Abstract
AexT is an extracellular ADP ribosyltransferase produced by the fish pathogen Aeromonas salmonicida subsp. salmonicida. The protein is secreted by the bacterium via a recently identified type III secretion system. In this study, we have identified a further 12 open reading frames that possess high homology to genes encoding both structural and regulatory components of the Yersinia type III secretion apparatus. Using marker replacement mutagenesis of aopB, the A. salmonicida subsp. salmonicida homologue of yopB in Yersinia, we demonstrate that the bacterium translocates the AexT toxin directly into the cytosol of cultured fish cells via this type III secretion pathway. An acrV mutant of A. salmonicida subsp. salmonicida displays a calcium-blind phenotype, expressing and secreting significant amounts of AexT even in the presence of CaCl2 concentrations as high as 10 mM. This acrV mutant is also unable to translocate AexT into the cytosol of fish cells, indicating AcrV is involved in the translocation process. Inactivation of either the aopB or acrV gene in A. salmonicida subsp. salmonicida (resulting in an inability to translocate AexT) is accompanied by a loss of cytotoxicity that can be restored by trans complementation. Finally, we present data indicating that preincubation of the wild-type bacteria with antibodies directed against recombinant AcrV-His protein provides fish cells protection against the toxic effects of the bacterium.
Aeromonas salmonicida subsp. salmonicida is an important fish pathogen causing a systemic and fatal disease in members of the family Salmonidae. Despite the wide distribution and economic significance of this pathogen, very little is known about the mechanisms it uses to cause disease. A number of potential virulence factors of A. salmonicida subsp. salmonicida have been identified. These factors include extracellular proteins, such as hemolysins (13) and proteases (35), and surface-exposed structures, including the A-layer protein (7), lipopolysaccharide (17), and type IV pili (19). However, the role these factors play in the disease process remains unclear. This study focuses on the recently identified ADP ribosyltransferase toxin A.salmonicida exoenzyme T, or AexT, which has recently been shown to play a role in the cytotoxicity of A. salmonicida subsp. salmonicida to fish cells (4).
AexT possesses significant sequence homology with proteins secreted via the well-characterized type III secretion systems (TTSSs) of Pseudomonas aeruginosa and the pathogenic Yersinia species. For example, sequence homology over the entire length of AexT is found with exoenzyme S (ExoS; 63% similarity and 58% identity) and exoenzyme T (ExoT; 68% similarity and 63% identity) of P. aeruginosa. The N-terminal domain of AexT reveals homology with the type III effector protein YopE of both Yersinia pseudotuberculosis (37% similarity and 34% identity) and Yersinia pestis (33% similarity and 27% identity).
It was this high degree of sequence similarity between AexT and the ExoS and YopE proteins that prompted us to screen A. salmonicida subsp. salmonicida for the presence of a TTSS. The identification of eight open reading frames (ORFs) in this bacterium that are homologous to TTSS genes found within Yersinia species and P. aeruginosa (6) was recently reported. We have shown that marker replacement mutagenesis of one of these genes, ascV (a homologue of the well-characterized yscV gene [formerly designated lcrD] of pathogenic Yersinia species) prevents secretion of the AexT toxin, indicating that the cell secretes this protein via the TTSS.
Here, we report further sequencing and characterization of the A. salmonicida subsp. salmonicida TTSS. We have identified an additional 12 ORFs that comprise analogues of the virB and lcrGVH-yopBD operons in Yersina. These operons encode structural components of the secretion and translocation apparatus, as well as regulatory molecules. Using marker replacement mutagenesis of the yopB homologue, aopB, we present evidence that indicates that A. salmonicida subsp. salmonicida translocates the AexT toxin directly into the cytosol of target cells via the type III pathway we have identified. This is the first report of type III-dependent protein translocation by this organism. Furthermore, we show that translocation of AexT can be prevented by inactivation of the lcrV homologue acrV and that this is accompanied by a loss of cytotoxicity to fish cells cultured in vitro. Finally, preincubation of the bacterium with anti-AcrV antibodies protects fish cells against the toxic effects of the bacterium. These findings lend valuable insight into the virulence mechanisms used by this pathogen and suggest that the TTSS (in particular AcrV) may be a potential target for improved vaccines to control furunculosis in fish.
For brevity, A. salmonicida subsp. salmonicida will be referred to as A. salmonicida from this point on.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
A summary of the bacterial strains and plasmids used in this study is provided in Table 1. Escherichia coli strains were routinely grown in Luria-Bertani (LB) agar or broth at 37°C. A. salmonicida strains were cultured on tryptic soy agar (Becton Dickinson) or Luria-Bertani agar plates at 18°C unless otherwise indicated. Liquid cultures of A. salmonicida were grown in tryptic soy broth (TSB; Becton Dickinson). For growth in Ca2+-rich medium, TSB was supplemented with 5 or 10 mM CaCl2. When indicated, antibiotics were added to the culture media at the following final concentrations: for E. coli, 100 μg of ampicillin/ml, 20 μg of tetracycline/ml, and 50 μg of kanamycin/ml; for A. salmonicida, 20 μg of rifampin/ml, 50 μg of ampicillin/ml, and 40 μg of kanamycin/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Properties | Source or reference |
|---|---|---|
| A. salmonicida | ||
| JF2267 | Virulent isolate; acrV+aopB+ | 4 |
| JF2646 | Rifr derivative of JF2267 | 6 |
| JF2684 | ΔacrV::Km; derived from JF2646 | This study |
| JF2724 | ΔaopB::Km; derived from JF2646 | This study |
| JF2953 | ΔaopB::Km/pMMB66HEaopB+; derived from JF2724 | This study |
| JF2956 | ΔacrV::Km/pMMB66HEacrV+; derived from JF2684 | This study |
| E. coli | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB laclqZΔM15 Tn10 (Tetr)] | 5 |
| BL21(DE3) | F′ dcm ompT hsdS(rB− mB−) gal λ(DE3) | 29 |
| S17-1 | thi pro hsdR hsdM+ recA [RP4 2-Tc::Mu-Km::Tn7 (Tpr Smr) Tra+] | 28 |
| Plasmids | ||
| pBSK | Bluescript II SK−; cloning and sequencing | Stratagene |
| pGEM-T | Cloning vector | Promega |
| pETHIS-1 | Expression vector | Novagen |
| pSSVI186 | Kanamycin resistance determinant on 1.3-kb PstI fragment from Tn903 | 33 |
| pSUP202sac | Mobilizable suicide vector; confers SucS (Mob+sacRB+ Tetr) | 32 |
| pMMB66HE | Broad-host-range expression vector (Ampr) | 10 |
DNA isolation, manipulation, and PCR amplification.
