cap (VP1, VP2, VP3) protein expression. Shown are Western blot hybridization blots of transfected cell lysates (P0). “U” designates an untransfected cell lysate, while “wt” denotes a wild-type AAV2-infected lysate.
Full nucleotide sequence determination of several selected molecular clones was performed. The cap sequences obtained via SSLRCA were identical to the original sequences derived by nested PCR, which were previously reported (7). rep gene sequences from samples T88, T70, T71, S17, T32, and T40 were highly homologous to each other and to AAV2 (>98% amino acid identity). Sequence analysis of the 5′ and 3′ ITR regions revealed the presence of ITR D regions from either direction, characteristic of a double-D ITR structure. The ITR sequence for multiple infectious clones (T88-41, T88-79, T70-19, T70-43, T70-64, T71-1, T71-5, T71-10) was determined, and all except one contained a complete double-D ITR structure; both “flip” and “flop” forms were represented. Interestingly, the single imperfect double-D ITR infectious clone (T88-62) possessed an A region duplication with a partial deletion of the sequence between the two A elements (Fig. 5). In addition, double-D ITR structures from several noninfectious molecular clones (T88-16, T88-78B) were determined and shown to possess significant internal deletions in A, B, and C hairpin regions (Fig. 6).
FIG. 5.
Structure of the AAV ITR junctions from selected molecular clones. Shown schematically are ITR structures observed in several infectious molecular clones, with the palindromic ITR sequences designated as D, A, B, B', C, C', A', and D', in accord with standard nomenclature. The complete ITR sequence was determined for clones T88-41, T88-62, and T88-79. Clones 41 and 62 contain a complete double-D ITR (in both flip and flop orientations). T88-62 contains an imperfect double-D ITR junction with two A' regions, both of which contain small deletions. In addition, three T70 clones (no. 18, 42, and 64) and three T71 clones (no. 1, 5, and 10) also possessed a complete double-D structure that was identical to that of T88-79. All ITR sequences were identical to the published AAV2 ITR sequence.
FIG. 6.
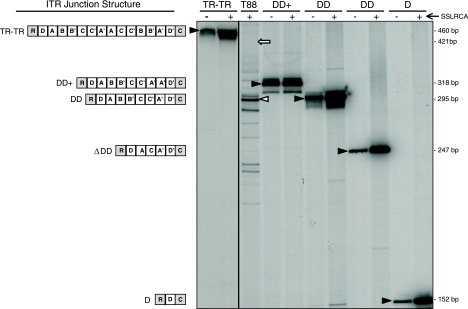
Episomal AAV contains double-D ITR structures in vivo. Various circular AAV templates were subjected to SSLRCA. The input templates (−) and the amplified products (+) were end-labeled and analyzed by acrylamide gel electrophoresis. Schematics of the ITR structure of various templates are shown to the left of the autoradiograph, with the various ITR regions denoted as A, B, C, and D. The ITR junction is oriented with respect to the AAV rep (R) and cap (C) genes. Black arrowheads designate the specific input (−) and SSLRCA (+) bands. The ITR-ITR template contains two individual end-joined ITRs. The DD+ template was derived from clone T88-62, DD from clone T88-41, ΔDD from clone T88-16, and D from clone T88-78B. In all cases, the input template was amplified correctly by SSLRCA, as evidenced by identical band migration before and after SSLRCA. When SSLRCA products from tonsil sample T88 were analyzed in this fashion, several distinct bands were identified. The most prevalent band (48% of the overall signal; white arrowhead) corresponded in size to the complete DD template. The predicted size of an end-joined ITR-ITR junction from tonsil sample T88 is represented by a white arrow.
Double-D ITR structures present in tissues.
Based on ITR sequence analysis, we concluded that infectious molecular clones contained a complete double-D ITR element. Conversely, limited ITR sequence analysis of noninfectious clones suggested that the deletion of internal ITR regions was responsible for the lack of infectivity, since these clones expressed the expected Rep and Cap proteins (data not shown). Consequently, we were interested to know if double-D ITR junction formation in vivo yielded predominately complete nonrearranged structures or if the ITR deletions were imprecise and only a subset contained the complete double-D ITR.
