Abstract
Deciphering antibody specificities that constrain human immunodeficiency virus type 1 (HIV-1) envelope (Env) diversity, limit virus replication, and contribute to neutralization breadth and potency is an important goal of current HIV/AIDS vaccine research. Transplantation of discrete HIV-1 neutralizing epitopes into HIV-2 scaffolds may provide a sensitive, biologically functional context by which to quantify specific antibody reactivities even in complex sera. Here, we describe a novel HIV-2 proviral scaffold (pHIV-2KR.X7) into which we substituted the complete variable region 3 (V3) of the env gene of HIV-1YU2 or HIV-1Ccon to yield the chimeric proviruses pHIV-2KR.X7 YU2 V3 and pHIV-2KR.X7 Ccon V3. These HIV-2/HIV-1 chimeras were replication competent and sensitive to selective pharmacological inhibitors of virus entry. V3 chimeric viruses were resistant to neutralization by HIV-1 monoclonal antibodies directed against the CD4 binding site, coreceptor binding site, and gp41 membrane proximal external region but exhibited striking sensitivity to HIV-1 V3-specific monoclonal antibodies, 447-52D and F425 B4e8 (50% inhibitory concentration of [IC50] <0.005 μg/ml for each). Plasma specimens from 11 HIV-1 clade B- and 10 HIV-1 clade C-infected subjects showed no neutralizing activity against HIV-2 but exhibited high-titer V3-specific neutralization against both HIV-2/HIV-1 V3 chimeras with IC50 measurements ranging from 1:50 to greater than 1:40,000. Neutralization titers of B clade plasmas were as much as 1,000-fold lower when tested against the primary HIV-1YU2 virus than with the HIV-2KR.X7 YU2 V3 chimera, demonstrating highly effective shielding of V3 epitopes in the native Env trimer. This finding was replicated using a second primary HIV-1 strain (HIV-1BORI) and the corresponding HIV-2KR.X7 BORI V3 chimera. We conclude that V3 is highly immunogenic in vivo, eliciting antibodies with substantial breadth of reactivity and neutralizing potential. These antibodies constrain HIV-1 Env to a structure(s) in which V3 epitopes are concealed prior to CD4 engagement but do not otherwise contribute to neutralization breadth and potency against most primary virus strains. Triggering of the viral spike to reveal V3 epitopes may be required if V3 immunogens are to be components of an effective HIV-1 vaccine.
Infection by human immunodeficiency virus type 1 (HIV-1) is followed by the rapid development of a virus-specific antibody response that results in diagnostic antibody seroconversion approximately 3 to 6 weeks later (14, 23). Neutralizing antibodies (NAbs) reactive with the external region of the gp120/41 envelope (Env) glycoprotein of primary virus strains first appear in the plasma approximately 12 to 16 weeks after virus transmission (83, 97). Such antibodies are directed at the most exposed epitopes on the Env surface of transmitted/early founder viruses (49, 90) and they are invariably strain specific (25, 83, 97). Within 3 to 6 months of infection, these NAb responses reach high titers and effect potent virus neutralization that is detectable in vitro by classical neutralization assays (1, 35, 57, 66, 67, 75, 77, 80, 83, 97) and in vivo by the rapid evolution of NAb escape mutants (25, 57, 83, 86, 87, 97). The same is true for the kinetics of HIV-1-specific cytotoxic T-lymphocyte (CTL) recognition and escape, which are even faster than for NAbs (6, 7, 46, 52). Thus, it is not uncommon for the replicating virus quasispecies to escape completely from multiple CTL and NAb epitopes distributed across the HIV-1 proteome within 4 months of virus transmission (6, 7, 25, 46, 49, 52, 83, 91, 97; also unpublished data). Much later in infection, in a small minority of subjects, broadly reactive antibodies that neutralize heterologous primary virus strains develop (10, 19, 61, 73). What role NAbs of narrow or broad neutralizing specificity play in virus containment in vivo and in disease outcome is presently unclear.
To better understand what contributions NAbs make to virus containment in natural HIV-1 infection and potentially in vaccinated subjects, attention has turned to defining epitope specificities of NAbs in polyclonal human serum (19, 35, 61, 62, 68, 86, 87). There is evidence that early in infection, NAbs are generally directed against the surface-exposed hypervariable loop structures of the gp120 ectodomain, especially variable region 1 (V1), V2, and possibly V4 (25, 40, 65, 68, 78, 86, 86, 101). This accounts for the strain-specific neutralizing reactivity of these early responses and the ability of HIV-1 to rapidly escape NAb-mediated elimination by any of several molecular mechanisms including epitope variation, conformational masking, and glycan shielding (10, 73). A second set of HIV-1 Env-specific antibodies elicited early in infection that also have neutralizing potential are antibodies directed against the coreceptor-binding surface of gp120 (18, 35). These antibodies are termed CD4-induced (CD4i) because their target epitopes are formed only after the binding of CD4 to gp120 and structural rearrangement of the inner domain and bridging sheet of gp120. CD4i antibodies are made in most (>90%) HIV-1-infected subjects and often reach high titers in the plasma (18). Because the coreceptor binding region of HIV-1 gp120 is functionally and antigenically conserved, CD4i antibodies exhibit far greater cross-reactivity than those that target most variable regions. This cross-reactivity extends to different HIV-1 viruses not only within a clade but also between clades and even between HIV-1 and HIV-2 (18, 91). However, CD4i NAbs exhibit low (if any) potency against primary virus strains because prior to Env-CD4 engagement their epitopes are not formed, and subsequent to CD4 engagement, their epitopes are sterically concealed at the virus-cell interface, preventing antibody access (56). A third set of NAbs develops much later in infection and then only in a small subset of individuals. These are potent, broadly reactive NAbs whose target epitopes include the CD4-binding site (CD4bs) of gp120, the membrane-proximal external region (MPER) of gp41, or conserved carbohydrate epitopes on gp120 (10, 73). Examples of this set of NAbs include the human monoclonal antibodies (MAbs) b12, 2F5, 4E10, and 2G12. These human MAbs, however, are atypical in that they exhibit long third complementarity-determining regions of the heavy chain, heavy chain dominant recognition, reactivity with host lipids, or a heavy chain variable region domain-swapped dimeric structure (11, 12, 38, 70, 92, 105). The scarcity of potent, broadly neutralizing human MAbs is consistent with the infrequency of potent, broadly neutralizing polyclonal antibodies in patients with chronic HIV-1 infection (10, 73).
NAbs specific for gp120 V3 constitute a fourth group of antibodies whose roles in HIV-1 natural history and as potential contributors to vaccine-elicited antibody protection are not yet fully explored. The HIV-1 V3 is a disulfide-bound structure that emanates from the outer domain of gp120 and determines target cell specificity through direct interactions with the chemokine coreceptor (generally CCR5 or CXCR4) (16, 17, 37, 42, 44, 84, 85). It is also thought to play a role in antibody evasion by conferring neutralization resistance through its quaternary interactions with neighboring protomers within the Env trimer complex (42, 55, 94, 100, 103). As a consequence of these critical roles in virus-coreceptor binding and antibody evasion, the HIV-1 V3 is highly conserved in loop length and, to a degree, in primary amino acid sequence and secondary and tertiary structure (78, 106). Thus, while tolerant of some sequence diversity, V3 is more conserved than are other variable regions of the glycoprotein such as V1, V2, V4, and V5. The antigenic and immunogenic properties of V3 have been the subject of substantial prior investigation. Early work focused on laboratory-adapted, T-cell line derived (TCLA) viruses that exhibited CXCR4 tropism. The V3 region of such viruses was found to be highly immunogenic and to be a sensitive target of V3-specific NAbs (33, 45, 69, 71, 81, 88). Certain viruses of the CCR5 phenotype, including the brain-derived HIV-1YU2 molecular clone (60), were also found to be moderately sensitive to V3 NAbs as were some other HIV-1 strains that were variably laboratory adapted (5, 58, 72). Conversely, most primary (non-laboratory adapted) CCR5-tropic viruses, including molecularly derived transmitted/early founder viral Envs from more than 50 subjects acutely infected with clade B virus (49), were found to be resistant to neutralization by V3-specific NAbs, including the human MAbs 447-52D and F425 B4e8. If, however, the V3 region of primary HIV-1 viruses was exchanged for the V3 region of a neutralization-sensitive HIV-1 strain (e.g., HIV-1SF162), then the resulting HIV-1 Env V3 chimeras were rendered sensitive to V3-specific NAbs (53, 54).
Given the importance of V3 in virus biology and immune evasion, important questions remain concerning its immunogenicity and antigenicity in vivo. For example, how immunogenic is the V3 region of different HIV-1 clades in natural human infection? How cross-reactive are V3-specific NAbs with viruses from the same or different clades? Do V3-specific NAbs, along with CD4i antibodies, constrain the HIV-1 Env to a CD4-dependent phenotype? Do V3-specific NAbs contribute to the neutralization breadth of polyclonal human serum against different primary virus strains in natural infection? And do V3-specific antibodies have the potential to contribute to vaccine efficacy? To address these questions, investigators have pursued different strategies to detect HIV-1 V3 NAbs. One approach has been to isolate human MAbs from infected individuals for analysis of V3-specific neutralizing activity (26-29, 53). This has the advantage of generating replenishable reagents for definitive structure-function studies, but it has the disadvantage that it cannot illuminate the relative contributions of V3 antibodies compared with other antibody specificities to neutralization by complex polyclonal serum. A second strategy has been to use V3 (or other) epitope-containing peptides to remove or enrich NAb activities in patient serum (19, 62, 68). This approach, however, is limited to NAbs with linear epitopes that have sufficient affinity for peptide binding outside the context of the native glycoprotein trimer. A third strategy has been to make site-directed mutations in NAb epitopes in Env glycoproteins and to use these proteins as probes in binding, absorption, or infectivity experiments (54, 61, 62, 72). A fourth approach has been to make reciprocal exchanges between homologous regions of different HIV-1 Env glycoproteins to look for gain or loss of NAb sensitivity (54, 68, 76, 86, 87, 89). The last three approaches share a common limitation in that V3 neutralizing activity is analyzed against a backdrop of other HIV-1 gp120/41 neutralizing reactivities, making a clear dissection of V3-specific NAb activity challenging.