Plasmids were routinely prepared from E. coli strains using the QIA Prep Spin Miniprep kit (Qiagen) according to the supplied instructions. Total DNA from A. salmonicida was extracted by the guanidium hydrochloride method (23).
Primer walking was carried out using the Vectorette system (Genosys) according to the manufacturer's instructions.
PCR amplification was performed with either a PE9600 or PE2400 automated thermocycler with MicroAmp tubes (Applied Biosystems). The reactions were carried out in a 50-μl reaction mixture containing 10 mM Tris-HCl, pH 8.3; 1.75 mM MgCl2; 50 mM KCl; 170 μM (each) dATP, dCTP, dGTP, and dTTP; 0.25 μM forward and reverse primers; 2.5 U of DNA polymerase containing proofreading capability (Expand Long Template PCR System; Roche Diagnostics); and ∼20 ng of template DNA. The PCR conditions were as follows: 3 min at 94°C, followed by 40 cycles of 30 s at 94°C, 30 s at the primer-annealing temperature, and up to 4 min at 68°C. An extension step of 5 min at 68°C was carried out following the last cycle in order to ensure full-length synthesis of the fragments.
DNA sequencing and sequence data analysis.
DNA sequencing was performed with the dRhodamine Terminator cycle-sequencing kit (Applied Biosystems) according to the manufacturer's protocol using either T3 and T7 primers or custom-synthesized internal primers (Microsynth). Details of all oligonucleotide sequences used are available upon request. All sequences were determined on both strands. Reaction products were analyzed on a Prism 3100 Genetic Analyzer (Applied Biosystems).
Sequence alignment and editing were performed using the software Sequencher (Gene Codes Corp.). Comparisons of DNA sequences and their corresponding amino acid sequences with EMBL-GenBank and National Biomedical Research Foundation databases were performed using BLAST (1). The molecular mass and theoretical isoelectric point (pI) of the TTSS proteins were calculated using ProtParam (11).
Marker replacement mutagenesis.
The aopB and acrV genes were inactivated by marker replacement mutagenesis utilizing the kanamycin cassette from pSSVI186 (33). In the case of acrV, a 507-bp fragment from the gene was excised using the restriction enzymes BglII and AatII (Roche Diagnostics) and replaced with the kanamycin cassette that had been previously excised on a 1.3-kb BglII-AatII fragment. For aopB, the kanamycin cassette from pSSVI186 was excised and blunt ended using S1 nuclease (Promega) and subsequently ligated into the two NheI sites within the aopB gene, thereby removing a 627-bp fragment. The inactivated acrV or aopB and flanking genes were cloned into the mobilizable suicide vector pSUP202sac (32). The resulting plasmids, pSUP202sac-aopB::Km and pSUP202sac-acrV::Km, were transformed into E. coli S17-1 (28) for subsequent conjugation into A. salmonicida. In order to provide a means for selection against E. coli, spontaneous rifampin-resistant clones of A. salmonicida subsp. salmonicida JF2267 were isolated following growth of the organism on rifampin agar plates (40 μg/ml) for two passages. A single clone was selected (strain JF2646) (6) and filter mated (28) with E. coli S17-1 (carrying either pSUP202sac-aopB::Km or pSUP202sac-acrV::Km) for 3 days at 15°C. Double-crossover mutants were selected for directly by growth on tryptic soy agar plates containing 15% (wt/vol) sucrose, 40 μg of kanamycin/ml, and 20 μg of rifampin/ml at 15°C for 7 days. PCR was then used to ensure that the correct mutations were present in A. salmonicida.
In order to complement the ΔaopB and ΔacrV mutations, each gene was amplified by PCR before being cloned into the broad-host-range plasmid pMMB66HE (10). The resulting plasmids were transformed into E. coli strain S17-1 and then introduced into A. salmonicida strain JF2684 or strain JF2724 by conjugation as described above. Clones containing the recombinant plasmids were selected for on media containing 50 μg of ampicillin/ml and 40 μg of kanamycin/ml, and the presence of the wild-type (wt) genes was verified by PCR. Expression of the cloned genes was induced by the addition of isopropyl-β-d-thiogalactoside to a final concentration of 1 mM.
Fish cell infections with A. salmonicida.
Epithelioma papulosum cyprini cells (EPC cells; epitheloid cells cultured from carp skin [ECACC 93120820]) and rainbow trout (Oncorhynchus mykiss) gonad cells (RTG-2 cells [ATCC CCL-55]) were grown in 75-cm2 tissue culture flasks (Techno Plastic Products AG) at 20°C in minimum essential medium (GibcoBRL) supplemented with 2 mM l-glutamine, 1× nonessential amino acids, 3 g of sodium bicarbonate/liter, and 10% fetal bovine serum. One day before infection, the cells were trypsinized and seeded in 24-well culture plates. Monolayered EPC cells (8 × 105 [6 × 105 for RTG-2 cells] per 2-cm2 well in 1 ml of supplemented medium) were infected with A. salmonicida cells suspended in phosphate-buffered saline (PBS), pH 7.4, at a multiplicity of infection of 20:1 or 2:1 (bacteria to fish cells). The addition of PBS, pH 7.4, to fish cells was used as a negative control in all assays. In order to assay the protective effect of anti-AcrV antibodies, the bacterial cells were incubated with immunoglobin G (IgG) purified from preimmune or anti-AcrV immune serum for 30 min at 18°C prior to infection of the fish cells. Five hours following infection at 18°C, all of the cells were photographed under a green-filtered phase-contrast microscope (Axiovert 100; Zeiss).
AexT translocation.