To answer this question, SSLRCA was performed directly on tissue genomic DNA because ITR rearrangements have been observed to occur during passage in bacteria. Preceding in vivo analysis of ITR forms, we characterized the fidelity of the SSLRCA assay by using several complex ITR structures, the first of which possessed two complete ITRs joined at their ends (Fig. 6). Following amplification, the ITR-ITR construct (input) and SSLRCA-amplified material were end-labeled and fractionated on an acrylamide gel. The end-joined ITR-ITR template was amplified faithfully, with no deletions evident based on identical mobility through the gel (Fig. 6, lane “TR-TR”). Additionally, when T88 molecular clones possessing four distinct double-D structures (Fig. 6, lanes “DD+,” “DD,” “ΔDD,” and “D”) were similarly analyzed, all SSLRCA-amplified material was replicated faithfully.
After the preliminary analyses of these known substrates, SSLRCA was then performed on genomic DNA isolated directly from tonsil-adenoid sample T88 (Fig. 6, lane “T88”). Multiple discrete species were easily identified, and when these amplified fragments were compared on the same gel to the known tonsil T88 molecular clones, the predominant in vivo form corresponded in size to the complete double-D structure (Fig. 6, white arrowhead). Quantitative densitometry revealed that 48% of the total signal corresponded to intact double-D ITR structures. Significantly, there was no specific band present at the predicted size of an end-joined ITR-ITR structure (Fig. 6, white arrow), suggesting that in this particular sample, this was not an in vivo form. Moreover, the presence of numerous ITR species (some larger and many smaller) indicated that double-D formation is imprecise.
DISCUSSION
Molecular details regarding wild-type AAV replication, gene expression, and genome integration have been largely elucidated through the use of in vitro mammalian cell culture infection systems. These studies spawned a life cycle paradigm that holds site-specific virus integration as a central tenet. Integration in this model is mediated by the virally encoded large Rep proteins (Rep78 or Rep68) and involves recognition of cognate binding sites within the virus and host genome (GAGC tetra-nucleotide repeats). While targeted integration into chromosome 19 can be readily detected in cultured cells, it is driven by high MOIs and is actually a rare event that is estimated to occur once in every 1,000 productive transduction events in vitro (16). Only a single study to date has looked at experimental AAV infection in the primate host (14). The authors encountered difficulty in detecting the presence of the virus postinoculation in the oropharynx or respiratory tract, even in the presence of coinfecting adenovirus. This complexity illustrates the challenges associated with virus recovery and sampling effects when using large animal models. Studies using AAVS1 transgenic mice have documented targeted integration (32, 35), illustrating that there are probably inherent differences between cell culture and in vivo models of infection.
We took an alternative approach to understanding molecular aspects of AAV natural infection by characterizing the molecular form of AAV genomes in human tissues. The SSLRCA method enabled the isolation of episomal molecular clones, from which structure function data were derived.
Our analyses led to four fundamental observations. First, based on our previous analysis (38), the predominant in vivo forms were extrachromosomal circles. Second, episomal circles possessed complete or partially deleted double-D ITR structures. Third, infectious DNA clones possessed complete double-D structures. Finally, the cellular mechanism for circularization appeared to be imprecise and resulted in multiple double-D ITR forms within a single tissue (but preferentially yielded the complete double-D element). The observation that almost 50% of the circular forms within a human tonsil sample were perfect double-D structures suggested sufficient recombinational specificity to maintain ITR continuity in a significant proportion of viral genomes to enable subsequent virus rescue. Additionally, the fact that infectious molecular clones were also adenovirus DNA negative (7) argues that the episomal forms amplified were not simply circular replicative forms.
