Abstract
Herpes simplex virus 1 (HSV-1) regulatory protein ICP27 has been reported to bind viral RNA and to interact with the nuclear export adaptor Aly/REF and the major cellular mRNA nuclear export receptor TAP/NXF1. Using in situ hybridization and in vitro export assays, we show here that poly(A)+ RNA was retained in the nucleus of cells infected with viral ICP27 mutants that either cannot bind RNA or that do not interact with TAP/NXF1. Microarray analysis of nuclear and cytoplasmic RNA fractions demonstrated that efficient export of the majority of viral transcripts requires that ICP27 be able to bind RNA and to interact with TAP/NXF1. We conclude that ICP27 is the major export adaptor for HSV-1 mRNA and that it links bound transcripts to the TAP/NXF1 export receptor.
Herpes simplex virus 1 (HSV-1) regulatory protein ICP27 is a multifunctional protein that acts at both the transcriptional and posttranscriptional level. Among its posttranscriptional activities, ICP27 has been implicated in the export of viral mRNA because it has been shown to bind several viral transcripts both in the nucleus and cytoplasm of infected cells (30) and to interact with the cellular mRNA export proteins Aly/REF (3, 4, 13) and TAP/NXF1 (3). ICP27 binds RNA through an RGG box RNA binding motif (23, 30), which is required for RNA binding in vivo (30) and which is sufficient for RNA binding in vitro (23). ICP27 also encodes three predicted KH domains (34); however, these domains have not been shown to be required for RNA binding by ICP27.
ICP27 is predominantly nuclear at early times after infection; however, beginning about 4 h after infection, ICP27 shuttles between the nucleus and cytoplasm (3, 4, 24, 26, 33). The export of ICP27 to the cytoplasm occurs through the TAP/NXF1 export receptor, with which ICP27 interacts directly (3). Both the N and C termini of ICP27 are involved in the interaction with TAP/NXF1, and mutations in these regions confine ICP27 to the nucleus (3). ICP27 also interacts with the cellular mRNA adaptor protein Aly/REF (4, 13), which interacts directly with TAP/NXF1 (29, 37). The region required for ICP27's interaction with Aly/REF spans the nuclear localization signal and does not overlap with the N-terminal leucine-rich region or the C-terminal zinc-finger-like region, both of which are required for interaction with TAP/NXF1, yet, surprisingly, ICP27 export to the cytoplasm cannot be mediated through Aly/REF. ICP27 mutants with lesions in the N or C terminus cannot shuttle to the cytoplasm even though these mutants can bind to Aly/REF (3). Thus, export of ICP27 results from its interaction with TAP/NXF1.
Here, we asked whether ICP27 is required for the export of the majority of HSV-1 transcripts. By using viral ICP27 mutants with deletions in the regions required for RNA binding or interaction with TAP/NXF1, we found that at late times after infection, nuclear export of bulk poly(A)+ RNA and the majority of HSV-1 mRNAs required an ICP27 protein capable of binding RNA and interacting with TAP/NXF1.
MATERIALS AND METHODS
Cells, viruses, and recombinant plasmids.
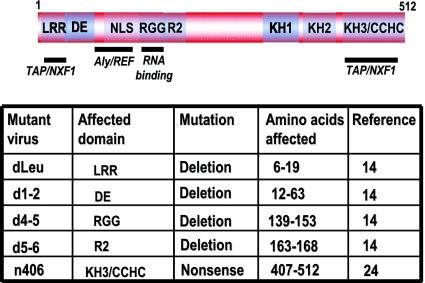
HeLa R19 cells were grown on minimal essential medium containing 10% newborn calf serum. HSV-1 strains KOS and 27-LacZ were previously described (32). Mutant HSV-1 strains dLeu, d1-2, d4-5, d5-6, and n406 were generously provided by Steve Rice (15, 27, 28). Green fluorescent protein (GFP) was expressed from the plasmid pEGFP-C1 (Clontech). pEGFP-TAP was kindly provided by Eliza Izaurralde (1), and pEGFP-TAPΔC was described previously (3).
Virus infection and transfection procedures.
Cells were infected with wild-type or mutant virus for 8 h at a multiplicity of infection of 10 and incubated at 37°C. Transfection of plasmid DNA was performed by using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol. Cells were infected 24 h after transfection.
In situ hybridization.
Cells grown on coverslips in 24-well dishes were fixed in 3.7% formaldehyde after infection and then overlaid with 70% EtOH and stored at 4°C. For poly(A)+ RNA hybridizations, cells were rehydrated for 5 min at room temperature in 15% formamide in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and then overlaid with 40 μl hybridization solution (15% formamide, 10% dextran sulfate, 40 μg yeast tRNA, 0.02% bovine serum albumin, 5 ng biotinylated oligo[dT] [Promega], RNAsin, 0.5 M dithiothreitol, 2× SSC) and incubated at 37°C for 90 min. Cells were washed twice for 30 min at 37°C in wash solution (15% formamide, 2× SSC, 0.1% NP-40) and then immunostained. The probe for glycoprotein C (gC) RNA (5′-CGTGGAGGTGGGCTTGGGGGTTGTT-3′) was designed and synthesized by Exiqon and contains a biotin tag at the 5′ end and locked nucleic acids. Hybridization with this probe was performed as described above except with 60% formamide, 50 ng probe/well, and overnight hybridization. When indicated, cells were treated with 10 μg RNase A for 30 min at 37°C prior to hybridization.
Immunofluorescence microscopy.