To investigate the presence of AexT in the eukaryotic cytosol following infection with A. salmonicida, all cells (bacterial and fish cells) were manually scraped from the wells using cell scrapers (Nunc) and were separated from the culture medium by centrifugation for 5 min at 21,000 × g in a benchtop microcentrifuge at 4°C. The cell pellets were washed once with 10 mM Tris-HCl (pH 7.0)- 200 mM NaCl- 5 mM EDTA- 10% glycerol before being resuspended in the same buffer containing 0.1% Triton X-100 and 1 mM phenylmethylsulfonyl fluoride. The cells were then incubated on ice for 10 min and centrifuged to separate the Triton-insoluble and -soluble fractions.
Production of polyclonal anti-AcrV antibodies.
To generate polyclonal antibodies directed against AcrV, we first amplified the acrV gene using the primers AsacrVt-R (atgcggccgcAAATTGCGCCAAGAATGTCG) and AsacrVt-L (gggaattcGATGAGCACAATCCCTGACTAC) (nucleotides in lowercase are extensions containing recognition sites for the restriction enzymes NotI and EcoRI, respectively). The resulting PCR product was cloned into the pGEM-T vector (Promega) and transformed into E. coli strain XL-1 Blue. Recombinant plasmids were then digested with EcoRI and NotI, and the DNA fragments were inserted into the expression vector pETHIS-1 (27). The cloned insert was sequenced to ensure correct fusion with the vector's poly-His codons before being transformed into E. coli BL21(DE3) cells (29) for expression. Induction and subsequent purification of the recombinant protein was performed as described previously (4). Polyclonal antibodies against AcrV were obtained by immunization of a rabbit with purified AcrV-His as described for other polyhistidine-tailed proteins (3). Preimmune sera and anti-AcrV antisera were decomplemented for 30 min at 56°C before the isolation of IgG. IgG was isolated using HiTrap affinity columns (Amersham Pharmacia Biotech) according to the manufacturers' instructions. The proteins were then precipitated using ammonium sulfate and dialyzed overnight against 0.1 M NaHCO3- 0.25 M NaCl, pH 8.3.
SDS-PAGE and immunoblot analyses.
Proteins were separated on 8% or 12% acrylamide gels by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) according to the method of Laemmli (14). Once separated, the proteins were electroblotted onto nitrocellulose membranes (Bio-Rad Laboratories). The membranes were blocked overnight in 1% milk buffer. In order to detect AexT or AcrV, the membranes were incubated with rabbit polyclonal anti-AexT or anti-AcrV IgG (4) (diluted 1:1,000 in milk buffer), followed by incubation with a phosphate- or peroxidase-labeled conjugate (goat anti-rabbit IgG heavy and light chains [Kirkegaard and Perry]) diluted 1:2,000 in milk buffer. The proteins were visualized using the chromogenic substrate 5-bromo-4-chloro-3-indolylphosphate- nitroblue tetrazolium (Sigma) or detected by enhanced chemiluminescense using the LumiGlow regant (Kirkegaard and Perry) according to the supplied instructions.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this communication have been submitted to the EMBL Nucleotide Sequence Database under the accession numbers AJ516008 and AJ516009.
RESULTS
Nucleotide sequence analysis.
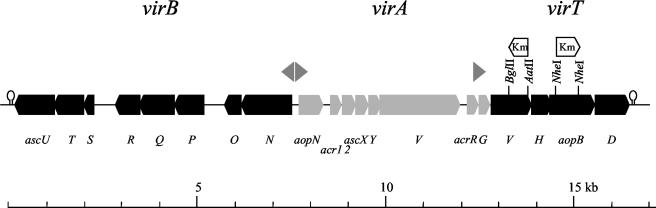
Eight complete ORFs that are homologous to the TTSS genes comprising the virA locus of Yersinia species (6) were recently identified in a virulent field strain of A. salmonicida (strain JF2267) (4) (Fig. 1). These genes were identified following screening of the bacterium with a probe against the yscV gene (formerly called lcrD) of Yersinia entercolitica, a gene that encodes the archetype for a family of inner membrane proteins found in every known TTSS. Using primer walking, we have now cloned and sequenced regions both directly upstream and downstream of the virA locus. The results of this work have led to the identification of a further 12 ORFs which exhibit high sequence similarity to TTSS genes of Yersinia species and P. aeruginosa (Fig. 1).
FIG. 1.
Genetic map of the type III secretion genes found in A. salmonicida strain JF2267. The solid boxes represent genes identified in this study. Potential promoter sequences, represented by arrowheads, are found preceding the ascN, aopN, and acrG genes. (In the case of ascN, two potential promoter sequences have been identified.) The restriction sites used in the marker replacement of the acrV and aopB genes and the direction of the kanamycin cassette replacement are indicated. All genes have been designated based solely on sequence similarity to Yersinia (ysc, lcr, and yop) TTSS genes.
Eight of the newly identified ORFs are homologous to genes comprising the virB locus of Yersinia species. In A. salmonicida strain JF2267, these genes are found upstream of the virA locus (Fig. 1) and are predicted to encode structural components of the type III secretion apparatus. In keeping with the nomenclature used for their Yersinia counterparts (ysc for yersinia secretion), we have given these genes the designation asc (for aeromonas secretion). This gene locus is preceded by two potential promoter regions directly downstream of the ascN gene (Fig. 1). The first is situated ∼75 bp from the ascN ATG start codon and contains putative −10 (TGAAATC) and −35 (TTTTCT) boxes. The second is found 95 bp from the start codon and also contains potential −10 (TAAAAAG) and −35 (TTTTTA) boxes. We have also identified a potential stem-loop structure upstream of the ascU gene that could function as a transcription termination site (ΔG, −5.2 kcal/mol) (Fig. 1).
Downstream of the virA locus, we have identified an additional four ORFs that are homologous to the genes comprising the Yersinia lcrGHV-yopBD locus. Again, in keeping with established nomenclature, we have designated these A. salmonicida genes acr (for aeromonas calcium response) and aop (for aeromonas outer proteins). This locus also contains a potential promoter sequence positioned upstream of acrG (6) and a potential stem-loop structure located downstream of aopD (ΔG, −12.3 kcal/mol) (Fig. 1). All of the members of the lcrGHV-yopBD locus in Yersinia are thought to play a role in the process of translocating effector molecules into the eukaryotic cytosol. For this reason, we have assigned this gene locus the designation virT (for translocation).