These data have significant implications for our understanding of the AAV life cycle. The presence of the double-D ITR structure has been demonstrated to be the sole cis element required for AAV propagation (42). Upon double-D ITR resolution (which occurs in the presence of a coinfecting helper virus), a no-end substrate is generated, and subsequent nicking at one of the two available terminal resolution sites by the large Rep proteins would allow AAV DNA replication to proceed via leading-strand synthesis and infectious virus formation. The direct implication is that AAV can proceed through its life cycle in the complete absence of an integration intermediate in vivo, and in fact, this has been shown in cell culture (30). Additionally, we and other investigators have reported identical double-D ITR elements for recombinant AAV vectors in muscle tissue, suggesting that AAV ITRs contain all the cis sequence information necessary to undergo rep-independent circularization (11, 37, 42). Thus, these data make the case for an additional cis ITR property, namely, circularization. It is logical that the viral ITR has evolved to promote accurate circularization via homologous recombination to preserve a critical structural element (double-D ITR) that appears essential for viral infectivity upon rescue following helper virus infection.
The mechanism of ITR circularization is unknown, but data using self-complementary vectors suggest that it occurs through a double-stranded intermediate, following second-strand synthesis (23). In cell culture, ITR circularization is a rapid process, and higher-order forms (concatemers) appear to form through the interaction of double-D ITR elements on separate circles (10, 11). Additional research using cells in culture has focused on characterizing those cellular enzymes involved in the circularization process. To date, multiple DNA double-strand break repair enzymes are implicated and include ATM, Mre11, DNA-PKcs, KU 70/86, and Rad 50, 51, and 52 proteins (8, 12, 39, 41). These recombination proteins could be envisioned to either promote circularization (e.g., Mre11 and ATM) or act to inhibit integration (e.g., DNA-PKcs). The multiple-sized double-D ITR structures observed in tonsil sample T88 are consistent with either homologous recombination or nonhomologous end-joining repair pathways facilitating ITR-ITR ligation. The failure to detect complete head-to-head ITR junctions and the presence of complete double-D isoforms argue that recombination via nonhomologous end joining may not be the favored pathway of ITR circularization but could still play a role in ITR-ITR ligation. In fact, it might be beneficial for AAV to use multiple pathways for circularization, thereby avoiding free hairpin ends that can be sensed as abnormal by the cell and, under specific circumstances, trigger apoptotic stimuli (31).
Under conditions of high virus inputs, the circularized viral form may act as an integration intermediate. Two observations are consistent with this supposition. First, most proviral integrants are arranged in head-to-tail arrays, which are thought to require a circular template and limited DNA replication to generate the arrays. Second, efficient AAVS1 integration in cell culture is heavily dependent on the MOI (28). Thus, at low MOI (<100 to 1,000), integration in cell culture is a rare event, and the viral genomes are lost during subsequent passage. It is unclear why a threshold MOI is needed for targeted integration; perhaps a relatively high intranuclear concentration of circular AAV molecules is needed for juxtaposition and formation of viral ITR/AAVS1 complexes. Whether similar input viral loads are necessary in vivo for integration is unknown, but we have observed no detectable integration into the AAVS1 locus by wild-type AAV genomes in excess of 400 copies per cell in nonhuman primate tissues (unpublished observation).
Although we were unable to isolate infectious molecular clones from episomal AAV genomes from all AAV-positive tissue samples (T17, T41, and LG15) by using SSLRCA, we were nonetheless unable to detect AAVS1 integration in these same tissue samples by using a very sensitive nested PCR assay (five integration events in 1.67 × 105 cell equivalents) (38). Thus, it seemed that integration within the AAVS1 locus was unlikely. Clearly, an integration event could have occurred at other sites within the genome, but we documented only a single event in sample T71 after use of a sensitive linear amplification-mediated PCR assay (one integration event in 1 × 103 cell equivalents) (38). Our inability to isolate infectious molecular clones seems more likely due to the fact that the AAV copy numbers in these tissues were at or below the limit of sensitivity for SSLRCA.