Our laboratory has pursued a different strategy for detecting epitope-specific NAb responses based on the observation that while the tertiary and quaternary structures of HIV-1 and HIV-2 Env gp120/gp41 trimers are similar (15), the primary amino acid sequences of these glycoproteins share only ∼40% homology and are largely non-cross-reactive with respect to surface-accessible NAb epitopes (8, 18, 98). We hypothesized that by employing the structurally related native HIV-2 Env glycoprotein as a molecular scaffold on which to display discreet HIV-1 neutralizing epitopes in a biologically functional context, we could obtain a clear distinction of HIV-1 epitope-specific NAb reactivities even in complex human or animal anti-HIV-1 immune serum. A particular advantage to this approach is that the assay endpoint is virus neutralization, and thus only those antibodies specific for the HIV-1 epitope of interest and capable of neutralizing the functional Env trimer are detected. We first demonstrated the feasibility of this approach by creating HIV-2/HIV-1 Env chimeras modified by site-directed mutagenesis or by larger exchanges of portions (or all) of the gp41 MPER sequences containing epitopes for 2F5, 4E10, and Z13 (34; F. Bibollet-Ruche, H. Li, J. M. Decker, et al., presented at the Keystone Symposium on HIV Vaccines, Keystone, CO, 2006; F. Bibollet-Ruche, H. Li, J. M. Decker, et al., presented at the AIDS Vaccine 2005 Conference, Montreal, Canada, 2005) and by making site-directed mutations in the HIV-2 gp120 bridging sheet to facilitate binding of HIV-1 CD4i antibodies (18). These infectious chimeras enabled us to detect HIV-1 MPER and coreceptor (CD4i)-specific NAbs in patients with HIV-1 infection and in humans and animals vaccinated with HIV-1 Env immunogens (18, 19, 24, 34, 35, 61; F. Bibollet-Ruche et al., presented at the Keystone Symposium on HIV Vaccines, Keystone, CO, 2006; F. Bibollet-Ruche et al., presented at the AIDS Vaccine 2005 Conference, Montreal, Canada, 2005; J. M. Decker, F. Bibollet-Ruche, H. Li, et al., presented at the Keystone Symposium on HIV Vaccines, Keystone, CO, 2006). Here, we extend this work by describing a strategy for creating HIV-2/HIV-1 V3 chimeras, by determining the phenotypic properties of these chimeras using a battery of HIV-2 and HIV-1 Env-specific ligands, and by demonstrating the utility of the chimeras for detecting V3-specific NAbs in plasma from humans infected by subtype B or subtype C HIV-1.
(This work was completed in partial fulfillment of the requirements for the Ph.D. degree [K.L.D.]).
MATERIALS AND METHODS
Molecular cloning and mutagenesis. (i) Construction of pHIV-2KR.P1.
Overlapping HIV-2KR half-genomic constructs (5′-pRTsac; 3′-pKTM) were obtained as a gift from D. Looney. The HIV-2KR long terminal repeat (LTR) sequence was amplified from the pKTM plasmid using primers that incorporate MluI and ApaI restriction sequences flanking the U3 and U5 regions (ATTACGCGTTGGATGGGATGTATTACAGTGAG and TTTGGGCCCTAACTTGCTTCTAACTGGCAGCT; Operon Biotechnologies, Inc., Huntsville, AL) using an Expand High Fidelity PCR System according to the manufacturer's instructions (Roche Applied Science, Indianapolis, IN). Amplification products were subsequently digested with MluI and ApaI and cloned into the pCR XL TOPO vector (Invitrogen Life Technologies, Carlsbad, CA) to yield pHIV-2KR.LTR. pRTsac, pKTM, and pHIV-2KR.LTR constructs were digested with AgeI (unique restriction site located within the R region of the LTR sequence) and SacI (unique restriction site located within the vpr gene sequence) and ligated to yield a full-length pHIV-2KR.P1 proviral construct.
(ii) Construction of pHIV-2KR.X4.
Unique silent restriction sites were introduced using a QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) into the env gene of the pKTM construct using the following primer pairs: XhoI restriction sequence, GAGTATAATGGACTCGAGAAATCAGCTA and TAGCTGATTTCTCGAGTCCATTATACTC; SnaBI restriction sequence, GCGCCCAATACGTAACTGTTTTCTATGGC and CCATAGAAAACAGTTACGTATTGGGCGCA; XmaI restriction sequence, CTTTACACAACCCGGGAAAGGTTCAGATGCAGAA and CTGCATCTGAACCTTTCCCGGGTTGTGTAAAG; and XbaI restriction sequence, GTAGTACAATTACTATCTAGACTTAGAAAGG and CCTTTCTAAGTCTAGATAGTAATTGTACTAC. The resulting modified 3′ half-genome was designated pKTMKR.X4. pRTsac, pKTMKR.X4, and pHIV-2KR.LTR constructs were digested with AgeI and SacI and ligated to yield a full-length pHIV-2KR.X4 proviral construct. Additionally, a subclone containing a 1.3-kb N-terminal env fragment incorporating the XhoI, SnaBI, and XmaI restriction sequences was created in the pGEM-T Easy cloning vector according to the manufacturer's protocol (Promega, Madison, WI) and is designated pGEM HIV-2KR.X4 (forward primer, GGGACTCGGGATATGTTATGAACGG; reverse primer, CCTTATATGGCACGGTGCATAATTGC).
(iii) Construction of pHIV-2KR.X7 MN V3.
Molt 4/8 cells chronically infected with a chimeric virus containing an HIV-2KR genome and an HIV-1MN V3 loop substitution (pHIV-2KR-MNV3) were obtained as a gift from D. Looney and cultured in RPMI 1640 medium (Gibco/Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT) and 1% l-glutamine and penicillin-streptomycin (PS; Gibco/Invitrogen, Carlsbad, CA) at 37°C with 5% CO2. Genomic DNA was isolated according to the DNeasy protocol (Qiagen, Inc., Valencia, CA); chimeric env sequences were amplified by bulk PCR according to the Expand High Fidelity PCR System protocol (Roche Applied Science, Indianapolis, IN) and cloned into an HIV-2 screening vector, pBlueScript HIV-27312A, developed by our laboratory. Fourteen of the molecularly cloned pHIV-2KR-MNV3 env genes were sequenced, and the nonsynonymous substitutions E204K, T360A, and Y668H were identified in all clones. One pHIV-2KR-MNV3 env clone was identical to the parental pHIV-2KR.P1 env except for three changes (E204K, T360A, and Y668H) and contained a V3 region exactly matching that of HIV-1MN (accession number M17449). This env gene was amplified using primers containing the SnaBI and XbaI restriction sites and was subsequently cloned into the full-length HIV-2KR.X4 backbone to yield pHIV-2KR.X7 MN V3. Additionally, a subclone containing a 1.3-kb N-terminal env fragment was created in the pGEM-T Easy cloning vector according to the manufacturer's protocol (Promega, Madison, WI) and is designated pGEM HIV-2KR.X7 MN V3 (forward primer, GGGACTCGGGATATGTTATGAACGG; reverse primer, CCTTATATGGCACGGTGCATAATTGC).
(iv) Construction of pHIV-2KR.X7.
E204K and T360A nonsynonymous substitutions were incorporated into the pGEM HIV-2KR.X4 subclone using a QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) following the manufacturer's protocol. Primer sequences are as follows: E204K change, ACATCAGTCATCACAAAGTCATGTG and CACATGACTTTGTGATGACTGATG; T360A change, TTGAGGAACCAACGACGCACAGAAAATTAAC andGTTAATTTTCTGTGCGTCGTTGGTTCCTC. The resultant subclone, pGEM HIV-2KR.X7, was then digested by XhoI and XmaI restriction and ligated into the pHIV-2KR.X7 MN V3 proviral clone to yield a full-length construct containing three gp160 adaptations (E204K, T360A, and Y668H) and a wild-type HIV-2KR V3 sequence, designated HIV-2KR.X7.
(v) Construction of HIV-2KR.X7 YU2 V3 and HIV-2KR.X7 Ccon V3.
Substitution of additional HIV-1 V3 sequences was completed using a modified version of the QuikChange Site-Directed Mutagenesis protocol (Stratagene, La Jolla, CA). 5′ Phosphorylated (Phos) primers were designed to anneal to the HIV-2KR backbone immediately flanking the HIV-2KR.X7 V3 region and containing the desired HIV-1 V3 substitutions as follows: HIV-1YU2 V3, GAGCATTGTATACAACAGGAGAAATAATAGGAGATATAAGACAAGCACATTGCTGGTTCGGAGGTG and (Phos)TCCCTGGTCCTATATTTATACTTTTTCTTGTATTGTTGTTGGGTCTTGTACAATGCATTGTGAGATTAT; HIV-1Ccon V3, ACATTCTATGCAACAGGAGACATAATAGGAGATATAAGACAAGCACATTGCTGGTTCGGAGGTGATTGG and (Phos)TTGTCCTGGTCCTATCCTTATACTTTTTCTTGTATTATTGTTGGGTCTTGTACAATGCATTGTGAG. Fifteen pmol of each primer and 100 ng of template pGEM HIV-2KR.X7 MN V3 plasmid were used for each PCR mutagenesis reaction. PCR conditions were as follows: 95°C for 1 min (1 cycle); 95°C for 50 s, 50°C for 50 s, and 68°C for 6 min (18 cycles); and 68°C for 7 min (1 cycle). PCR products were digested with 10 U of DpnI (New England Biolabs, Ipswich, MA) at 37°C for 1 h to cleave template DNA and heat inactivated at 72°C for 30 min. Amplification products were ligated to yield a chimeric env construct with appropriate V3 substitutions and subsequently cloned into the full-length pHIV-2KR.X7 backbone using the XhoI and XmaI restriction sequences.