The relevant characteristics of the predicated A. salmonicida TTSS proteins (based on the ORFs identified) are given in Table 2. The greatest degree of sequence similarity was found between these proteins and their corresponding Yersinia homologues. The exceptions to this were the aopB, aopD, ascP, and ascR genes, which display marginally higher sequence similarity to TTSS genes of P. aeruginosa (on the order of 2 to 3% higher identity and similarity). The acrH gene also possesses greater sequence homology with its P. aeruginosa counterpart; however, in this case, the differences are greater (64% identity and 76% similarity to pcrH of P. aeruginosa versus 57% identity and 79% similarity to lcrH of Yersinia).
TABLE 2.
Characteristics of predicted A. salmonicida TTSS proteins
| A. salmonicida protein | No. of residues | Theoretical pI | Molecular mass (kDa) | Yersinia homologue | % Identity | % Similarity |
|---|---|---|---|---|---|---|
| AcrV | 361 | 5.1 | 40.2 | LcrV | 37 | 53 |
| AcrH | 167 | 4.3 | 18.6 | SycD/LcrH | 57 | 79 |
| AopB | 397 | 6.7 | 41.1 | YopB | 42 | 57 |
| AopD | 298 | 8.7 | 32.5 | YopD | 41 | 55 |
| AscN | 445 | 5.8 | 14.4 | YscN | 88 | 91 |
| AscO | 133 | 11.8 | 14.3 | YscO | 47 | 64 |
| AscP | 260 | 4.8 | 28.5 | YscP | 30 | 44 |
| AscQ | 308 | 4.6 | 33.6 | YscQ | 43 | 58 |
| AscR | 217 | 5.7 | 24.2 | YscR | 77 | 83 |
| AscS | 88 | 6.0 | 9.6 | YscS | 78 | 89 |
| AscT | 262 | 5.2 | 28.5 | YscT | 56 | 67 |
| AscU | 352 | 9.0 | 39.6 | YscU | 69 | 82 |
AexT is translocated in a type III-dependent manner.
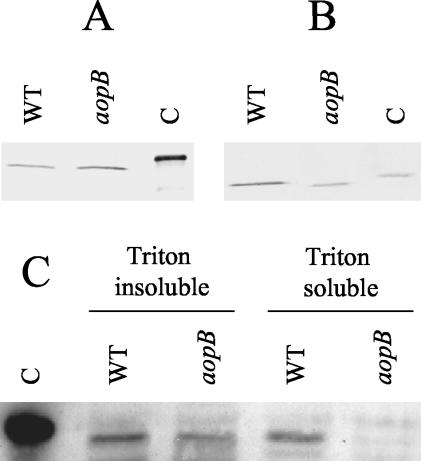
It was previously shown that secretion of the AexT protein by A. salmonicida is inhibited by inactivation of the ascV gene, indicating that the toxin is secreted via the bacterium's TTSS (6). However, there has been no evidence to indicate that the bacterium translocates this toxin directly into the cytosol of target cells. In order to address this issue, we utilized marker replacement mutagenesis to construct an isogenic ΔaopB mutant of A. salmonicida. In Yersinia, the AopB homologue, YopB, plays a role in the translocation process by assisting in the formation of pores in the eukaryotic cell membrane (12, 21). Because of this, we speculated that if AexT is translocated into the cytosol of target cells via the TTSS, this process would be inhibited in an aopB mutant. It was first necessary to ensure that the ΔaopB mutant we had constructed was still able to secrete AexT. To accomplish this, we incubated A. salmonicida strain JF2646 (a rifampin-resistant derivative of the wt A. salmonicida isolate, strain JF2267, used in the marker replacement mutagenesis procedure; see Materials and Methods) or the ΔaopB derivative, A. salmonicida strain JF2724, overnight in liquid culture. The supernatant was then collected, and the proteins were separated by SDS-PAGE. Western blotting was performed using monospecific polyclonal antibodies against AexT. AexT was found in the culture supernatants of both strain JF2646 and strain JF2724, indicating that marker replacement of the aopB gene did not interfere with the bacterium's ability to secrete AexT (Fig. 2A). As a further control, we also examined the culture supernatant for the presence of AcrV, a protein thought to be involved in the translocation process. As with AexT, secretion of AcrV was not affected by the marker replacement mutation in aopB (Fig. 2B).
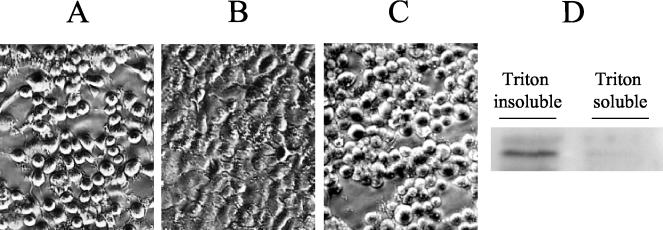
FIG. 2.
Type III-dependent translocation of AexT. (A and B) Bacterial cultures were grown overnight in TSB medium, and the presence of the proteins AexT (A) and AcrV (B) was analyzed by SDS-PAGE and Western blotting using polyclonal antibodies. WT, A. salmonicida strain JF2646 (aopB+); aopB, isogenic ΔaopB mutant, strain JF2724. Lane C in panel A contains recombinant AexT-His protein, whereas lane C in panel B contains recombinant AcrV-His. (C) After infection of EPC cells with A. salmonicida strain JF2646 (WT) or A. salmonicida strain JF2724 (aopB), cytosolic fractions of EPC were prepared using Triton X-100 lysis. The Triton-insoluble (containing bacterial cells, unlysed EPC cells, and nuclei) and -soluble (containing EPC cytosolic proteins and membrane proteins) fractions were analyzed by SDS-PAGE and Western blotting using polyclonal anti-AexT antibodies. Lane C contains recombinant AexT-His protein.