Cells were grown on coverslips and transfected and/or infected as indicated in the figure legends. At 8 h after infection, cells were fixed in 3.7% formaldehyde and immunofluorescent staining was performed as previously described (8, 16, 31) with SC35 hybridoma supernatant (9) or monoclonal antibodies to ICP27 (catalog no. P1119; Virusys), ICP4 (catalog no. P1114; Virusys), or Aly/REF (11G5; Sigma). Biotinylated in situ hybridization probes were detected with streptavidin-Texas red antibody (GE Healthcare). When indicated, cells were treated with 10 μg/ml actinomycin D at 7 h after infection for 1 h before fixation. GFP fluorescence was viewed directly. Cells were viewed with a Zeiss Axiovert S100 microscope, and images were pseudocolored and merged using Adobe Photoshop. Cells in all immunofluorescence images shown are representative of more than 75% of the cells visualized and are shown at a higher magnification to allow observation of intracellular structures.
In vitro export assay.
Cells were grown on coverslips and infected for 8 h. In vitro nuclear export assays were performed as previously described (3). At each time point as indicated below, cells were rinsed with phosphate-buffered saline and fixed in 3.7% formaldehyde.
Microarray analysis and quantitative real-time PCR (real-time qPCR).
Cell monolayers were infected for 8 h, and RNA was harvested with Trizol (Invitrogen) from either whole-cell lysates (total RNA) or nuclear and cytoplasmic fractions (40). Fractionation was confirmed by Western blot analysis of β-tubulin and histone proteins. RNA was extracted with chloroform and precipitated in ethanol. Poly(A)+ RNA was selected by using a MicroPoly(A)Purist kit (Ambion) according to the manufacturer's instructions. Synthesis of cDNA and subsequent hybridization to arrays of HSV-1 transcript-specific probes was performed as previously described (38). For real-time PCR analysis, each RNA sample (200 ng) was reverse transcribed by using an iScript cDNA synthesis kit (Bio-Rad) and amplified in iQ Sybr green Supermix (Bio-Rad) using primers specific to VP16 (F5′GTACGCCGAGCAGATGATG-3′ and R5′-TCCGTTGACGAACATGAAGG-3′), gB (F5′-GCAGGAAGTGGACGAGATG-3′ and R5′-GTACGCCGAGCAGATGATG-3′), and gC (F5′-GTTCACCACCGTCTCTACC-3′ and R5′-CGAGAACGTCACGGAGTC-3′). The relative quantities of each transcript were determined based on a standard curve generated from plasmids encoding each gene.
RESULTS
Poly(A)+ RNA is retained in the nucleus in cells infected with viral ICP27 mutants that cannot bind RNA or interact with TAP/NXF1.
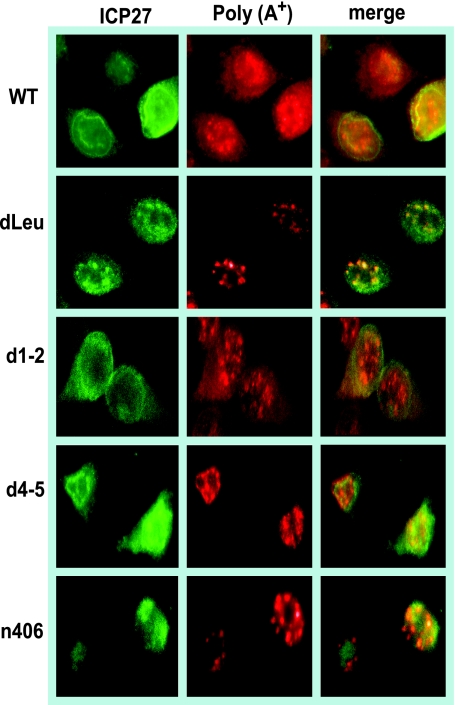
To investigate the export of mRNA during HSV-1 infection, in situ hybridization was performed on cells that were infected with wild-type HSV-1 and with ICP27 mutants with mutations in functional regions of the protein that have been shown to affect its ability to bind RNA or to be exported to the cytoplasm. The regions of ICP27 involved in RNA export are shown in Fig. 1, as is a complete description of each of the mutant viruses used in this study. Poly(A)+ RNA was hybridized with an oligo(dT) probe (Fig. 2). To maximize the observation of viral RNA, cells were infected for 8 h, at which time viral transcription is very robust compared to cellular transcription (35). In wild-type HSV-1-infected cells, both poly(A)+ RNA and ICP27 were seen throughout the nucleus and cytoplasm, indicating efficient mRNA export and ICP27 shuttling (Fig. 2). A similar result was seen in a control infection with mutant d1-2. The lesion in this mutant maps just outside of the region required for TAP/NXF1 interaction (Fig. 1) (3). In contrast, both ICP27 and poly(A)+ RNA were confined to the nucleus in infections with two mutants that cannot interact with TAP/NXF1, N-terminal mutant dLeu and C-terminal mutant n406 (Fig. 2). In mutant d4-5-infected cells, ICP27 was seen to shuttle to the cytoplasm because this mutation does not affect the interaction with TAP/NXF1; however, poly(A)+ RNA was nuclear. The RGG box RNA binding domain is deleted in d4-5. This result indicates that ICP27 must be able to bind RNA for viral RNA export to the cytoplasm.
FIG. 1.