(vi) Molecular cloning.
pHIV-2KR.X4, pHIV-2KR.X7, pHIV-2KR.X7 MN V3, pHIV-2KR.X7 YU2 V3, and pHIV-2KR.X7 Ccon V3 plasmid DNA was transformed into XL2-Blue MRF′ Ultracompetent cells (Stratagene, La Jolla, CA), plated on LB agar plates supplemented with 5 μg/ml kanamycin and cultured overnight at 30°C. Single colonies were selected and grown overnight in liquid LB broth containing 5 μg/ml kanamycin at 30°C with shaking at 225 rpm, followed by plasmid isolation. Each molecular clone was confirmed by nucleotide sequencing.
Virus stocks.
A total of 3 × 106 293T cells were seeded into 10 cm2 tissue culture dishes and cultured overnight in complete Dulbecco's modified Eagle medium (DMEM; Gibco/Invitrogen, Carlsbad, CA) supplemented with 10% FBS and 1% PS (D10 medium) at 37°C with 5% CO2. The following day, 6 μg of proviral DNA was transfected into 293T cells using Fugene 6 (Roche Applied Science, Indianapolis, IN) as specified by the manufacturer and cultured in D10 medium at 37°C for 48 h. Supernatants containing infectious virus were harvested, cleared of cellular debris by low-speed centrifugation, and used directly or stored at −80°C for future use.
Single-cycle entry assay.
Virus stocks were prepared as described above. The day prior to the assay, 6 × 104 TZM-bl cells (catalog item 8129; NIH AIDS Research and Reference Reagent Program) were seeded into each well of a 24-well tissue culture plate (BD Falcon, Franklin Lakes, NJ) and cultured overnight in D10 medium at 37°C with 5% CO2. The day of assay, D10 medium was removed from the TZM-bl monolayer and replaced with 150 μl of DMEM supplemented with 1% FBS and 1% PS. A total of 150 μl of cleared 293T cell culture supernatant containing infectious virus was then transferred to the TZM-bl reporter cells and incubated for 2 h in the presence of 40 μg/ml DEAE-dextran (Sigma-Aldrich, St. Louis, MO). D10 medium (500 μl) was added, and cells were cultured for an additional 48 h. Luciferase expression was quantified according to the Luciferase Assay System protocol (Promega, Madison, WI). Additionally, 293T transfection supernatants were assayed for HIV-2 Gag protein p27 by simian immunodeficiency virus (SIV) p27 antigen enzyme-linked immunosorbent assay (Zeptometrix Corporation, Buffalo, NY) following the manufacturer's instructions.
Virus titration.
For these studies, 1 × 104 TZM-bl reporter cells were seeded in D10 medium and cultured in 96-well tissue culture plates (BD Falcon, Franklin Lakes, NJ) overnight at 37°C with 5% CO2. The following day, D10 medium was removed from the cell monolayers and replaced with 50 μl of DMEM, 6% FBS, and 1% PS (D6 medium) containing DEAE-dextran hydrochloride at 80 μg/ml (2× final concentration). Serial fivefold serial dilutions of virus stock were prepared in D10 medium. Fifty microliters of virus stock dilution was then transferred to the TZM-bl cells, bringing the final volume to 100 μl and the final DEAE-dextran concentration to 40 μg/ml. Cells were incubated with virus for 48 h at 37°C, lysed, and analyzed for luciferase production using a Luciferase Assay System (Promega, Madison, WI). Future neutralization and inhibition experiments were performed using approximately 1 × 105 relative light units (RLU)/well (96-well format) of virus stock.
Western blot analysis.
Virus stocks were prepared as described above. Forty-eight hours following transfection, 293T supernatants containing infectious virus were cleared of cellular debris by low-speed centrifugation, filtered through 0.22-μm-pore-size disposable vacuum conical filters (Millipore, Billerica, MA), and transferred to 1- by 3.5-in. ultracentrifuge tubes (catalog number 344058; Beckman Coulter, Inc., Fullerton, CA). Two milliliters of 20% sucrose (vol/vol, in water) was then added below the supernatants, and the samples were subjected to ultracentrifugation at 27,000 rpm for 2 h at 4°C in an Optima XL-100k Ultracentrifuge fitted with an SW 28 rotor (Beckman Coulter, Inc., Fullerton, CA). Supernatants were decanted, and pelleted virus was resuspended in 80 μl of phosphate-buffered saline (Gibco/Invitrogen, Carlsbad, CA). Solubilized pellets were heat inactivated at 56°C for 1 h. Virus pellet preps were normalized by immunoblotting for the Gag protein p27 using human anti-HIV-2 polyclonal serum to ensure that an equivalent amount of virus was analyzed in Western blotting experiments. For each sample, an equivalent amount of purified virus, as determined by p27 content, was combined with NuPage sample buffer and reducing agent (Invitrogen, Carlsbad, CA), heated at 99°C for 5 min, loaded into a NuPage 4 to 12% Bis-Tris acrylamide gel (Invitrogen, Carlsbad, CA), and subjected to gel electrophoresis. Proteins were transferred to a polyvinylidene difluoride membrane (Bio-Rad Laboratories, Hercules, CA) following the NuPage transfer protocol (Invitrogen, Carlsbad, CA) and incubated with blocking buffer (5% nonfat dry milk in phosphate-buffered saline) for 1 h at room temperature. Western blots were probed with guinea pig anti-HIV-2 gp120 polyclonal serum (1:1,000; produced in-house) for 1 h at room temperature, followed by a second blocking step, and incubation with a goat anti-guinea pig immunoglobulin G (IgG) conjugated to horseradish peroxidase (1:5,000; SouthernBiotech, Birmingham, AL) for 1 h at room temperature. Immunoblots were then incubated with ECL Western Blotting Detection Reagent (GE Healthcare Life Sciences, Piscataway, NJ) following the manufacturer's protocol and developed. The amount of processed gp120 Env protein was quantified by band densitometry using the Bio-Rad Quantity One software package (Bio-Rad Laboratories, Hercules, CA).
Determination of coreceptor usage.
In order to determine the coreceptor tropism of the HIV-2KR.X7/HIV-1 V3 chimeras, infectivity assays were repeated in the presence of TAK-779 (catalog number 4983; NIH AIDS Research and Reference Reagent Program) and AMD3100 (catalog number 8128; NIH AIDS Research and Reference Reagent Program). For these studies, 1 × 104 TZM-bl reporter cells were seeded and cultured in 96-well tissue culture plates overnight. The following day, the culture medium was removed, and the cells were incubated with D6 medium containing 10 μM TAK-779 and/or 1.2 μM AMD3100 at 37°C for 1 h. Fifty microliters of stock virus supernatant containing 80 μg/ml DEAE-dextran hydrochloride was then added and cultured with the cells for 48 h at 37°C with 5% CO2. Luciferase expression was quantified according to the Luciferase Assay System protocol (Promega, Madison, WI).
Antibodies and plasmas.
MAb b12 was provided by D. Burton (The Scripps Research Institute, La Jolla, CA). MAbs 2F5 (1475; contributed by Hermann Katinger), 4E10 (10091; contributed by Hermann Katinger), and F425 B4e8 (7626; contributed by Marshall Posner and Lisa Cavacini) were obtained from the NIH AIDS Research and Reference Reagent Program. The CD4i MAbs 17b, 19e, 21c, E51, 4.12D, 48d, ED47, and ED49 were provided by J. Robinson (Tulane University Medical Center, New Orleans, LA). 447-52D was provided by S. Zolla-Pazner (New York University Medical Center, New York, NY). The HIV-1 clade B plasmas were obtained from therapy-naïve, chronically infected subjects from the United States, and HIV-1 clade C plasmas were from therapy-naïve, chronically infected subjects in a South African blood bank cohort. IgG was purified by protein G column affinity separation (Pierce-Thermo Fisher Scientific, Rockford, IL).
Neutralization assays.
For neutralization assays, 1 × 104 TZM-bl cells were seeded into 96-well tissue culture plates and cultured overnight in D10 medium. Six fivefold serial MAb, inhibitor, or plasma dilutions were prepared in D6 medium. Virus 293T stock supernatants were prepared as described previously and diluted to approximately 1 × 105 RLU/well in D10 medium containing 80 μg/ml DEAE-dextran hydrochloride. Equal parts MAb and virus dilutions were then combined and incubated at 37°C for 1 h. Medium was removed from the TZM-bl cells, and 100 μl of the virus-MAb mixture was applied to the cells and incubated for an additional 48 h at 37°C with 5% CO2. Control wells included a virus-only control (no MAb) and a medium-only control (no MAb and no virus). Luciferase expression was quantified according to the Luciferase Assay System protocol (Promega, Madison, WI). For plasma neutralization assays, all wells except the starting plasma dilution were supplemented with 2% (for a 1:50 starting dilution) or 10% (for a 1:10 starting dilution) normal human plasma.
Peptide and fusion protein absorption.