Having shown that the ΔaopB mutant, JF2724, was still able to secrete AexT, we inoculated cultures of EPC cells with either the wt strain A. salmonicida strain JF2646 or strain JF2724 at a multiplicity of infection of 20:1. Following inoculation with the bacteria, the EPC cell cultures were incubated for 5 h at 18°C. After this time, all of the cells were scraped from the wells and separated from the culture supernatant by centrifugation. The cell pellet was washed once, and then cytosolic fractions of the EPC cells were prepared using the detergent Triton X-100. The Triton X-100-soluble fraction (containing cytosolic eukaryotic proteins, as well as membrane proteins) was then separated from the insoluble fraction (containing bacteria, unlysed eukaryotic cells, and nuclei). The proteins present in both fractions (Triton X-100 soluble and insoluble) were separated by SDS-PAGE, and Western blotting was performed using monospecific polyclonal anti-AexT antibodies. The results are presented in Fig. 2C. AexT can be detected in both the Triton X-100-insoluble and -soluble fractions of EPC cells that were infected with the wt A. salmonicida strain JF2646. This result suggests that AexT is translocated into the cytosol of the EPC cells (eukaryotic cells). In contrast, when the EPC cells were infected with the ΔaopB mutant, strain JF2724, AexT was present only in the Triton-insoluble fraction; it was not found in the soluble fraction, which contained cytosolic EPC proteins (Fig. 2C), indicating that the aopB gene plays a role in the translocation of AexT. Therefore, translocation of AexT occurs in a type III-dependent manner.
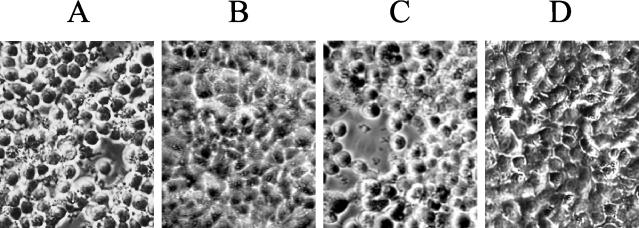
We were interested to see whether preventing AexT translocation by inactivation of aopB affected the virulence of the bacterium. EPC cells were again inoculated with either the wt strain, JF2646, or the ΔaopB mutant, strain JF2724. Five hours following infection, the cells were examined using phase-contrast microscopy. Cells infected with the wt bacterium (JF2646) had become rounded and detached from the plastic support (Fig. 3A). In contrast, cells infected with the ΔaopB mutant had not displayed any significant morphological changes (Fig. 3B). trans complementation of the ΔaopB mutant with plasmid pMMB66HEaopB+, in which the wt aopB gene is under the control of the tac promoter, restored cytotoxicity (strain JF2953 [Fig. 3C]). Cells that were inoculated with PBS only were used as a negative control in the assay and displayed no morphological changes (Fig. 3D).
FIG. 3.
Cytotoxicity of A. salmonicida to EPC cells. Phase-contrast micrographs of EPC cells 5 h after infection with A. salmonicida strain JF2646 (aopB+) (A); the isogenic ΔaopB mutant, strain JF2724 (B); strain JF2953 (ΔaopB/pMMB66HEaopB+) (C); or PBS only (D). The multiplicity of infection was 20:1.
Role of AcrV in AexT secretion and translocation.
After having established that AexT is translocated into the cytosol of EPC cells, we were interested to see how the translocation process is affected by inactivation of the acrV gene. This gene is predicated to encode AcrV, a homologue of the LcrV protein of Yersinia species. This protein is of considerable interest, as it has long been known that antiserum raised against LcrV provides mice with passive immunity against Y. pestis (15, 20, 25, 30, 31). More recent studies have suggested that LcrV plays a role in the translocation of effector molecules and that anti-LcrV antibodies interfere with this process (22, 26).
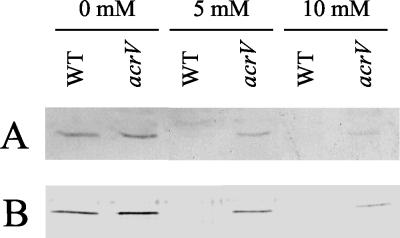
We again utilized marker replacement mutagenesis and constructed an isogenic ΔacrV mutant in A. salmonicida strain JF2646. We then examined how this mutation affects the expression and secretion of AexT under low-calcium conditions. A. salmoncidica strain JF2646 (wt) and strain JF2684 (ΔacrV mutant) were grown overnight in liquid culture in the presence or absence of millimolar concentrations of CaCl2. The cells and culture supernatants were separated by centrifugation, and the proteins were separated by SDS-PAGE. Western blotting was then performed, again using polyclonal antibodies against AexT. The results are shown in Fig. 4. As expected, AexT can be found in both the cell pellet (Fig. 4A) and the culture supernatant (Fig. 4B) of the wt cells grown under low-calcium conditions. However, expression and secretion was prevented when the medium was supplemented with calcium ions. In contrast, however, AexT was found in the cell pellet (Fig. 4A) and culture supernatant (Fig. 4B) of the ΔacrV mutant in the absence or presence of calcium ions. Even CaCl2 concentrations of 10 mM did not prevent AexT expression or secretion, although the amount of toxin produced was reduced as the calcium concentration rose.
FIG. 4.
Type III-dependent expression and secretion of AexT. Expression (A) and secretion (B) of AexT under low-calcium conditions are shown. Bacterial cultures were grown overnight in TSB medium supplemented with millimolar concentrations of CaCl2 as indicated. The presence of AexT in the cell pellets (A) and culture supernatants (B) was analyzed by SDS-PAGE and Western blotting using polyclonal anti-AexT antibodies. WT, A. salmonicida strain JF2646 (acrV+); acrV, isogenic ΔacrV mutant, strain JF2684.