A schematic representation of the ICP27 protein. The N-terminal leucine-rich region (LRR) and C-terminal zinc-finger-like region (CCHC) required for interaction with TAP/NXF1 are shown, as is the Aly/REF interaction region that overlaps the nuclear localization signal (NLS) and the RGG box motif required for RNA binding. An acidic region in the N-terminal portion of the protein (DE), a second arginine-rich region (R2), and three predicted KH domains are also shown. The ICP27 viral mutants used in this study are listed along with the affected domains and the sites of the lesions.
FIG. 2.
Poly(A)+ RNA was retained in the nucleus in infections with ICP27 mutants unable to bind RNA or interact with TAP/NXF1. HeLa cells were either mock infected or infected with wild-type (WT) HSV-1 KOS or ICP27 mutant dLeu, d1-2, d4-5, or n406 for 8 h. In situ hybridization was performed with a biotinylated oligo(dT) probe and visualized by fluorescent staining with a streptavidin-Texas red-conjugated antibody, followed by immunofluorescent staining with monoclonal antibody to ICP27 (green).
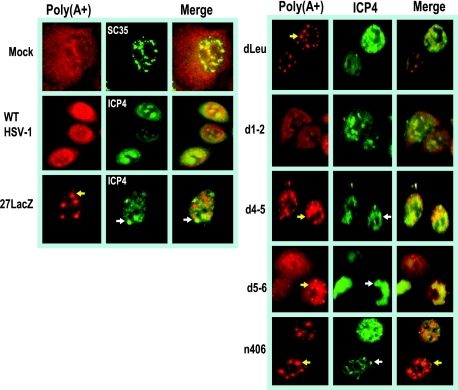
It was noted that in dLeu- and n406-infected cells, poly(A)+ RNA appeared to be in nuclear clusters or enlarged speckles (Fig. 2). We have shown previously that viral transcription is greatly reduced in dLeu- and n406-infected cells because these mutants cannot interact with RNA polymerase II and they fail to recruit RNA polymerase II to viral transcription-replication compartments, which are poorly formed (8). To determine the localization of poly(A)+ RNA with respect to these compartments, in situ hybridization was performed in conjunction with immunofluorescent staining for ICP4 as a marker for HSV-1 transcription-replication compartments (18, 39). In wild-type-infected cells, full-blown replication compartments were visualized in the ICP4-stained cells and poly(A)+ RNA was seen to partially colocalize (Fig. 3). In addition, cytoplasmic poly(A)+ RNA was seen, as expected. Similar results were seen in d1-2- and d5-6-infected cells. These mutants have lesions outside of the regions involved in RNA export (Fig. 1) and were used as controls to demonstrate that deletions elsewhere in the protein do not affect the export activity of ICP27. In d4-5-infected cells, ICP4-containing replication compartments were fully formed and poly(A)+ RNA colocalized with these compartments. This finding again demonstrated that poly(A)+ RNA remained in the nucleus in cells infected with this RGG box deletion mutant. A somewhat different pattern was observed in infections with the null mutant 27-LacZ and with mutants dLeu and n406. Viral transcription is reduced in these mutants, and replication compartments are poorly formed and resemble prereplication sites. Poly(A)+ RNA was seen to form speckled structures that did not colocalize with ICP4 staining (Fig. 3). There was a resemblance of these structures to splicing speckles, as seen in the mock-infected sample stained with an antibody to a splicing protein, SC35, which is a marker for splicing speckles (9). In studies that will be published elsewhere, we provide evidence that HSV-1 infection disrupts cellular mRNA export, resulting in the confinement of cellular poly(A)+ RNA to splicing speckles (L. A. Johnson, L. Li, and R. M. Sandri-Goldin, unpublished results). Thus, the poly(A)+-speckled patterns seen in dLeu- and n406-infected cells may represent cellular transcripts.
FIG. 3.
Poly(A)+ RNA was retained in the nucleus in infections with ICP27 viral mutants. HeLa cells were either mock infected or were infected with wild-type (WT) HSV-1 or the ICP27 viral mutant indicated for 8 h. In situ hybridization was performed as described in the legend to Fig. 2. Immunofluorescent staining was subsequently performed with hybridoma supernatant to SC35 or monoclonal antibody to ICP4. Arrows point to poly(A)+ RNA in speckled structures (red) and ICP4 prereplication or replication compartments (green).
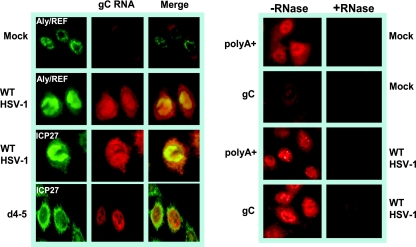
To verify the results that we found with poly(A)+ RNA with a specific viral transcript, we also performed in situ hybridization with a probe to the transcript of the gC gene (Fig. 4). In wild-type-infected cells, both gC RNA and ICP27 were found throughout the nucleus and cytoplasm. Nuclear gC RNA was enriched in ICP4 containing viral transcription and replication compartments but was not detected in the speckled structures that were observed with the oligo(dT) probe. This further suggests that the poly(A)+ RNA in these SC35-rich speckles likely represents cellular mRNAs that are sequestered in stalled spliceosomal complexes. In d4-5-infected cells, ICP27 was efficiently exported to the cytoplasm while gC RNA was retained in the nucleus (Fig. 4). This confirms that ICP27 must be able to bind RNA via its RGG box to facilitate export. It should be noted that because of the high GC content of the gC probe, there was some background hybridization visible in the cytoplasm of mock-infected cells that was likely due to cross-hybridization with GC-rich rRNA. To be certain that the nuclear hybridization signals we observed were due to RNA and not genomic DNA, both mock- and wild-type HSV-1-infected cells were treated with RNase. Only background signals were observed in the RNase-treated cells (Fig. 4, right panels).