Taken together, our data compel us to take a fresh look at the role of integration in the AAV life cycle. On the one hand, we have shown that AAV DNA does not appear to integrate with any measurable frequency at the AAVS1 locus in human tissues (38). Importantly, we have now shown that integration is not required to generate infectious viral genomes. These observations have been confirmed in AAV-infected nonhuman primates (our unpublished observations). Furthermore, others have shown that in cell culture, only 0.1% of infectious AAV particles integrate into AAVS1 (28, 29). Thus, these considerations make it difficult to argue for a prominent role of virus integration in AAV natural infection. On the other hand, integration would clearly be of benefit in actively dividing cell populations, and perhaps this might occur at higher frequencies under “favorable” intracellular conditions (e.g., sufficient Rep expression and high virus inputs). Testing of this latter hypothesis will await controlled nonhuman primate infection studies.
In conclusion, our data convincingly suggest that the natural AAV life cycle, in contrast to that of retroviruses, does not have DNA integration as an obligatory component. Rather, AAV genomes appear to persist in tissues as monomeric and concatemeric circles. Importantly, the molecular form (double-D ITR) of these extrachromosomal circles probably permits virus rescue and replication upon helper virus infection, and it allows for a viral life cycle that does not require integration for propagation in the human host. Given that recombinant AAV vectors contain wild-type ITRs, it now seems likely that recombinant vector genomes behave in an analogous manner to wild-type genomes, and this has been borne out in the literature (11, 27, 37, 38).
Acknowledgments
We thank Douglas McCarty for helpful discussions and critical reading of the manuscript. We also thank Robin Bolek for expert technical assistance.
This work was funded by the National Institutes of Health.
Footnotes
Published ahead of print on 19 November 2008.
REFERENCES
- 1.Atchison, R. W., B. C. Casto, and W. M. Hammon. 1965. Adenovirus-associated defective virus particles. Science 149754-756. [DOI] [PubMed] [Google Scholar]
- 2.Atchison, R. W., B. C. Casto, and W. M. Hammon. 1966. Electron microscopy of adenovirus-associated virus (AAV) in cell cultures. Virology 29353-357. [DOI] [PubMed] [Google Scholar]
- 3.Blacklow, N. R., M. D. Hoggan, A. Z. Kapikian, J. B. Austin, and W. P. Rowe. 1968. Epidemiology of adenovirus-associated virus infection in a nursery population. Am. J. Epidemiol. 88368-378. [DOI] [PubMed] [Google Scholar]
- 4.Blacklow, N. R., M. D. Hoggan, and W. P. Rowe. 1968. Serologic evidence for human infection with adenovirus-associated viruses. J. Natl. Cancer Inst. 40319-327. [PubMed] [Google Scholar]
- 5.Blacklow, N. R., M. D. Hoggan, M. S. Sereno, C. D. Brandt, H. W. Kim, R. H. Parrott, and R. M. Chanock. 1971. A seroepidemiologic study of adenovirus-associated virus infection in infants and children. Am. J. Epidemiol. 94359-366. [DOI] [PubMed] [Google Scholar]
- 6.Buller, R., J. Janik, E. Sebring, and J. Rose. 1981. Herpes simplex virus types 1 and 2 completely help adenovirus-associated virus replication. J. Virol. 40241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C. L., R. L. Jensen, B. C. Schnepp, M. J. Connell, R. Shell, T. J. Sferra, J. S. Bartlett, K. R. Clark, and P. R. Johnson. 2005. Molecular characterization of adeno-associated viruses infecting children. J. Virol. 7914781-14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, V. W., D. M. McCarty, and R. J. Samulski. 2006. Host cell DNA repair pathways in adeno-associated viral genome processing. J. Virol. 8010346-10356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clark, K. R. 2002. Recent advances in recombinant adeno-associated virus vector production. Kidney Int. 61(Suppl. 1)9-15. [DOI] [PubMed] [Google Scholar]