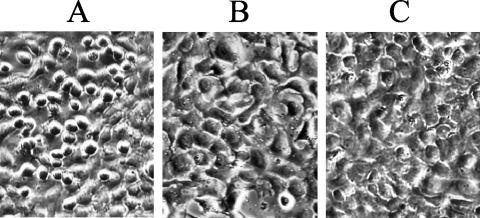
We next examined the cytotoxicity of the ΔacrV mutant. EPC cells were infected with either A. salmonicida strain JF2646 (wt), strain JF2684 (ΔacrV mutant), or strain JF2956 (in which the ΔacrV mutation is complemented in trans with plasmid pMMB66HEacrV+), and the cultures were incubated for 5 h at 18°C. As expected, cells infected with the wt bacterium displayed significant rounding and retraction (Fig. 5A), whereas cells infected with the ΔacrV mutant displayed no morphological changes despite the high numbers of bacterial cells in the culture (Fig. 5B). trans complementation of ΔacrV restored the bacterium's cytotoxic effect (Fig. 5C). Infected cells were harvested and fractionated using the Triton X-100 procedure. Again, the fractions were analyzed by SDS-PAGE and Western blotting. AexT was detected in the Triton X-100-insoluble and -soluble fractions when EPC cells were infected with the wt strain JF2646. However, the toxin was detected only in the insoluble fraction (containing bacterial cells and unlysed EPC cells) when the ΔacrV mutant was used to infect the EPC cells (Fig. 5D). These results confirm that expression of AexT is not prevented when the acrV gene is inactivated but indicate that AexT cannot be translocated under these conditions. Therefore, AcrV, like AopB, plays a role in AexT translocation.
FIG. 5.
Role of AcrV in A. salmonicida cytotoxicity. (A to C) Phase-contrast micrographs of EPC cells 5 h after infection with A. salmonicida strain JF2646 (acrV+) (A), isogenic ΔacrV mutant strain JF2684 (B), or strain JF2953 (ΔacrV/pMMB66HEacrV+) (C). The multiplicity of infection was 20:1. (D) After infection with strain JF2684, cytosolic fractions of EPC cells were prepared using Triton X-100 lysis. The Triton-insoluble and -soluble fractions were analyzed by SDS-PAGE and Western blotting using polyclonal anti-AexT antibodies.
Anti-AcrV antibodies protect against A. salmonicida.
Having found that inactivation of acrV prevents AexT translocation and significantly reduces the bacterium's pathogenicity, we were interested in determining whether masking of the AcrV protein by use of anti-AcrV antibodies could serve to reduce the toxicity of the bacterium. We repeated the infection assay using EPC cells; however, we preincubated wt A. salmonicida strain JF2267 with IgG purified from preimmune serum (Fig. 6A) or with IgG purified from anti-AcrV antiserum (Fig. 6B). The results indicate that preincubation with the anti-AcrV IgG offered a significant degree of protection against the toxic effects of the bacterium when a multiplicity of infection of 2:1 was used. Five hours following infection, the EPC cells infected with the bacteria incubated in the presence of preimmune IgG had begun to undergo cell rounding (Fig. 6A), whereas those incubated with the anti-AcrV IgG remained unchanged (Fig. 6B). This protection, however, waned over time, as all cultures incubated overnight showed significant rounding up and cell lysis. In addition, very little protective effect was seen when the multiplicity of infection was increased 10-fold to 20:1.
FIG. 6.
Anti-AcrV antibodies protect against A. salmonicida cytotoxicity. (A and B) Phase-contrast micrographs of EPC cells 5 h following infection with the wt isolate A. salmonicida strain JF2267 in the presence of preimmune IgG (A) or anti-AcrV IgG (B). (C) Cells treated with anti-AcrV IgG in PBS only as a control. The multiplicity of infection was 2:1.
In order to see whether the protective effect could be extended to a different host cell, the experiment was repeated using RTG-2 cells, which, unlike EPC cells (originating from carp epithelium), are derived from rainbow trout gonad cells. The results obtained from this experiment were similar to those obtained with EPC cells; the anti-AcrV IgG provided a significant degree of protection against infection with the bacterium; however, IgG purified from preimmune serum did not (not shown).
DISCUSSION
Studies of the pathogenicity of A. salmonicida have long been hampered by an inability to identify major virulence determinants produced by the bacterium. Several studies have found that knockout mutations of potential virulence factors do not reduce the pathogenicity of the organism or have only minimal effects. For example, marker replacement mutagenesis of either the satA or aspA gene (encoding the glycerophospholipid-cholesterol acyltransferase GCAT and the serine protease AspA, respectively) did not result in any significant decrease in virulence compared to that of the wt bacterium (32). Inactivation of the tapA gene (ecoding a protein with high homology to type IV pilus subunits) resulted in only a 2.5-fold difference in the 50% lethal dose compared to the wt (19). The identification of a functional TTSS, which plays a central role in the pathogenesis of many gram-negative organisms, is therefore an important step in the search for significant virulence determinants of A. salmonicida.
In this communication, we have presented results that indicate that the TTSS of A. salmonicida is responsible for the translocation of the ADP-ribosylating toxin AexT. Inactivation of the yopB homologue (which in Yersinia has been shown to be involved in type III-dependent translocation [12, 21]) prevents translocation of AexT into the eukaryotic cytosol. Furthermore, when translocation of AexT is prevented in this manner, the pathogenicity of the bacterium to fish cells is significantly reduced. Previous studies have found that AexT itself is required for cytotoxicity of A. salmonicida to cultured fish cells; infection of RTG-2 cells with an isogenic knockout mutant of aexT does not result in the characteristic retraction and rounding up usually seen following infection with the wt bacterium (4). This result suggests that AexT is a virulence determinant of A. salmonicida. Therefore, one can speculate that the inability of the ΔaopB mutant (constructed in this study) to cause damage to fish cells is a direct result of its inability to translocate the AexT toxin. However, there may well be other effector molecules translocated by the A. salmonicida TTSS. In Yersinia, the TTSS is responsible for the translocation of six different effector molecules (YopH, YopE, YopT, YpkA/YopO, YopP/YopJ, and YopM) that act to inhibit signaling cascades, prevent phagocytosis, and counteract the proinflammatory response (for a recent review, see reference 8). In the case of P. aeruginosa, four such effector molecules are known (ExoS, ExoT, ExoU, and ExoY). It is therefore, reasonable to speculate that AexT is not the sole effector molecule targeted by the TTSS of A. salmonicida. Consistent with this hypothesis is the recent identification of a potential yopPJ homologue on a small plasmid found in A. salmonicida strain JF2267 (GenBank accession no. AJ508382). Work has yet to be done to determine if this and indeed other type III effector molecules are expressed by A. salmonicida and, if so, what role these molecules play in the disease process.