FIG. 4.
Viral gC RNA was retained in the nucleus in cells infected with the RGG box deletion mutant d4-5. HeLa cells were either mock infected or infected with wild-type (WT) HSV-1 or mutant d4-5 for 8 h. Cells were hybridized with a biotin-labeled, locked-nucleic acid probe specific to the gC transcript. The probe was visualized with a streptavidin-Texas red-conjugated antibody. Staining with antibody to Aly/REF (to mark nuclei) or ICP27 followed. Right panels: mock-infected and wild-type HSV-1-infected cells were either treated or were not treated with RNase as indicated and then hybridized with an oligo(dT) or gC probe.
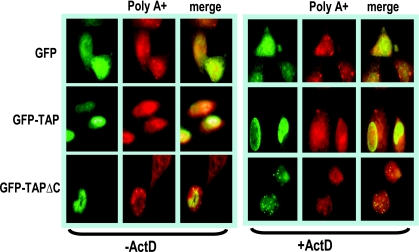
To further verify that TAP/NXF1 is the major export receptor for poly(A)+ RNA in HSV-infected cells, transfections were performed with a dominant-negative TAP/NXF1 mutant termed TAPΔC (Fig. 5). This mutant lacks the C-terminal region of TAP/NXF1 that is required for its interaction with nucleoporins, and as a consequence, the mutant TAP/NXF1 protein cannot move through the nuclear pore complex. It acts as a dominant negative since it can still interact with export adapter proteins and its cofactor p15 but cannot escort its cargo to the cytoplasm. We have previously reported that transfection with this TAP/NXF1 mutant prevents the export of ICP27 (3). Poly(A)+ RNA was exported to the cytoplasm in wild-type-HSV-1-infected cells that were transfected with a GFP-expressing plasmid or with GFP-TAP used as controls (Fig. 5). In the cell transfected with GFP-TAPΔC, poly(A)+ RNA was confined to the nucleus, whereas in the cell next to it, not expressing GFP-TAPΔC, poly(A)+ was exported to the cytoplasm (Fig. 5). To determine if RNA could be chased into the cytoplasm in GFP-TAPΔC-expressing cells, actinomycin D was added to infected cells at 7 h after infection and cells were fixed 1 h later. Poly(A)+ RNA was still confined to the nucleus in GFP-TAPΔC-expressing cells (Fig. 5), indicating that the absence of poly(A)+ RNA in the cytoplasm is due to an export defect and not a decrease in mRNA stability.
FIG. 5.
RNA is exported through the TAP/NXF1 pathway during HSV-1 infection. HeLa cells were transfected with plasmids encoding GFP, GFP-TAP, or GFP-TAPΔC. Twenty-four hours later, cells were infected with wild-type HSV-1 for 8 h. Right panels: at 7 h after infection, actinomycin D was added to infected cells to stall transcription and cells were fixed 1 h later. In situ hybridization was performed with a biotinylated oligo(dT) probe as described in Materials and Methods.
We conclude that during HSV-1 infection, viral RNA is exported to the cytoplasm through the TAP/NXF1 export receptor and that ICP27 must be able to bind RNA and to interact with TAP/NXF1 for export of poly(A)+ RNA and gC RNA.
ICP27 must have an intact RGG box and TAP/NXF1 interaction regions for RNA export during HSV-1 infection.
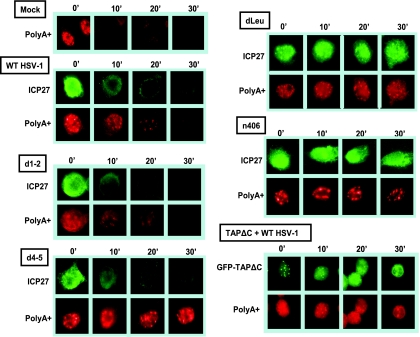
To determine if the rate of export of poly(A)+ RNA to the cytoplasm is correlated to the export of ICP27, we performed in vitro export assays coupled with in situ hybridization. Cells were mock-infected or infected with wild-type HSV-1 or with ICP27 mutant viruses for 8 h, at which time cells were placed on ice and permeabilized with digitonin. The in vitro export assays were performed as we described previously (3), and at 10-min intervals, samples were fixed and in situ hybridization was performed with an oligo(dT) probe, followed by immunostaining for ICP27. In these assays, RNA and protein that are not exported from the nucleus during the course of the assay are visualized. In mock-infected cells, export of cellular RNA was rapid, with poly(A)+ RNA being depleted from the nucleus in 10 min. In wild-type-HSV-1-infected cells, the nucleus was nearly devoid of ICP27 protein and poly(A)+ in 20 min, and a similar result was seen with the d1-2 control (Fig. 6). In d4-5-infected cells, ICP27 was quickly exported from the nucleus, which is consistent with previously published results showing that ICP27 does not need to bind RNA in order to shuttle (3). However, poly(A)+ RNA was confined to the nucleus (Fig. 6), again demonstrating that while the d4-5 mutant ICP27 can be exported, it cannot bind RNA and thus cannot stimulate the export of poly(A)+ RNA from the nucleus. In dLeu- and n406-infected cells, both ICP27 and poly(A)+ RNA remained nuclear throughout the assay. TAPΔC expression blocked poly(A)+ RNA export in wild-type-HSV-1-infected cells, again showing that poly(A)+ RNA in HSV-1-infected cells is exported through the TAP/NXF1 pathway (Fig. 6). These results clearly establish that RNA export during HSV-1 infection requires ICP27 binding to RNA and interaction with TAP/NXF1.