Linear peptides comprised of the 24 N-terminal V3 residues of HIV-1YU2 (V3YU2) and HIV-1JR-FL (V3JR-FL) were provided by R. Pantophlet (The Scripps Research Institute, La Jolla, CA). A scrambled 23-amino-acid V3 linear peptide was supplied by H. Liao (Duke University Medical Center, Durham, NC). Inhibition experiments were performed as described above for MAb and plasma neutralization assays, except that MAb/plasma dilutions were preincubated with peptide (final concentration, 50 μg/ml) for 30 min at 37°C prior to incubation with virus. Fusion proteins consisted of a rabbit Fc molecule with a complete HIV-1JR-FL or HIV-1Ccon V3 loop fused to the N terminus (Fc-V3 FP, where FP is fusion protein), a rabbit Fc molecule presenting the base and stem of the HIV-1JR-FL and a Gly-Ala-Gly linker substitution at the V3 tip (Fc-V3p8027 FP), or a control fusion proteins that consisted of a rabbit Fc protein alone (48; also A. Pinter and A. Salomon, unpublished data). Fusion protein inhibition studies were performed as described above for the peptide inhibition assays with Fc-V3 FP at a final concentration of 10 μg/ml. Peptide and Fc-V3 FP concentrations were kept constant in all wells including control wells.
Statistical analyses.
Median reciprocal 50% inhibitory concentration (IC50) neutralization titers were compared by the Wilcoxon rank sum test using the SAS, version 9.1, software package. A P value of 0.05 was considered significant for these studies.
RESULTS
Construction of an HIV-2KR shuttle vector and HIV-2/HIV-1 V3 chimeras.
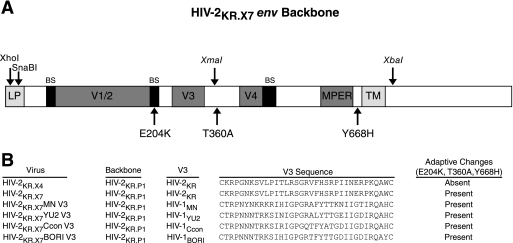
Mamounas and colleagues previously reported that the V3 loop of HIV-1MN could be exchanged for the V3 region of HIV-2, resulting in an infectious chimeric virus (64). However, that work was limited to the introduction of an X4-tropic HIV-1 V3 sequence (MN) into an HIV-2 3′ half-genome (pRT-MNV3) and required cotransfection of an adjoining 5′ half-genome into electroporated Molt 4/8 cells. Progeny virus then underwent undefined adaptive changes ultimately leading to efficient replication in human cells. This strategy for HIV-2/HIV-1 V3 chimera design was not subsequently pursued as an immunogen or as a diagnostic reagent. Based on our success in developing infectious HIV-2/HIV-1 chimeras that expressed antigenically preserved CD4i and MPER epitopes (18, 19, 34, 34, 61; F. Bibollet-Ruche et al., presented at the Keystone Symposium on HIV Vaccines, Keystone, CO, 2006; F. Bibollet-Ruche et al., presented at the AIDS Vaccine 2005 Conference, Montreal, Canada, 2005), we tested several full-length replication-competent HIV-2 or SIV proviruses as potential scaffolds for HIV-1 V3 chimeras and found that the HIV-2KR provirus permitted functional transplantation. Thus, starting with overlapping molecular clones of HIV-2KR (pKTM and pRTsac), we performed a three-way ligation of contiguous viral sequences into a plasmid into which we had inserted HIV-2KR LTR sequences (pHIV-2KR.LTR). This resulted in a plasmid containing a complete HIV-2 provirus that we designated pHIV-2KR.P1. We found this proviral clone to be replication competent in human CD4+ T-cell lines H9, Jurkat, PM1, and Molt 4/8 (data not shown). To provide a convenient shuttle system for exchanging HIV-1 V3 sequences into the corresponding region of HIV-2, we modified pHIV-2KR.P1 to contain XhoI, XmaI, XbaI, and SnaBI restriction sites at positions indicated in Fig. 1A without altering the amino acid coding sequence. We modified the resulting cassette vector, pHIV-2KR.X4, further by site-directed mutagenesis to create nonsynonymous substitutions at positions E204K, T360A, and Y668H in Env (corresponding to positions Q208, T363, and Y686 in HXB-2 [82]), giving rise to the vector pHIV-2KR.X7. The last three substitutions were introduced because we found these mutations to represent adaptive changes under strong positive selection pressure in samples from the original cultures of HIV-2KR-MNV3 (64). Proviral clones containing the complete V3 sequences from HIV-1MN (accession number M17449) (36), HIV-1YU2 (accession number M93258) (59), or HIV-1Ccon (Los Alamos Sequence Database [http://www.hiv.lanl.gov/]) were then constructed as described in the Materials and Methods section and depicted in Fig. 1B. The env genes of all clones were sequenced to confirm that only the intended changes were present in the final chimeric proviral constructs.
FIG. 1.
Construction of HIV-2KR.X7 Env scaffold and HIV-2/HIV-1 V3 chimeras. (A) The parental pHIV-2KR.P1 env backbone was modified to include unique silent restriction sequences for XhoI, SnaBI (located within the leader peptide region [LP]), XmaI (located in the C3 coding region), and XbaI (located 3′ of the transmembrane region [TM]) to create the parental shuttle vector pHIV-2KR.X4. Nonsynonymous mutations (see Materials and Methods) were introduced at amino acid positions 204 (E204K), 360 (T360A), and 668 (Y668H) to create the final scaffold vector pHIV-2KR.X7. BS, bridging sheet. (B) V3 sequences for HIV-1MN, HIV-1YU2, HIV-1Ccon, and HIV-1BORI were incorporated into the pHIV-2KR.X7 env cassette as described in Materials and Methods to generate the chimeric proviruses pHIV-2KR.X7 MN V3, pHIV-2KR.X7 YU2 V3, pHIV-2KR.X7 Ccon V3, and pHIV-2KR.X7 BORI V3.
Infectivity and replication competence of HIV-2KR.X7/HIV-1 V3 chimeras.
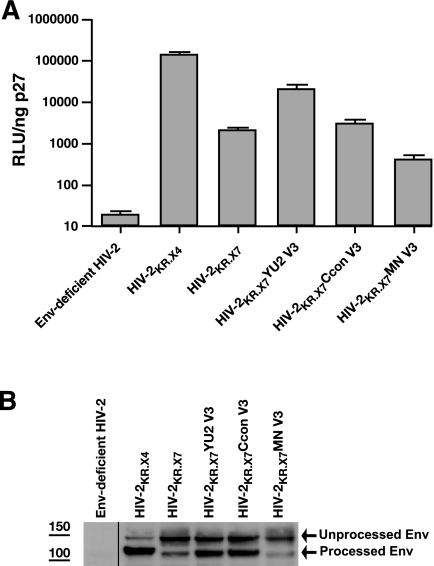
The ability of the HIV-2KR.X7/HIV-1 V3 chimeric Env proteins to confer virus entry into target cells was assessed using a TZM-bl reporter cell assay (96, 97). All virus stocks were generated in 293T cells, and virus inocula were normalized by Gag antigen (p27) content. An HIV-2 Env-deficient virus gave negligible luminescence readings and served as a negative control. Not surprisingly, the parental virus HIV-2KR.X4 was most efficient at virus entry (Fig. 2A). HIV-2KR.X7 was less infectious than HIV-2KR.X4, although still within a range observed for primary HIV-2 and HIV-1 strains and previously described HIV-2/HIV-1 chimeras (18, 34, 49). The V3 chimeric viruses HIV-2KR.X7 YU2 V3, HIV-2KR.X7 Ccon V3, and HIV-2KR.X7 MN V3 were infectious and again exhibited a broad range in infectivity compared with the isogenic HIV-2KR.X7 scaffold and the HIV-2KR.X4 parental strain. Like HIV-2KR.X4 and HIV-2KR.X7, the three HIV-2KR.X7/HIV-1 V3 chimeras were replication competent in one or more human cell types, including peripheral blood mononuclear cells or the cell lines H9, Jurkat, PM1, or Molt 4/8. Interestingly, the HIV-2KR.X7 MN V3 virus, which was only marginally infectious in the TZM-bl reporter cell assay, was highly replicative in Molt 4/8 and Jurkat cells (data not shown).
FIG. 2.
Infectivity and Env processing of HIV-2KR.X7/HIV-1 V3 chimeras. (A) Proviral constructs were transfected into 293T cells to generate infectious virus stocks. Virus infectivity was assessed by luciferase production (RLU) in the TZM-bl single-cycle entry assay (96, 97). Luciferase readout was normalized by HIV-2 p27 antigen quantification. Data are presented as the mean and standard deviation of three independent experiments. Env-deficient HIV-2 was tested as a negative control. (B) Virus stocks were prepared by 293T transfection. At 48 h after transfection, virus was harvested from culture supernatants, pelleted, solubilized, and subjected to sodium dodecyl sulfate-gel electrophoresis and immunoblotting with a guinea pig anti-HIV-2 gp120 polyclonal antibody. Envelope-deficient HIV-2 was included as a negative control. The positions of the gp160 precursor glycoprotein and processed gp120 are identified by arrows.
Envelope processing in wild-type and chimeric viruses.
Infectivity of the different wild-type and chimeric viruses in the TZM-bl reporter cell assay varied by over 100-fold (Fig. 2A). We postulated that these differences might be explained in part by altered processing or content of Env gp120/gp41 on the virion surface. Figure 2B shows the immunoreactivity of purified virus particles with a guinea pig anti-HIV-2 gp120 polyclonal serum. Visual inspection of the gels showed that Env processing to mature virion-associated gp120 was most efficient for the parental virus HIV-2KR.X4; intermediate for HIV-2KR.X7 YU2 V3, HIV-2KR.X7 Ccon V3, and HIV-2KR.X7; and least efficient for HIV-2KR.X7 MN V3; furthermore this processing was correlated with viral infectivity in the TZM-bl reporter cell assay. To formally address this relationship, we quantified viral infectivity by chemiluminescence (RLU) (Fig. 2A) and gp120 Env content by band densitometry (Fig. 2B) and found a significant correlation (ρ = 0.87; P = 0.05). Because the HIV-2KR.X7 MN V3 virus was substantially less infectious in the TZM-bl reporter assay than the other two chimeras, we elected to exclude this virus from further analysis.