We have presented evidence that indicates that the acrV gene of A. salmonicida plays an important role in the regulation of AexT expression and secretion. Marker replacement mutagenesis of the acrV gene results in AexT expression and secretion in both the absence and presence of Ca2+, indicating that this mutation causes deregulation of the TTSS. The observation that the ΔacrV mutant secretes AexT under low-calcium conditions is not unexpected, as previous studies carried out with Yersinia indicate that knockout mutations of lcrV (the acrV homologue) do not prevent the secretion of effector Yop proteins (24). Pettersson et al. (22) also found that an lcrV mutant secreted considerable amounts of all Yop proteins when grown in calcium-depleted medium at 37°C; however, the amounts of Yop proteins secreted were smaller than those secreted by the wt strain. In contrast, our results indicate that secretion of AexT under low-calcium conditions is slightly higher in the ΔacrV mutant than in the wt cells (Fig. 4B). Furthermore, the isogenic ΔacrV mutant still secretes appreciable amounts of AexT, even when calcium is present at high concentrations. Such a response has also been observed in P. aeruginosa, where a pcrV mutant was found to have a calcium-blind phenotype, secreting type III proteins in the presence or absence of a calcium chelator (26).
We have found that, in addition to playing a role in the regulation of AexT secretion, AcrV also functions in the translocation of the toxin. Cytosolic fractions of EPC cells infected with the ΔacrV mutant did not contain AexT (Fig. 5D), whereas those infected with the wt bacteria did (Fig. 2C). This result is consistent with studies carried out with Yersinia indicating that knockout mutations of the acrV homologue, lcrV, prevent targeting of effector Yop proteins into the cytosol of eukaryotic cells (18, 22). Similarly, a knockout mutant of pcrV has been shown to prevent the translocation of ExoY in P. aeruginosa (26).
While it now seems clear that AcrV and its homologues play roles in the translocation of effector molecules, contradictory results exist in reference to the role of anti-LcrV antibodies. Fields et al. (9) have demonstrated that LcrV-specific antibodies (or antibody fragments) do not protect HeLa cells from the cytotoxic effect caused by translocation of the YopE cytotoxin of Y. pestis. However, analogous experiments carried out by Pettersson et al. (22) indicate that the reverse is true. In their studies with Y. pseudotuberculosis, the authors demonstrated that anti-LcrV antibodies blocked translocation of YopE. Support for these findings has come from studies with P. aeruginosa that indicate that anti-PcrV IgGs reduce the amount of ExoY translocated into the macrophage cell line J774. Yet another study, carried out with the macrophage cell line J774.A1, has shown that anti-LcrV antibodies provide protection from Y. pestis-induced cytotoxicity, presumably by blocking Yop translocation (34). Furthermore, the same study has shown that when Yop translocation is prevented in this manner, the bacterium's resistance to phagocytosis is dramactically reduced (34). Our results have shown that IgG directed against recombinant AcrV prevented the characteristic cell rounding and retraction of EPC cells normally induced by the wt strain JF2267 (aexT+), suggesting that the IgG prevents AexT translocation. However, this effect was observed only when a low multiplicity of infection (2:1) was used. Furthermore, the protective effect decreased over time, suggesting that despite the lower translocation levels, the cytotoxic effect of A. salmonicida was able to build up gradually over time.
The protective effect of anti-AcrV antibodies against A. salmonicida is of considerable interest. Infection by this bacterium causes severe losses in farm production of salmonid fish, and current vaccines used to protect against A. salmonicida display wide variations in efficacy. Furthermore, large amounts of antibiotics are often used in both open and closed waters in order to treat this disease. Recent studies with the AcrV homologue LcrV have shown that recombinant LcrV can be used to successfully immunize mice against the plague (2, 16). Similar studies have shown that immunization with recombinant PcrV provided mice with protection against lethal doses of P. aeruginosa (26). Based on these findings and our own data indicating that anti-AcrV antibodies protect against the toxic effect of A. salmonicida in vitro, we speculate that AcrV could be utilized to great effect in the production of an improved vaccine for the prevention of furunculosis in farmed fish.
Acknowledgments
We thank Guy Cornelis and Jaime Mota, University of Basel, for helpful discussions and for assistance with the cell fractionation experiments. We are also grateful to Shelia MacIntyre, University of Reading, for providing us with plasmid pSUP202sac and to Lea Lagcher, University of Berne, for cultivation of fish cells.