FIG. 6.
Export of ICP27 correlated with export of RNA during HSV-1 infection. HeLa cells were mock infected or infected with wild-type (WT) HSV-1 or the viral mutant indicated for 8 h. Cells transfected with GFP-TAPΔC plasmid were infected with wild-type HSV-1 for 8 h beginning 24 h after transfection. Cells were placed on ice, and nuclear export assays were performed as described previously (3). Cells were fixed at 10-min intervals after the start of the export assays, as indicated. In situ hybridization was performed on the fixed cells with an oligo(dT) probe. Cells were subsequently stained with monoclonal antibody to ICP27 (green). The oligo(dT) probe was visualized with streptavidin-Texas red-conjugated antibody. GFP was visualized directly for GFP-TAPΔC-expressing cells.
The majority of viral transcripts require ICP27 for efficient export.
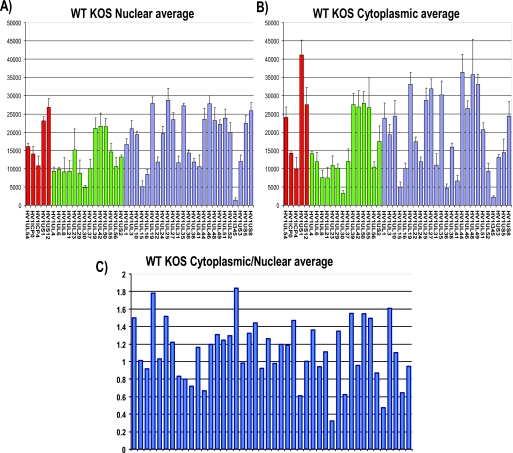
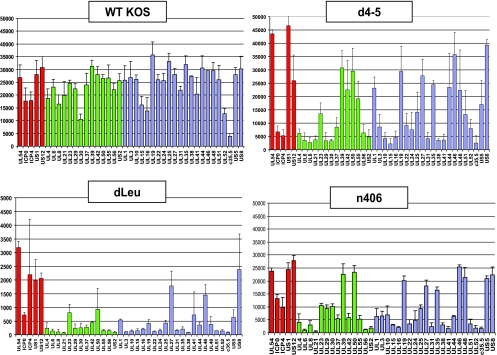
To determine whether specific viral transcripts or most viral transcripts require ICP27 for their export to the cytoplasm, we performed microarray analysis using HSV-1 transcript-specific probes that have been described previously (16, 38). Nuclear and cytoplasmic fractionation was performed on infected-cell lysates at 8 h after infection, and poly(A)+ selection was carried out on the nuclear and cytoplasmic RNA fractions. The nuclear and cytoplasmic averages from three separate experiments are shown for wild-type KOS (Fig. 7A and B). Results for immediate-early transcripts, early transcripts, and late transcripts are shown. Cytoplasmic/nuclear ratios were calculated using the cytoplasmic and nuclear averages for each transcript (Fig. 7C). A value of 1 indicates that the transcript was equally abundant in the nucleus and cytoplasm, whereas a value greater than 1 indicates that the transcript was more abundant in the cytoplasm and is being actively exported.
FIG. 7.
Microarray analysis of the cytoplasmic/nuclear ratio for HSV-1 transcripts. (A, B) HeLa cells were infected with wild-type (WT) HSV-1 KOS for 8 h. Cells were lysed, and nuclear and cytoplasmic fractions were prepared as described previously (30). RNA was isolated from each fraction, and poly(A)+ RNA was selected and reverse transcribed as described previously (38). The cDNA from each fraction was hybridized to HSV-1-specific microarray chips as described previously (38) and quantified by using Array Vision software. Experiments were performed in triplicate, and the average expression value for each transcript from each fraction was calculated and plotted, with the y axis representing the intensity of the light scattering signal and the x axis representing individual HSV-1 transcripts. Error bars show standard deviations. Red bars represent immediate early transcripts, green bars show early transcripts, and blue bars show late transcripts. (C) The cytoplasmic/nuclear ratio for each transcript was calculated from the average values for each transcript from the cytoplasmic and nuclear fractions. The ratio for each transcript was plotted on the y axis, with 1.0 indicating equal abundance in the cytoplasm and nucleus. A ratio greater than 1.0 indicates that a transcript was more abundant in the cytoplasm than in the nucleus. The x axis represents individual transcripts in the same order as in panels A and B.