Selective inhibition of HIV-2KR.X7/HIV-1 V3 chimeras by receptor, coreceptor, and gp41 fusion inhibitors.
Since virus-cell membrane attachment and fusion represent essential, highly ordered steps in virus entry, we asked if HIV-2KR.X7/HIV-1 V3 chimeras were inhibited by CD4 receptor, chemokine coreceptor, and gp41 fusion protein inhibitors in a manner comparable to that of the parental HIV-2KR virus strain and other primary HIV-2 and HIV-1 viruses. HIV-2KR.X4, HIV-2KR.X7, HIV-2KR.X7 YU2 V3, and HIV-2KR.X7 Ccon V3 were each inhibited efficiently by soluble CD4 (sCD4) in concentrations comparable to primary HIV-2 strains, which as a group are more sensitive to sCD4 than are HIV-1 viruses (Table 1) (18, 49). CD4 binding is followed by Env-coreceptor engagement, and the primary determinants of coreceptor tropism reside in residues located within V3 (37, 43, 44). We thus tested if virus entry was blocked selectively by coreceptor antagonists and if coreceptor usage of the chimeric viruses corresponded with that of the virus from which the V3 originated. HIV-2KR and HIV-1NL4.3 are X4-tropic viruses, and HIV-1YU2 and HIV-1Ccon are R5 tropic; each served as a control. To determine coreceptor usage, the selective coreceptor antagonists TAK-779 (a CCR5 antagonist) and AMD3100 (a CXCR4 antagonist) were used to compete with virus entry in TZM-bl reporter cells (104). The results, shown in Fig. 3A, demonstrate that entry of HIV-2KR.X7 YU2 V3 and HIV-2KR.X7 Ccon V3 was completely inhibited by TAK-779 but not by AMD3100. Conversely, entry of HIV-2KR.X4 and HIV-2KR.X7 was inhibited by AMD3100 but not by TAK-779. HIV-1NL4.3 and HIV-1YU2 were inhibited by AMD3100 and TAK-779, respectively. These findings indicate that the chimeric viruses maintain the CD4-dependent, R5-tropic phenotype of their parental HIV-1 Env proteins and that the coreceptor specificity for these viruses is determined solely by the V3 region. To determine if the HIV-2KR.X7 HIV-1 V3 chimeric viruses were selectively and efficiently neutralized by inhibitors of gp41-mediated fusion, peptide homologues of the HIV-2/HIV-1 heptad repeat region 2 (T1249) and the HIV-1 (only) heptad repeat region 2 (T20) (22) were tested against these viruses and against relevant HIV-2KR and HIV-1YU2 controls. HIV-2KR and its derivatives including the HIV-2KR.X7/HIV-1 V3 chimeras were each neutralized efficiently by T1249 but not at all by T20 (Table 1). Moreover, the chimeric viruses exhibited typical sigmoidally shaped inhibition curves, with IC50 values for T1249 similar to the value of the parental virus HIV-2KR.X4 (Fig. 3B) and of HIV-1 viruses (22). Altogether, the findings suggest that the HIV-2KR.X7/HIV-1 V3 chimeras maintain their selective binding properties for CD4, coreceptor, and gp41 ligands and that the molecular events underlying their entry into cells are preserved in a biologically relevant fashion comparable to wild-type HIV-1 and HIV-2 viruses.
TABLE 1.
Neutralization titers of HIV-2, HIV-1, and HIV-2/HIV-1 V3 chimeras
| Antibody or inhibitor | Ligand binding region | IC50 (μg/ml)a
|
|||||
|---|---|---|---|---|---|---|---|
| HIV-2KR.P1 | HIV-2KR.X4 | HIV-2KR.X7 | HIV-2KR.X7 YU2 V3 | HIV-2KR.X7 Ccon V3 | HIV-1YU2 | ||
| IgG1 b12 | CD4bs | >10 | >10 | >10 | >10 | >10 | 3.1 (0.60) |
| 2F5 | HIV-1 MPER | >10 | >10 | >10 | >10 | >10 | >10 |
| 4E10 | HIV-1 MPER | >10 | >10 | 2.0 (0.47) | 0.93 (0.11) | 1.0 (0.42) | >10 |
| 447-52D | HIV-1 V3 | >10 | >10 | >10 | 0.001 (0.0005) | 0.03 (0.02) | 4.15 (0.35) |
| F425 B4e8 | HIV-1 V3 | >10 | >10 | >10 | 0.004 (0.001) | 0.48 (0.19) | >10 |
| E51 | HIV-1 CD4i | >10 | >10 | >10 | >10 | >10 | >10 |
| 17b | HIV-1 CD4i | >10 | >10 | >10 | >10 | >10 | >10 |
| 48d | HIV-1 CD4i | >10 | >10 | >10 | >10 | >10 | ND |
| 21c | HIV-1 CD4i | >10 | >10 | >10 | >10 | >10 | >10 |
| 4.12D | HIV-1 CD4i | >10 | >10 | >10 | >10 | >10 | >10 |
| 19e | HIV-1 CD4i | >10 | >10 | >10 | >10 | >10 | >10 |
| ED47 | HIV-1 CD4i | >10 | >10 | >10 | >10 | >10 | >10 |
| ED49 | HIV-1 CD4i | >10 | >10 | >10 | >10 | >10 | >10 |
| T1249 | gp41 | ND | 0.04 (0.03) | 0.23 (0.13) | 0.02 (0.002) | 0.10 (0.09) | 0.08 (0.02) |
| T20 | gp41 | ND | >2.5 | >2.5 | >2.5 | >2.5 | 0.22 (0.08) |
| sCD4 (nM) | CD4bs | 116 (54) | 49 (39) | 0.23 (0.11) | 2.3 (0.9) | 2.3 (1.2) | 10.5 (0.71) |
All neutralization titers (IC50s) are presented as the mean of at least four independent determinations with standard deviation noted in parentheses. ND, not done.
FIG. 3.
Coreceptor tropism of HIV-2KR.X7/HIV-1 V3 chimeras and fusion inhibition by T1249. (A) TZM-bl reporter cells were incubated with 10 μM TAK-779 (CCR5 antagonist), 1.2 μM AMD3100 (CXCR4 antagonist), medium only (untreated), or a combination of both coreceptor inhibitors for 30 min prior to the addition of infectious virus stock. TZM-bl cells express high levels of surface CD4, CCR5, and CXCR4, rendering them susceptible to infection by both CCR5- and CXCR4-tropic viruses. Entry was assessed by luciferase production (RLU) measured 48 h after infection. Values are presented as percent infectivity compared to the untreated control. HIV-1NL4.3 is a CXCR4-tropic control virus, and HIV-1YU2 is a CCR5-tropic control virus. Data are presented as the mean and standard deviation of 4 to 12 determinations. (B) Serial dilutions of T1249 were combined with an equal volume of infectious virus stock, incubated at 37°C for 1 h, and transferred to TZM-bl reporter cells. Virus entry was measured by luciferase production 48 h later and normalized to luciferase expression in the absence of T1249. Inhibition curves for HIV-2KR.X4, HIV-2KR.X7 YU2 V3, and HIV-2KR.X7 Ccon V3 are shown. IC50 values are presented in Table 1. Data represented are the mean and standard deviation of three independent experiments.
Neutralization of HIV-2KR.X7/HIV-1 V3 chimeras by HIV-1 MAbs.