This work was supported by the research fund of the Institute of Veterinary Bacteriology, University of Berne.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, G. W., Jr., S. E. Leary, E. D. Williamson, R. W. Titball, S. L. Welkos, P. L. Worsham, and A. M. Friedlander. 1996. Recombinant V antigen protects mice against pneumonic and bubonic plague caused by F1-capsule-positive and -negative strains of Yersinia pestis. Infect. Immun. 64:4580-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun, M., P. Kuhnert, J. Nicolet, A. P. Burnens, and J. Frey. 1999. Cloning and characterization of two bistructural S-layer-RTX proteins from Campylobacter rectus. J. Bacteriol. 181:2501-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, M., K. Stuber, Y. Schlatter, T. Wahli, P. Kuhnert, and J. Frey. 2002. Characterization of an ADP-ribosyltransferase toxin (AexT) from Aeromonas salmonicida subsp. salmonicida. J. Bacteriol. 184:1851-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques 5:376-378. [Google Scholar]
- 6.Burr, S. E., K. Stuber, T. Wahli, and J. Frey. 2002. Evidence for a type III secretion system in Aeromonas salmonicida subsp. salmonicida. J. Bacteriol. 184:5966-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu, S., S. Cavaignac, J. Feutrier, B. M. Phipps, M. Kostrzynska, W. W. Kay, and T. J. Trust. 1991. Structure of the tetragonal surface virulence array protein and gene of Aeromonas salmonicida. J. Biol. Chem. 266:15258-15265. [PubMed] [Google Scholar]
- 8.Cornelis, G. R. 2002. The Yersinia Ysc-Yop ′type III' weaponry. Nat. Rev. Mol. Cell. Biol. 3:742-752. [DOI] [PubMed] [Google Scholar]
- 9.Fields, K. A., M. L. Nilles, C. Cowan, and S. C. Straley. 1999. Virulence role of V antigen of Yersinia pestis at the bacterial surface. Infect. Immun. 67:5395-5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fürste, J. P., W. Pansegrau, R. Frank, H. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 11.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. (Erratum, 189: 283, 1990.) [DOI] [PubMed] [Google Scholar]
- 12.Hakansson, S., K. Schesser, C. Persson, E. E. Galyov, R. Rosqvist, F. Homble, and H. Wolf-Watz. 1996. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 15:5812-5823. [PMC free article] [PubMed] [Google Scholar]
- 13.Hirono, I., and T. Aoki. 1993. Cloning and characterization of three hemolysin genes from Aeromonas salmonicida. Microb. Pathog. 15:269-282. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Lawton, W. D., R. L. Erdman, and M. J. Surgalla. 1963. Biosynthesis and purification of V and W antigen in Pasteurella pestis. J. Immunol. 91:179-184. [DOI] [PubMed] [Google Scholar]
- 16.Leary, S. E., E. D. Williamson, K. F. Griffin, P. Russell, S. M. Eley, and R. W. Titball. 1995. Active immunization with recombinant V antigen from Yersinia pestis protects mice against plague. Infect. Immun. 63:2854-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, K. K., and A. E. Ellis. 1990. Glycerophospholipid:cholesterol acyltransferase complexed with lipopolysaccharide (LPS) is a major lethal exotoxin and cytolysin of Aeromonas salmonicida: LPS stabilizes and enhances toxicity of the enzyme. J. Bacteriol. 172:5382-5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, V. T., C. Tam, and O. Schneewind. 2000. LcrV, a substrate for Yersinia enterocolitica type III secretion, is required for toxin targeting into the cytosol of HeLa cells. J. Biol. Chem. 275:36869-36875. [DOI] [PubMed] [Google Scholar]
- 19.Masada, C. L., S. E. LaPatra, A. W. Morton, and M. S. Strom. 2002. An Aeromonas salmonicida type IV pilin is required for virulence in rainbow trout Oncorhynchus mykiss. Dis. Aquat. Organ. 51:13-25. [DOI] [PubMed] [Google Scholar]
- 20.Motin, V. L., R. Nakajima, G. B. Smirnov, and R. R. Brubaker. 1994. Passive immunity to yersiniae mediated by anti-recombinant V antigen and protein A-V antigen fusion peptide. Infect. Immun. 62:4192-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neyt, C., and G. R. Cornelis. 1999. Insertion of a Yop translocation pore into the macrophage plasma membrane by Yersinia enterocolitica: requirement for translocators YopB and YopD, but not LcrG. Mol. Microbiol. 33:971-981. [DOI] [PubMed] [Google Scholar]
- 22.Pettersson, J., A. Holmstrom, J. Hill, S. Leary, E. Frithz-Lindsten, A. Von Euler-Matell, E. Carlsson, R. Titball, A. Forsberg, and H. Wolf-Watz. 1999. The V-antigen of Yersinia is surface exposed before target cell contact and involved in virulence protein translocation. Mol. Microbiol. 32:961-976. [DOI] [PubMed] [Google Scholar]
- 23.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 24.Sarker, M. R., C. Neyt, I. Stainier, and G. R. Cornelis. 1998. The Yersinia Yop virulon: LcrV is required for extrusion of the translocators YopB and YopD. J. Bacteriol. 180:1207-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato, K., R. Nakajima, F. Hara, T. Une, and Y. Osada. 1991. Preparation of monoclonal antibody to V antigen from Yersinia pestis. Contrib. Microbiol. Immunol. 12:225-229. [PubMed] [Google Scholar]
- 26.Sawa, T., T. L. Yahr, M. Ohara, K. Kurahashi, M. A. Gropper, J. P. Wiener-Kronish, and D. W. Frank. 1999. Active and passive immunization with the Pseudomonas V antigen protects against type III intoxication and lung injury. Nat. Med. 5:392-398. [DOI] [PubMed] [Google Scholar]
- 27.Schaller, A., R. Kuhn, P. Kuhnert, J. Nicolet, T. J. Anderson, J. I. MacInnes, R. P. A. M. Segers, and J. Frey. 1999. Characterization of apxIVA, a new RTX determinant of Actinobacillus pleuropneumoniae. Microbiology 145:2105-2116. [DOI] [PubMed] [Google Scholar]
- 28.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 29.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 30.Une, T., and R. R. Brubaker. 1984. Roles of V antigen in promoting virulence and immunity in Yersiniae. J. Immunol. 133:2226-2230. [PubMed] [Google Scholar]
- 31.Une, T., R. Nakajima, and R. R. Brubaker. 1987. Roles of V antigen in promoting virulence in Yersiniae. Contrib. Microbiol. Immunol. 9:179-185. [PubMed] [Google Scholar]
- 32.Vipond, R., I. R. Bricknell, E. Durant, T. J. Bowden, A. E. Ellis, M. Smith, and S. MacIntyre. 1998. Defined deletion mutants demonstrate that the major secreted toxins are not essential for the virulence of Aeromonas salmonicida. Infect. Immun. 66:1990-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viret, J. F. 1993. Meganuclease I-SceI as a tool for the easy subcloning of large DNA fragments devoid of selection marker. BioTechniques 14:325-326. [PubMed] [Google Scholar]
- 34.Weeks, S., J. Hill, A. Friedlander, and S. Welkos. 2002. Anti-V antigen antibody protects macrophages from Yersinia pestis-induced cell death and promotes phagocytosis. Microb. Pathog. 32:227-237. [DOI] [PubMed] [Google Scholar]
- 35.Whitby, P. W., M. Landon, and G. Coleman. 1992. The cloning and nucleotide sequence of the serine protease gene (aspA) of Aeromonas salmonicida ssp. salmonicida. FEMS Microbiol. Lett. 78:65-71. [DOI] [PubMed] [Google Scholar]