- 10.Duan, D., P. Sharma, J. Yang, Y. Yue, L. Dudus, Y. Zhang, K. J. Fisher, and J. F. Engelhardt. 1998. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. J. Virol. 728568-8577. (Erratum, 73:861.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duan, D., Z. Yan, Y. Yue, and J. F. Engelhardt. 1999. Structural analysis of adeno-associated virus transduction circular intermediates. Virology 2618-14. [DOI] [PubMed] [Google Scholar]
- 12.Duan, D., Y. Yue, and J. F. Engelhardt. 2003. Consequences of DNA-dependent protein kinase catalytic subunit deficiency on recombinant adeno-associated virus genome circularization and heterodimerization in muscle tissue. J. Virol. 774751-4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erles, K., P. Sebokova, and J. R. Schlehofer. 1999. Update on the prevalence of serum antibodies (IgG and IgM) to adeno-associated virus (AAV). J. Med. Virol. 59406-411. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez, Y. J., J. Wang, W. G. Kearns, S. Loiler, A. Poirier, and T. R. Flotte. 1999. Latent adeno-associated virus infection elicits humoral but not cell-mediated immune responses in a nonhuman primate model. J. Virol. 738549-8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoggan, M. D., N. R. Blacklow, and W. P. Rowe. 1966. Studies of small DNA viruses found in various adenovirus preparations: physical, biological, and immunological characteristics. Proc. Natl. Acad. Sci. USA 551467-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hüser, D., S. Weger, and R. Heilbronn. 2002. Kinetics and frequency of adeno-associated virus site-specific integration into human chromosome 19 monitored by quantitative real-time PCR. J. Virol. 767554-7559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jay, F. T., C. A. Laughlin, and B. J. Carter. 1981. Eukaryotic translational control: adeno-associated virus protein synthesis is affected by a mutation in the adenovirus DNA-binding protein. Proc. Natl. Acad. Sci. USA 782927-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kern, A., K. Schmidt, C. Leder, O. J. Muller, C. E. Wobus, K. Bettinger, C. W. Von der Lieth, J. A. King, and J. A. Kleinschmidt. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 7711072-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotin, R. M., and K. I. Berns. 1989. Organization of adeno-associated virus DNA in latently infected Detroit 6 cells. Virology 170460-467. [DOI] [PubMed] [Google Scholar]
- 20.Kotin, R. M., J. C. Menninger, D. C. Ward, and K. I. Berns. 1991. Mapping and direct visualization of a region-specific viral DNA integration site on chromosome 19q13-qter. Genomics 10831-834. [DOI] [PubMed] [Google Scholar]
- 21.Kotin, R. M., M. Siniscalco, R. J. Samulski, X. D. Zhu, L. Hunter, C. A. Laughlin, S. McLaughlin, N. Muzyczka, M. Rocchi, and K. I. Berns. 1990. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 872211-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linden, R. M., P. Ward, C. Giraud, E. Winocour, and K. I. Berns. 1996. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA 9311288-11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarty, D. M., H. Fu, P. E. Monahan, C. E. Toulson, P. Naik, and R. J. Samulski. 2003. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 102112-2118. [DOI] [PubMed] [Google Scholar]
- 24.Mehrle, S., V. Rohde, and J. R. Schlehofer. 2004. Evidence of chromosomal integration of AAV DNA in human testis tissue. Virus Genes 2861-69. [DOI] [PubMed] [Google Scholar]
- 25.Muzyczka, N. 1992. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr. Top. Microbiol. Immunol. 15897- 129. [DOI] [PubMed] [Google Scholar]