To evaluate the utility of HIV-2KR.X7/HIV-1 V3 chimeras to detect HIV-1 V3-specific NAbs and to distinguish such antibodies from NAbs with other specificities, we examined the sensitivity of the parental and chimeric viruses to a panel of well-characterized HIV-1 MAbs having defined epitope specificity. Neutralization studies were performed by incubating virus stocks with serial dilutions of MAb starting at 10 μg/ml prior to transfer to TZM-bl cells. For this analysis, we first assessed the neutralization sensitivity of the wild-type HIV-2KR virus and its derivative HIV-2KR.X4 (Table 1 and Fig. 4). HIV-2KR.P1 and HIV-2KR.X4 were resistant to neutralization by all of the MAbs tested including those specific for the CD4bs, CD4i, MPER, or V3 epitopes although there was slight neutralization sensitivity to the MPER MAb 4E10. The 4E10 epitope is uniformly conserved on HIV-1 viruses of all subtypes and is present on many, but not all, strains of HIV-2 (H. Li, F. Bibollet-Ruche, J. M. Decker, et al., presented at the Keystone Symposium on HIV Vaccines, Keystone, CO, 2006). We next determined the neutralization profile of the third control virus, HIV-2KR.X7, to verify that the E204K, T360A, and Y668H mutations did not render this virus differentially sensitive to neutralization by HIV-1 MAbs (Fig. 4 and Table 1). Again, we found that HIV-2KR.X7 was resistant to neutralization by all of the HIV-1 MAbs tested except 4E10. We also tested HIV-2KR.X4 and HIV-2KR.X7 (and the two HIV-2KR.X7/HIV-1 V3 chimeras) for sensitivity to three HIV-2-specific neutralizing MAbs obtained as a gift from James Robinson (unpublished data). The MAbs recognize the HIV-2 V3 (6.10F), coreceptor binding region (1.4H), and an undefined epitope on gp120 (1.4B), and all three exhibited similar neutralization activities (IC50 of >10 μg/ml) against the two HIV-2 strains and the two HIV-2KR.X7/HIV-1 chimeras. These data, together with results presented above, suggest that the wild-type HIV-2KR.P1 virus, its derivatives HIV-2KR.X4 and HIV-2KR.X7, and the HIV-2KR.X7/HIV-1 V3 chimeras maintain functional and antigenic characteristics typical of what we have reported previously for primary HIV-2 viruses (18). Finally, we evaluated the two HIV-2KR.X7/HIV-1 V3 chimeras for sensitivity to V3 and non-V3 neutralizing HIV-1 MAbs. Figure 4 and Table 1 show that the HIV-2KR.X7 YU2 V3 and HIV-2KR.X7 Ccon V3 chimeras were resistant to all of the non-V3 MAbs except for 4E10, which inhibited both chimeras to an extent indistinguishable from the HIV-2KR.X7 control virus. However, both chimeras were remarkably sensitive to neutralization by the HIV-1 V3-specific MAbs 447-52D and F425 B4e8. For example, 447-52D and F425-B4e8 neutralized the HIV-2KR.X7 YU2 V3 chimera at an IC50 of 0.001 μg/ml and 0.004 μg/ml, respectively. The same 447-52D and F425-B4e8 MAbs neutralized the HIV-2KR.X7 Ccon V3 chimera at an IC50 of 0.03 μg/ml and 0.48 μg/ml, respectively. Importantly, HIV-2KR.X4 and HIV-2KR.X7 were each completely resistant to neutralization by both 447-52D and F425 B4e8. To explore further the sensitivity of the HIV-2KR.X7 YU2 (clade B) V3 chimera to other V3 specific MAbs, we tested its sensitivity to the MAbs 257-D and 268, which neutralize the TCLA strain HIV-1MN (30, 31; also data not shown), and 5F7 and 4G10, which target the atypical V3 of the TCLA strain HIV-1IIIB that contains a unique Gln-Arg insertion adjacent to the GPGX crown residues (95). The HIV-2KR.X7 YU2 V3 chimera was sensitive to 257-D and 268 (IC50 of 0.06 and 8.9 μg/ml, respectively) but not to 5F7 or 4G10 (95; also data not shown).
FIG. 4.
Neutralization of HIV-2KR.X7/HIV-1 V3 chimeras by HIV-1 MAbs. HIV-2KR.X7/HIV-1 V3 chimeras and control viruses were tested for neutralization susceptibility to HIV-1 MAbs targeting the CD4bs (b12), MPER (2F5, 4E10), and CD4i (17b, 19e, 21c, E51, 4.12D, ED47, ED49) epitopes and V3 (447-52D and F425 B4e8). Fivefold serial MAb dilutions were prepared at a starting concentration of 20 μg/ml, mixed with an equal volume of infectious virus stock to give the final concentrations shown, and incubated at 37°C for 1 h prior to transfer to TZM-bl reporter cells. Virus entry was measured by luciferase production 48 h after infection and normalized to luciferase expression in the absence of MAb. Non-V3-specific HIV-1 MAbs are represented by black lines. HIV-1 V3-specific MAbs (447-52D and F425 B4e8) are shown by red lines. IC50 neutralization values are presented in Table 1. Data represented are the mean and standard deviation of three independent experiments.
Epitope specificity of neutralization of HIV-2KR.X7/HIV-1 V3 chimeras by HIV-1 MAbs.
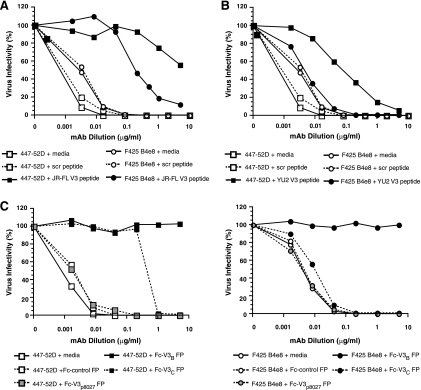
To confirm the V3 specificity of neutralization detected by the HIV-2KR.X7/HIV-1 V3 chimeras, we performed a series of competition experiments using linear peptides comprising the 24 N-terminal V3 residues of HIV-1JR-FL or HIV-1YU2 that include the epitopes of the 447-52D and F425 B4e8 MAbs or a fusion protein that presents the full-length V3 in its disulfide-bound form. Sequences of the V3 peptides used in these antibody absorption experiments are shown in Table 2. For competition studies, MAb was incubated with peptide or protein at 37°C for 30 min prior to the addition of HIV-2KR.X7 YU2 V3 and transfer to TZM-bl reporter cells. Figure 5A shows that a V3 linear peptide from HIV-1JR-FL (V3JR-FL) was able to absorb virtually all 447-52D neutralizing activity and a substantial proportion of F425 B4e8 neutralizing activity. An analogous peptide composed of the N-terminal residues of the HIV-1YU2 V3 (V3YU2) removed most of the 447-52D neutralizing activity but none of the F425 B4e8 reactivity (Fig. 5B). The decreased affinity of F425 B4e8 for the HIV-1YU2 peptide is most likely due to the presence of a leucine residue at position 317 in this peptide. Crystallographic descriptions of V3 peptide-F424 B4e8 complexes demonstrate that the aromatic side chain of Phe317, present in the HIV-1JR-FL peptide but absent in the V3YU2 peptide, serves to stabilize an unusual conformation adopted by Arg315 in the V3 crown that is required for high-affinity F425 B4e8 binding (2). A similar finding was reported by Dhillon and colleagues, who found that the linear peptide of HIV-1YU2 was not as effective as that of HIV-1JR-FL in competing for F425 B4e8 neutralization of HIV-1JR-FL (19). Scrambled V3 peptide and medium-only controls had no effect on 447-52D- and F425 B4e8-mediated neutralization of HIV-2KR.X7 YU2 V3 (Fig. 5A and B).
TABLE 2.
V3 peptides and fusion proteins used in absorption studies
| Protein | V3 derivation | Sequence |
|---|---|---|
| V3YU2 24-mer | HIV-1YU2 (clade B) | CTRPNNNTRKSINIGPGRALYTTG |
| V3JR-FL 24-mer | HIV-1JR-FL (clade B) | CTRPNNNTRKSIHIGPGRAFYTTG |
| Rabbit Fc-V3B FP | HIV-1JR-FL (clade B) | CTRPNNNTRKSIHIGPGRAFYTTGEIIGDIRQAHC |
| Rabbit Fc-V3C FP | HIV-1Ccon (clade C) | CTRPNNNTRKSIRIGPGQTFYATGDIIGDIRQAHC |
| Rabbit Fc-V3p8027 FP | HIV-1JR-FL (clade B) | CTRPNNNT--------GAG--------GDIRQAHC |
FIG. 5.
Epitope specificity of HIV-2KR.X7 YU2 V3 chimera neutralization by HIV-1 V3 MAbs. HIV-2KR.X7 YU2 V3 neutralization by 447-52D or F425 B4e8 is competed by V3JR-FL 24-mer peptide (A), V3YU2 24-mer peptide (B), or by Fc-V3B FP or Fc-V3C FP (C). V3 peptides and fusion protein (FP) competitors are shown in Table 2. For all experiments, fivefold serial dilutions of HIV-1 MAbs were combined with peptide or fusion protein and incubated at 37°C for 30 min. Virus was then added and incubated at 37°C for 1 h, and the mixture was transferred to TZM-bl reporter cells. The final peptide and fusion protein concentrations in all wells were 50 μg/ml and 10 μg/ml, respectively. Luciferase expression was assessed 48 h later and was normalized to that in the absence of antibody or inhibitor. Scrambled (scr) V3 peptide, fusion proteins lacking complete or partial V3 sequences, and medium-only controls were included.
Because competition for neutralizing activity of the MAbs by linear V3 peptides was not complete, we considered the possibility that linear peptides might not replicate V3 secondary and tertiary structure sufficiently to serve as effective competing antigens. Thus, we conducted similar absorption experiments using a chimeric protein containing a rabbit Fc backbone with the V3 residues from subtype B HIV-1JR-FL or subtype C consensus sequences fused to the N terminus of the Fc protein (A. Salomon and A. Pinter, unpublished). These fusion proteins are referred to as Fc-V3B FP and Fc-V3C FP, respectively. The Fc-V3B FP completely eliminated 447-52D- and F425 B4e8-mediated neutralization of HIV-2KR.X7 YU2 V3 (Fig. 5C). The Fc-V3C FP eliminated most 447-52D-mediated neutralization of HIV-2KR.X7 YU2 V3 but only a fraction of F425 B4e8-mediated neutralization. The greater efficiency of Fc-V3B FP than Fc-V3C FP in blocking 447-52D- and F425 B4e8-mediated neutralization is consistent with the HIV-1 B clade origin of these two human MAbs (13, 72, 108). The greater efficiency of Fc-V3C FP in blocking 447-52D neutralization than F425 B4e8 neutralization is consistent with the greater dependence of 447-52D on main chain interactions with the amino-terminal limb of V3 than F425 B4e8, which is more dependent on side chain interactions, especially with Arg315 in the V3 crown (2, 93). In most clade C viruses and in the Fc-V3C FP, Arg315 is replaced by Gln315 (Los Alamos Sequence Database [http://www.hiv.lanl.gov/]). Additionally, we tested a third fusion protein that consisted of a rabbit Fc backbone with the V3 sequence of HIV-1JR-FL fused to the N terminus, but in which 19 residues from the V3 crown (Arg-Lys-Ser-Ile-His-Ile-Gly-Pro-Gly-Arg-Ala-Phe-Tyr-Thr-Thr-Gly-Glu-Ile-Ile) are replaced by a Gly-Ala-Gly linker. This construct is referred to as Fc-V3p8027 FP and was employed to determine the proportion of the V3 reactivity that is directed at the V3 tip. As expected, we observed no reduction in the neutralization of HIV-2KR.X7 YU2 V3 by 447-52D and F425 B4e8 after absorption with the Fc-V3p8027 FP (Fig. 5C). A control rabbit Fc protein lacking HIV-1 V3 sequences and a medium-only control also had no effect on virus neutralization by 447-52D and F425 B4e8 (Fig. 5C). Fusion protein competition experiments were performed with 10 μg/ml (0.2 nM) Fc-V3 FP present in all wells. Peptide absorption experiments consisted of 50 μg/ml (20 nM) linear peptide in all wells. This corresponds to an approximately 100-fold molar excess of peptide relative to fusion protein for these studies. Divalency of the fusion proteins and stabilization in V3 loop conformations may contribute to their enhanced potency in binding V3-specific antibodies.