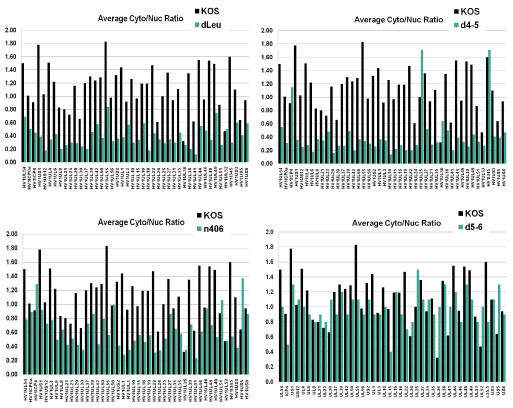
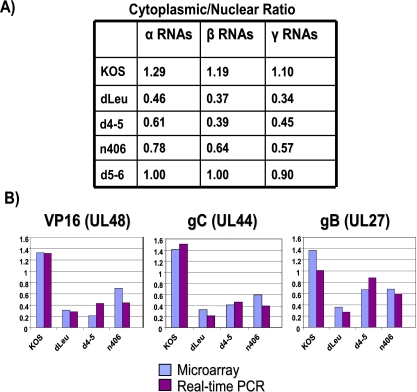
A similar analysis was performed on mutants dLeu, n406, and d4-5, and the results compared to those for wild-type KOS. For the mutant infections, again, each set of experiments was performed three times and average cytoplasmic values and average nuclear values were calculated for each transcript and compared to the ratios found with wild-type HSV-1 KOS (Fig. 8). There was an overall decrease in the cytoplasmic/nuclear ratio for early and late viral transcripts in dLeu, n406, and d4-5 infections compared to the results for infections with wild-type KOS, whereas the results for infections with the control mutant d5-6 were similar to those for wild-type HSV-1 (Fig. 8). The cytoplasmic/nuclear ratio was calculated for all immediate-early, early, and late RNAs for each mutant and was compared to the results for wild-type KOS (Fig. 9A). Decreases in export efficiency were seen for all kinetic classes in dLeu, d4-5, and n406 infections but were more reduced for early and late RNAs compared to the results for KOS (Fig. 9A). These results confirm that ICP27 is required for efficient export of the majority of viral transcripts. To validate these results for specific viral transcripts, real-time qPCR was performed for three late RNAs, those encoding VP16, gC, and gB (Fig. 9B). The cytoplasmic/nuclear ratio found with qPCR correlated well with the ratio found for these transcripts in the microarray analysis (Fig. 9B). Northern blots were also performed to confirm that the transcripts we were quantifying with the microarray hybridizations and qPCR were representative of the full-length mRNA and not degradation products (data not shown).
FIG. 8.
Export is inefficient for the majority of HSV-1 transcripts in ICP27 mutant virus infections. HeLa cells were infected with wild-type HSV-1 or ICP27 viral mutant dLeu, d4-5, n406, or d5-6 for 8 h. Nuclear and cytoplasmic poly(A)+ RNA was isolated, and cDNA was prepared as described in the legend to Fig. 7. Hybridization to HSV-1-specific microarray chips was performed, and results were quantified by using Array Vision software. Experiments were performed in triplicate, and the average cytoplasmic/nuclear ratio was calculated and plotted relative to that of wild-type KOS. A lower cytoplasmic/nuclear ratio (shown on the y axis) indicates the accumulation of a transcript in the nucleus.
FIG. 9.
Cytoplasmic/nuclear export ratios are reduced when ICP27 is unable to bind RNA or interact with TAP/NXF1. (A) The average cytoplasmic/nuclear ratio for all transcripts in each kinetic class was calculated for wild-type KOS and dLeu, d4-5, n406, and d5-6. (B) Real-time qPCR was performed on cDNA synthesized from cytoplasmic and nuclear fractions isolated from cells infected with KOS, dLeu, d4-5, or n406. Primers specific to late transcripts encoding VP16, gC, and gB were used to amplify the cDNA, and Bio-Rad iCycler software was used to quantify the Sybr green signal generated. Resulting threshold curve (CT) values were compared to a standard curve of threshold curve values from fivefold dilutions of plasmid DNA for each primer to determine the relative starting amount of RNA. The average cytoplasmic/nuclear ratio from three independent experiments was calculated and plotted adjacent to the ratio found for the transcript in the microarray analysis for each mutant (shown on the y axis).
To determine the overall effect of inefficient export on the relative abundance of each transcript in ICP27 mutant virus-infected cells, total poly(A)+ RNA was isolated at 8 h after infection and compared to the results for wild-type KOS (Fig. 10). As we reported previously (16), early and late transcript levels were significantly reduced in dLeu and n406 infections, likely reflecting both the transcriptional defect for these mutants, which are unable to interact with RNA polymerase II (8, 16), and reduced RNA export to the cytoplasm. There were also decreased levels of a number of early and late mRNAs in d4-5-infected cells (Fig. 10), which has not been shown to have reduced viral transcription. This may result from degradation of viral transcripts in the nucleus which are not able to be exported. Thus, the overall abundance of viral mRNA was reduced when viral RNA export was impaired and ICP27 appears to be critical for efficient viral RNA export.
FIG. 10.
HSV-1 transcripts were reduced in abundance in infections with ICP27 mutants dLeu, d4-5, and n406. HeLa cells were infected with wild-type (WT) KOS or with mutant dLeu, d4-5, or n406 as indicated for 8 h. RNA was harvested from total cell lysates, and poly(A)+ RNA was selected. Hybridizations were performed by using HSV-1-specific microarray chips, and data were analyzed as described in the legend to Fig. 7. Experiments were performed in triplicate; error bars show standard deviations. Immediate early transcripts are shown as red bars, early transcripts as green bars, and late transcripts as blue bars.
DISCUSSION
The export of mRNA from the nucleus to the cytoplasm for translation requires the deposition of multiple proteins on the mRNA that can direct the ribonucleoprotein (RNP) complex to the TAP/NXF1 nuclear export receptor, which possesses a domain that can interact with the FG repeats in nucleoporins to escort the RNP through the nuclear pore complex (1, 6). In mammalian cells, proteins that have been shown to be deposited on mRNA to facilitate export include Aly/REF (29, 37) and UAP56 (11, 19, 20). The association of proteins such as Aly/REF and UAP56 with newly transcribed mRNAs is essential for nuclear export. These proteins were first reported to be components of the exon junction complex that is deposited just upstream of exon-exon junctions to mark cellular, spliced mRNAs for export, mRNA surveillance, and translation initiation (2, 7, 14, 25). More-recent studies have shown that Aly/REF, which interacts directly with TAP/NXF1 (37), is recruited to mRNA by UAP56 (19), where Aly/REF can then associate with CBP80 at the 5′ end of the mRNA as part of the TREX (transcription-export) complex that directs mRNA to TAP/NXF1 (5, 22, 36). Therefore, these proteins are necessary for the export of both intronless and spliced cellular mRNAs.