- 26.Opie, S. R., K. H. Warrington, Jr., M. Agbandje-McKenna, S. Zolotukhin, and N. Muzyczka. 2003. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 776995-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Penaud-Budloo, M., C. Le Guiner, A. Nowrouzi, A. Toromanoff, Y. Cherel, P. Chenuaud, M. Schmidt, C. von Kalle, F. Rolling, P. Moullier, and R. O. Snyder. 2008. Adeno-associated virus vector genomes persist as episomal chromatin in primate muscle. J. Virol. 827875-7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Philpott, N. J., C. Giraud-Wali, C. Dupuis, J. Gomos, H. Hamilton, K. I. Berns, and E. Falck-Pedersen. 2002. Efficient integration of recombinant adeno-associated virus DNA vectors requires a p5-rep sequence in cis. J. Virol. 765411-5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Philpott, N. J., J. Gomos, K. I. Berns, and E. Falck-Pedersen. 2002. A p5 integration efficiency element mediates Rep-dependent integration into AAVS1 at chromosome 19. Proc. Natl. Acad. Sci. USA 9912381- 12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabinowitz, J. E., F. Rolling, C. Li, H. Conrath, W. Xiao, X. Xiao, and R. J. Samulski. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 76791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raj, K., P. Ogston, and P. Beard. 2001. Virus-mediated killing of cells that lack p53 activity. Nature 412914-917. [DOI] [PubMed] [Google Scholar]
- 32.Recchia, A., L. Perani, D. Sartori, C. Olgiati, and F. Mavilio. 2004. Site-specific integration of functional transgenes into the human genome by adeno/AAV hybrid vectors. Mol. Ther. 10660-670. [DOI] [PubMed] [Google Scholar]
- 33.Richardson, W. D., and H. Westphal. 1981. A cascade of adenovirus early functions is required for expression of adeno-associated virus. Cell 27133-141. [DOI] [PubMed] [Google Scholar]
- 34.Richardson, W. D., and H. Westphal. 1984. Requirement for either early region 1a or early region 1b adenovirus gene products in the helper effect for adeno-associated virus. J. Virol. 51404-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rizzuto, G., B. Gorgoni, M. Cappelletti, D. Lazzaro, I. Gloaguen, V. Poli, A. Sgura, D. Cimini, G. Ciliberto, R. Cortese, E. Fattori, and N. La Monica. 1999. Development of animal models for adeno-associated virus site-specific integration. J. Virol. 732517-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samulski, R. J., X. Zhu, X. Xiao, J. D. Brook, D. E. Housman, N. Epstein, and L. A. Hunter. 1991. Targeted integration of adeno-associated virus (AAV) into human chromosome 19. EMBO J. 103941-3950. (Erratum, 11:1228.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schnepp, B. C., K. R. Clark, D. L. Klemanski, C. A. Pacak, and P. R. Johnson. 2003. Genetic fate of recombinant adeno-associated virus vector genomes in muscle. J. Virol. 773495-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnepp, B. C., R. L. Jensen, C. L. Chen, P. R. Johnson, and K. R. Clark. 2005. Characterization of adeno-associated virus genomes isolated from human tissues. J. Virol. 7914793-14803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz, R., J. Palacios, G. Cassell, S. Adam, M. Giacca, and M. D. Weitzman. 2007. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J. Virol. 8112936-12945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, H., and M. Birnstiel. 1976. A simple method for DNA restriction site mapping. Nucleic Acids Res. 32387-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song, S., P. J. Laipis, K. I. Berns, and T. R. Flotte. 2001. Effect of DNA-dependent protein kinase on the molecular fate of the rAAV2 genome in skeletal muscle. Proc. Natl. Acad. Sci. USA 984084-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao, X., W. Xiao, J. Li, and R. J. Samulski. 1997. A novel 165-base-pair terminal repeat sequence is the sole cis requirement for the adeno-associated virus life cycle. J. Virol. 71941-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, C. C., X. Xiao, X. Zhu, D. C. Ansardi, N. D. Epstein, M. R. Frey, A. G. Matera, and R. J. Samulski. 1997. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J. Virol. 719231-9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhong, L., B. Li, C. S. Mah, L. Govindasamy, M. Agbandje-McKenna, M. Cooper, R. W. Herzog, I. Zolotukhin, K. H. Warrington, K. A. Weigel-Van Aken, J. A. Hobbs, S. Zolotukhin, N. Muzyczka, and A. Srivastava. 2008. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc. Natl. Acad. Sci. USA 1057827-7832. [DOI] [PMC free article] [PubMed] [Google Scholar]