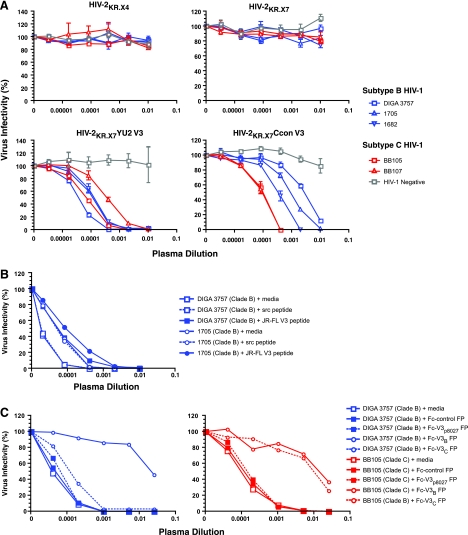
Neutralization of HIV-2KR.X7/HIV-1 V3 chimeras by polyclonal HIV-1-positive plasma.
Having established that the HIV-2KR.X7/HIV-1 V3 chimeras represent a sensitive and specific virological reagent to detect HIV-1 V3-specific neutralizing human MAbs, we next sought to determine if these reagents could detect anti-V3 NAbs in polyclonal HIV-1-reactive plasma from chronically infected individuals. For these experiments, we first tested the ability of plasmas from three subtype B HIV-1-infected patients, two subtype C HIV-1-infected patients, and a negative control individual to neutralize the HIV-2KR.X7/HIV-1 V3 chimeras and control viruses. Figure 6A shows that none of the six plasmas neutralized the HIV-2KR.X4 or HIV-2KR.X7 control viruses, indicating that cross-reactivity to HIV-2 V3 and non-V3 epitopes (including 4E10-like epitopes) is absent in these plasmas. Conversely, the three clade B plasmas neutralized the HIV-2KR.X7 YU2 V3 chimera (clade B V3) potently (reciprocal median IC50 titer of 6,850) and the HIV-2KR.X7 Ccon V3 chimera (clade C V3) quite effectively as well, albeit at lower titers (reciprocal median IC50 titer of 833). The clade C plasmas neutralized both HIV-2KR.X7/HIV-1 V3 chimeras potently (median IC50s of 8,047 and 8,383 for HIV-2KR.X7 YU2 V3 and HIV-2KR.X7 Ccon V3, respectively). Additionally, HIV-2KR.X7 YU2 V3 was neutralized by purified IgG from plasmas DIGA 3757 and 1682 at an IC50 titer of 2.0 μg/ml and 1.4 μg/ml, respectively, thus accounting for virtually all of the neutralizing activity in these plasmas (Table 3) (3). These findings, together with the selective inhibition of neutralization by V3-specific peptides and fusion proteins, demonstrate that the plasma neutralizing activities detected by the HIV-2KR.X7/HIV-1 V3 chimeras are IgG mediated. The negative control plasma exhibited no neutralizing activity against any of the viruses tested.
FIG. 6.
Epitope specificity of neutralization of HIV-2KR.X7/HIV-1 V3 chimeras by HIV-1 subtypes B and C plasmas. (A) Neutralization of HIV-2KR.X7/HIV-1 YU2 and Ccon V3 chimeras but not parental HIV-2 strains by plasma from subjects infected by HIV-1 subtype B and C. Neutralization of the HIV-2KR.X7 YU2 V3 chimera by HIV-1 clade B plasma is inhibited modestly by the V3JR-FL 24-mer peptide (B) and nearly completely by Fc-V3B FP (C, left). Neutralization of the HIV-2KR.X7 YU2 V3 chimera by HIV-1 clade C plasma is inhibited equally by Fc-V3B FP and Fc-V3C FP (C, right). V3 peptides and fusion protein (FP) competitors are shown in Table 2. For all experiments, fivefold serial dilutions of plasma were combined with peptide or fusion protein and incubated at 37°C for 30 min. Virus was then added and incubated at 37°C for 1 h, and the mixture transferred to TZM-bl reporter cells. The final peptide and fusion protein concentrations in all wells were 50 μg/ml and 10 μg/ml, respectively. Luciferase expression was assessed 48 h later and was normalized to that in the absence of plasma or inhibitor. Scrambled (scr) V3 peptide, fusion proteins lacking complete or partial V3 sequences, and medium-only controls had no effect on virus neutralization.
TABLE 3.
Neutralization titers of 11 subtype B and 10 subtype C plasmas against HIV-2, HIV-1, and HIV-2/HIV-1 V3 chimeras
| Plasma | Subtype | Reciprocal IC50a
|
||||
|---|---|---|---|---|---|---|
| HIV-2KR.X4 | HIV-2KR.X7 | HIV-2KR.X7 Ccon V3 | HIV-2KR.X7 YU2 V3 | HIV-1YU-2 | ||
| DIGA 3757 | B | <20 | <20 | 285 | 20,777 | 158 |
| 1705 | B | <20 | <20 | 833 | 6,849 | <20 |
| 1682 | B | <20 | <20 | 2,453 | 6,098 | 33 |
| YOAL 0522 | B | <20 | <20 | 1,306 | 3,804 | <20 |
| MIGE 1132 | B | <20 | <20 | 794 | 2,449 | <20 |
| RUTH 1145 | B | <20 | <20 | 740 | 873 | <20 |
| SMST 1012 | B | <20 | <20 | 3,102 | 9,259 | 47 |
| WATI 0885 | B | <20 | <20 | 45 | 331 | <20 |
| LENA 1029 | B | <20 | <20 | 674 | 3,189 | <20 |
| BELI 1233 | B | <20 | <20 | 3,529 | 6,080 | <20 |
| HIDO 1099 | B | <20 | <20 | 628 | 305 | <20 |
| BB10 | C | <20 | <20 | 4,713 | 96 | <20 |
| BB22 | C | <20 | <20 | 8,197 | 3,578 | 26 |
| BB25 | C | <20 | <20 | 4,847 | 3,347 | 21 |
| BB52 | C | <20 | <20 | 14,096 | 722 | 27 |
| BB70 | C | <20 | <20 | 21,618 | 15,104 | 147 |
| BB72 | C | <20 | <20 | 2,530 | 1,085 | <20 |
| BB83 | C | <20 | <20 | 42,369 | 936 | 20 |
| BB105 | C | <20 | <20 | 7,987 | 13,736 | <20 |
| BB106 | C | <20 | <20 | 13,062 | 198 | 37 |
| BB107 | C | <20 | <20 | 8,780 | 2,358 | 198 |
| HIV-1 negative | <20 | <20 | <20 | <20 | <20 | |
Neutralization titers are expressed as the reciprocal IC50 plasma dilution of a representative experiment. Experiments were repeated at least three times.
Epitope specificity of neutralization of HIV-2KR.X7/HIV-1 V3 chimeras by polyclonal HIV-1-positive plasma.
To confirm the V3 specificity of the neutralizing activity in polyclonal plasma, we tested the ability of the V3JR-FL linear peptide to compete with plasma antibodies for neutralization. The V3JR-FL 24-mer peptide was shown by the data given in Fig. 5 to compete more effectively against 447-52D and F425 B4e8 neutralization than did the V3YU2 24-mer peptide although neither competed as effectively as the rabbit Fc-V3B fusion protein. Figure 6B shows that the linear V3JR-FL peptide impaired the neutralizing activity of the two different polyclonal clade B plasmas for the YU2 chimera only minimally. This suggested to us that additional anti-V3 specificities besides those represented by 447-52D and F425 B4e8 might be present in the polyclonal plasma or that, as indicated by the data in Fig. 5, a linear V3 peptide is generally less efficient at competing with antibody than a V3 loop presented in a more native conformation. Thus, we tested the ability of the rabbit Fc-V3B FP and Fc-V3C FP to compete with NAbs in the polyclonal plasmas. Figure 6C shows that absorption with the Fc-V3B FP, but not the Fc-V3C FP, resulted in ∼300-fold (99.7%) reduction in the clade B plasma (DIGA 3757) IC50 neutralization titer against HIV-2KR.X7 YU2 V3. Absorption of the clade C plasma with either the Fc-V3C FP or the Fc-V3B FP resulted in ∼100-fold (99%) reduction in the IC50 neutralization titer against the same virus. We also performed competition experiments using the Fc-V3p8027 FP, which lacks the V3 crown residues. Our data show that the Fc-V3p8027 FP was ineffective at competing for antibody binding in the DIGA 3757 (clade B) and BB105 (clade C) plasmas and that these plasmas retained 100% of their neutralizing activity against the HIV-2KR.X7 YU2 V3 chimera relative to medium-only and rabbit Fc-only controls (Fig. 6C). These findings confirmed that most of the plasma neutralizing activity that was detected by the HIV-2KR.X7/HIV-1 V3 chimeras was V3 specific and dependent upon residues in the V3 crown that overlap the 447-52D and F425 B4e8 epitopes. This could be because V3 crown residues are part of the antibody binding epitope per se or because the GPGX beta-hairpin turn is required for juxtaposition of antibody contact sites elsewhere in the V3 loop.