Herpesvirus proteins in the family of multifunctional regulators typified by ICP27 have been found to interact with Aly/REF and/or UAP56. ICP27 interacts with both Aly/REF (3, 4) and UAP56 (L. A. Johnson and R. M. Sandri-Goldin, unpublished results), whereas CMV UL69 interacts with UAP56 (17) and Epstein-Barr virus SM protein and Kaposi's sarcoma-associated herpesvirus ORF57 interact with Aly/REF (12, 21). It has been proposed, therefore, that it is the interaction of the viral export protein with the cellular export adaptor proteins Aly/REF and UAP56 that directs viral RNA to the TAP/NXF1 pathway (13). To date, ICP27 is the only member of this family that has been shown to also interact directly with TAP/NXF1 (3). Therefore, we posited that ICP27 may in fact be the major export adaptor for HSV-1 RNA and that Aly/REF, UAP56, and, perhaps, other cellular export factors may simply act to increase the effectiveness of the process. These cellular export factors may also be important in compensating for the loss of ICP27's RNA binding and export activity during infection with ICP27 mutant viruses. We tested that hypothesis in this study by employing viral ICP27 mutants that could not interact with TAP/NXF1 or that cannot bind RNA. We found that ICP27 was required for the export of bulk poly(A)+ RNA, specific viral transcripts, and the majority of viral mRNAs during HSV-1 infection. Surprisingly, although Aly/REF is recruited to viral replication compartments by ICP27 and can interact directly with TAP/NFX1, it was not able to bridge the interaction of ICP27-bound RNA with TAP/NXF1 in infections with ICP27 mutants that were not able to interact with TAP/NXF1. Aly/REF has been shown to be dispensable for cellular mRNA export (10), and we recently found that Aly/REF is also dispensable for viral RNA export by using small interfering RNA knockdown (L. A. Johnson, L. Li, and R. M. Sandri-Goldin, unpublished results). Thus, Aly/REF's role may be auxiliary. The greatly reduced RNA export seen in d4-5 infections in which ICP27 cannot bind RNA but can still interact with Aly/REF also would suggest that Aly/REF plays a supporting role. This idea is further supported by the results of a previous study in which we found that Aly/REF was not bound to several late transcripts to which ICP27 was bound as measured by in vivo UV cross-linking-immunoprecipitation assays (3).
The results presented here suggest that ICP27 binds to the majority of viral RNAs and is essential for viral RNA export. Studies are under way to define the RNA binding specificity for ICP27. The studies described here provide strong evidence that acting as an mRNA export adaptor is a major role for this multifunctional regulatory protein.
Acknowledgments
We thank Steve Rice for providing viral mutants and Gayathri Devi-Rao for assistance with the microarray experiments.
This work was supported by National Institute of Allergy and Infectious Diseases (NIAID) grants AI61397 and AI21215.
Footnotes
Published ahead of print on 19 November 2008.
REFERENCES
- 1.Bachi, A., I. C. Braun, J. P. Rodrigues, N. Pante, K. Ribbeck, C. von Kobbe, U. Kutay, M. Wilm, D. Gorlich, M. Carmo-Fonseca, and E. Izaurralde. 2000. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6136-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan, C. C., J. Dostie, M. D. Diem, W. Feng, M. Mann, J. Rappsilber, and G. Dreyfuss. 2004. eIF4A3 is a novel component of the exon junction complex. RNA 10200-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, I. B., L. Li, L. Silva, and R. M. Sandri-Goldin. 2005. ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. J. Virol. 793949-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, I. B., K. S. Sciabica, and R. M. Sandri-Goldin. 2002. ICP27 interacts with the export factor Aly/REF to direct herpes simplex virus 1 intronless RNAs to the TAP export pathway. J. Virol. 7612877-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, H., K. Dufu, C. S. Lee, J. L. Hsu, A. Dias, and R. Reed. 2006. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 1271389-1400. [DOI] [PubMed] [Google Scholar]
- 6.Cullen, B. R. 2003. Nuclear RNA export. J. Cell Sci. 116587-597. [DOI] [PubMed] [Google Scholar]
- 7.Custodio, N., C. Carvalho, I. Condado, M. Antoniou, B. J. Blencowe, and M. Carmo-Fonseca. 2004. In vivo recruitment of exon junction complex proteins to transcription sites in mammalian cell nuclei. RNA 10622-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai-Ju, J. Q., L. Li, L. A. Johnson, and R. M. Sandri-Goldin. 2006. ICP27 interacts with the C-terminal domain of RNA polymerase II and facilitates its recruitment to herpes simplex virus-1 transcription sites, where it undergoes proteasomal degradation during infection. J. Virol. 803567-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu, X. D., and T. Maniatis. 1992. The 35-kDa mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3′ splice site. Proc. Natl. Acad. Sci. USA 891725-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatfield, D., and E. Izaurralde. 2002. REF/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J. Cell Biol. 159579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gatfield, D., H. LeHir, C. Schmitt, I. C. Braun, T. Kocher, M. Wilm, and E. Izaurralde. 2001. The DexH/D box protein HEL/UAP56 is essential for mRNA export in Drosophila. Curr. Biol. 111716-1721. [DOI] [PubMed] [Google Scholar]
- 12.Hiriart, E., G. Farjot, H. Gruffat, M. V. C. Nguyen, A. Sergeant, and E. Manet. 2003. A novel nuclear export signal and a REF interaction domain both promote mRNA export by the Epstein-Barr virus EB2 protein. J. Biol. Chem. 278335-342. [DOI] [PubMed] [Google Scholar]
- 13.Koffa, M. D., J. B. Clements, E. Izaurralde, S. Wadd, S. A. Wilson, I. W. Mattaj, and S. Kuersten. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 205769-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Hir, H., D. Gatfield, E. Izaurralde, and M. J. Moore. 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 204987-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lengyel, J., C. Guy, V. Leong, S. Borge, and S. A. Rice. 2002. Mapping of functional regions in the amino-terminal portion of the herpes simplex virus ICP27 regulatory protein: importance of the leucine-rich nuclear export signal and RGG box RNA-binding domain. J. Virol. 7611866-11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, L., L. A. Johnson, J. Q. Dai-Ju, and R. M. Sandri-Goldin. 2008. Hsc70 focus formation at the periphery of HSV-1 transcription sites requires ICP27. PLoS ONE 3e1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lischka, P., Z. Toth, M. Thomas, R. Mueller, and T. Stamminger. 2006. The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Mol. Cell. Biol. 261631-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukonis, C. J., and S. K. Weller. 1997. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J. Virol. 712390-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo, M. J., Z. Zhou, K. Magni, C. Christoforides, J. Rappsilber, M. Mann, and R. Reed. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413644-647. [DOI] [PubMed] [Google Scholar]
- 20.MacMorris, M., C. Brocker, and T. Blumenthal. 2003. UAP56 level affects viability and mRNA export in Caenorhabditis elegans. RNA 9847-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malik, P., D. J. Blackbourn, and J. B. Clements. 2004. The evolutionarily conserved Kaposi's sarcoma-associated ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 27933001-33011. [DOI] [PubMed] [Google Scholar]
- 22.Masuda, S., R. Das, H. Cheng, E. Hurt, N. Dorman, and R. Reed. 2005. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 191512-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mears, W. E., and S. A. Rice. 1996. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 707445-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mears, W. E., and S. A. Rice. 1998. The herpes simplex virus immediate-early protein ICP27 shuttles between the nucleus and cytoplasm. Virology 242128-137. [DOI] [PubMed] [Google Scholar]
- 25.Nott, A., H. LeHir, and M. J. Moore. 2004. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 18210-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phelan, A., and J. B. Clements. 1997. Herpes simplex virus type 1 immediate early protein IE63 shuttles between nuclear compartments and the cytoplasm. J. Gen. Virol. 783327-3331. [DOI] [PubMed] [Google Scholar]
- 27.Rice, S. A., and D. M. Knipe. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J. Virol. 641704-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rice, S. A., and V. Lam. 1994. Amino acid substitution mutations in the herpes simplex virus ICP27 protein define an essential gene regulation function. J. Virol. 68823-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodrigues, J. P., M. Rode, D. Gatfield, B. J. Blencowe, M. Carmo-Fonseca, and E. Izaurralde. 2001. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. USA 981030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandri-Goldin, R. M. 1998. ICP27 mediates herpes simplex virus RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sciabica, K. S., Q. J. Dai, and R. M. Sandri-Goldin. 2003. ICP27 interacts with SRPK1 to mediate HSV-1 inhibition of pre-mRNA splicing by altering SR protein phosphorylation. EMBO J. 221608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith, I. L., M. A. Hardwicke, and R. M. Sandri-Goldin. 1992. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology 18674-86. [DOI] [PubMed] [Google Scholar]
- 33.Soliman, T. M., R. M. Sandri-Goldin, and S. J. Silverstein. 1997. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J. Virol. 719188-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soliman, T. M., and S. J. Silverstein. 2000. Identification of an export control sequence and a requirement for the KH domains in ICP27 from herpes simplex virus type 1. J. Virol. 747600-7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spencer, C. A., M. E. Dahmus, and S. A. Rice. 1997. Repression of host RNA polymerase II transcription by herpes simplex virus type 1. J. Virol. 712031-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strasser, K., S. Masuda, P. Mason, J. Pfannstiel, M. Oppizzi, S. Rodriguez-Navarro, A. G. Rondon, A. Aguilera, K. Struhl, R. Reed, and E. Hurt. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417304-308. [DOI] [PubMed] [Google Scholar]
- 37.Stutz, F., A. Bachi, T. Doerks, I. C. Braun, B. Seraphin, M. Wilm, P. Bork, and E. Izaurralde. 2000. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6638-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, A., G. V. Devi-Rao, M. K. Rice, L. W. Gary, D. C. Bloom, R. M. Sandri-Goldin, P. Ghazal, and E. K. Wagner. 2004. Immediate-early expression of the herpes simplex virus type 1 ICP27 transcript is not critical for efficient replication in vitro or in vivo. J. Virol. 7810470-10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uprichard, S. L., and D. M. Knipe. 1997. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology 229113-125. [DOI] [PubMed] [Google Scholar]
- 40.Zhi, Y., K. S. Sciabica, and R. M. Sandri-Goldin. 1999. Self interaction of the herpes simplex virus type 1 regulatory protein ICP27. Virology 257341-351. [DOI] [PubMed] [Google Scholar]