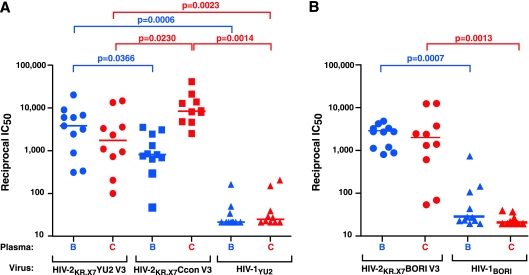
Breadth and potency of V3-specific NAbs in plasma of subjects infected with HIV-1 clade B or clade C against HIV-2KR.X7/HIV-1 V3 chimeras versus a V3 sequence-matched primary virus strain.
Finally, we sought to assess the breadth and potency of V3-specific NAbs in an expanded panel of HIV-1-positive plasmas against the HIV-2KR.X7/HIV-1 V3 chimeras compared with a V3 sequence-matched primary virus strain, HIV-1YU2. We tested a total of 21 plasmas obtained from individuals chronically infected with either HIV-1 subtype B or C against both HIV-2KR.X7/HIV-1 V3 chimeras and against a molecular clone-derived primary virus strain, HIV-1YU2, whose V3 loop sequence is identical to that of the HIV-2KR.X7 YU2 V3 chimera. The results, shown in Table 3 and Fig. 7, demonstrate that all of the plasmas contained high titers of V3-specific NAbs against the clade-matched chimeras. For example, 10 clade C plasmas neutralized the HIV-2KR.X7 Ccon (clade C) V3 chimera with reciprocal IC50 titers ranging from 2,530 to 42,369 (median titer, 8,488). Similarly, 11 clade B plasmas neutralized the HIV-2KR.X7 YU2 (clade B) V3 chimera with reciprocal IC50 titers ranging from 305 to 20,777 (median titer, 3,804). These plasmas also exhibited substantial cross-clade neutralizing activity with the 10 clade C plasmas neutralizing HIV-2KR.X7 YU2 V3 with reciprocal IC50 titers ranging from 96 to 15,104 (median titer, 1,722) and the 11 clade B plasmas neutralizing HIV-2KR.X7 Ccon (clade C) V3 chimera with reciprocal IC50 titers ranging from 45 to 3,529 (median titer, 794). The differences in neutralization titers within clade versus cross-clade for the B-clade and C-clade plasmas were in each case statistically significant (P = 0.0366 and P = 0.0230, respectively). The same HIV-1 clade B and clade C plasmas were then tested for neutralizing activity against the primary virus strain HIV-1YU2. Clade B plasmas exhibited IC50 titers ranging from <20 to 160 (median, <20), which were far less than against the clade-matched HIV-2KR.X7 YU2 V3 chimera (reciprocal IC50 titers of 305 to 20,777; median titer, 3,804; P = 0.0006). Similarly, clade C plasmas exhibited IC50 titers against the primary virus strain HIV-1YU2 ranging from <20 to 200 (median, 23), which again were far less than against the HIV-2KR.X7 YU2 V3 chimera (P = 0.0023) or the HIV-2KR.X7 Ccon V3 chimera (P = 0.0014).
FIG. 7.
Breadth and potency of V3-specific NAbs in plasma of clade B- and clade C-infected subjects. Eleven clade B plasmas (blue) and 10 clade C plasmas (red) were tested for V3-specific neutralizing activity against HIV-2KR.X7 YU2 V3, HIV-2KR.X7 Ccon V3, and HIV-1YU2 (A) and HIV-2KR.X7 BORI V3 and HIV-1BORI (B). Reciprocal IC50s and median values (horizontal lines) are plotted for each plasma-virus combination. IC50s for all plasmas tested against control HIV-2 viruses were all <1:20. Comparisons showing statistical significance are indicated.
These results suggested that individuals who are naturally infected by HIV-1 regularly develop high titers of V3-specific antibodies having broad within- and cross-clade V3 reactivity but that these antibodies lack neutralizing activity (i.e., potency) against primary HIV-1 viruses such as HIV-1YU2 because the V3 epitopes on primary viruses are concealed within the tertiary and quaternary confines of the native Env trimer (37, 47, 53, 54, 79). To confirm this hypothesis, we performed three additional analyses. First, we examined the sensitivity of a virus expressing the Env glycoprotein from a second molecularly cloned primary clade B strain, HIV-1BORI d9-4F8_1413 (46), compared with an HIV-2KR.X7 Env chimera containing HIV-1BORI d9-4F8_1413 V3 sequence to the V3 MAbs and to the 21 clade B and C plasmas. 447-52D and F425 B4e8 were unable to neutralize the primary HIV-1BORI virus at the highest concentrations tested (25 and 10 μg/ml, respectively) but neutralized the HIV-2KR.X7 BORI V3 chimera at concentrations 1,000-fold lower (<0.003 μg/ml for each) (Fig. 8). Similarly, the HIV-1-positive patient plasmas exhibited neutralization titers against the HIV-2KR.X7 BORI V3 chimera that were on average >100-fold higher than against the primary HIV-1BORI virus strain (Fig. 7b). Median IC50 neutralization titers against the HIV-2KR.X7 BORI V3 chimera were 2,941 and 2,033 for the clade B and clade C plasmas, respectively, but only 28 (P = 0.0007) and 20 (P = 0.0013) against HIV-1BORI. Second, in this and a companion study (49), we tested a total of 55 transmitted/early founder viral Env proteins from 47 subjects acutely infected with clade B virus for susceptibility to 447-52D and F425 B4e8 in the Env-pseudotype TZM-bl assay. These transmitted/early founder viral Envs were expressed from env genes that were molecularly derived from plasma virus that had never been cultivated ex vivo (49). Fifty-one of 55 (93%) of these transmitted/founder Envs were found to be resistant to neutralization by 447-52D and F425 B4e8 at the maximum concentrations tested (10 to 25 μg/ml). However, many of the same Envs showed sensitivity to 447-52D and F425 B4e8 when they were pretreated with submaximal ICs of sCD4 (Fig. 8B, C, and D) in order to trigger conformational changes in gp120 and expose V3 (42, 49, 63, 99). Third, we examined directly the contribution of V3-specific antibodies to the neutralization potencies of three broadly reactive clade B and two clade C plasmas against the primary virus strain HIV-1JR-FL (4). Each of the five broadly neutralizing plasmas tested exhibited high-titer V3-specific reactivity against the HIV-2KR.X7 YU2 chimera (IC50 of 1,235 to 8,137) and moderately high heterologous NAb titers against HIV-1JR-FL (IC50 of 206 to 1,007). However, when V3-specific antibodies were selectively depleted from these plasmas using the rabbit Fc-V3B FP, the plasmas retained nearly 100% of their neutralizing activities against HIV-1JR-FL, indicating that V3-specific antibodies in these patients did not contribute to heterologous neutralization of the primary virus. Altogether, the findings support the conclusion that the V3 region in primary HIV-1 Envs is highly conserved antigenically but is shielded from antibody recognition by the trimer complex.
FIG. 8.
Exposure of V3 neutralization epitopes in the native HIV-1 Env trimer. The exposure of V3 epitopes from the primary viruses HIV-1YU2, HIV-1BORI d9-4F8_1413, HIV-111006-11, and HIV-163068-05 (49) was tested by 447-52D and F425 B4e8 neutralization in three contexts: primary HIV-1 Env (open symbol; solid line), primary HIV-1 Env after sCD4 triggering (open symbol; dotted line), and in the HIV-2KR.X7/HIV-1 V3 Env scaffold (closed symbol). The HIV-2KR.X7 BORI V3 chimera (B) was constructed similarly to the HIV-2KR.X7 YU2 V3 chimera (A) (see Materials and Methods). Virus entry was measured by luciferase production 48 h after infection of TZM-bl reporter cells and normalized to luciferase expression in the absence of MAb.
DISCUSSION
Deciphering what antibody specificities constrain HIV-1 Env diversity (18, 21, 55, 63, 91), drive early Env evolution (25, 83, 97), restrict virus replication (80), and ultimately contribute to neutralization breadth and potency (19, 58, 61, 62) in naturally infected subjects and in vaccinees is an important goal. Previously, we along with others have demonstrated that HIV-2 Env could be used as a scaffold to present HIV-1 MPER and CD4i epitopes in the context of a functional glycoprotein and that such viruses could serve as sensitive and specific probes for HIV-1-elicited epitope-specific NAbs (18, 19, 34, 61, 102; F. Bibollet-Ruche et al., presented at the Keystone Symposium on HIV Vaccines, Keystone, CO, 2006; F. Bibollet-Ruche et al., presented at the AIDS Vaccine 2005 Conference, Montreal, Canada, 2005). In the present study, we focused on HIV-1 V3, which continues to attract substantial attention as a target of NAbs (2, 54, 72, 74, 76, 99, 107). Mamounas and colleagues first demonstrated that a chimeric virus containing an HIV-2 backbone and an HIV-1MN V3 loop was capable of infecting human T cells and showed susceptibility to HIV-1 V3-specific antiserum (64). However, that assay system consisted of two separate plasmids that required cotransfection along with unspecified adaptive changes including, but not limited to, mutations in V3 for efficient replication. We sought to extend this work by creating a full-length replication-competent HIV-2 provirus backbone containing an HIV-2 env gene modified so as to readily accept different HIV-1 V3 sequences and requiring no additional adaptive changes for efficient fusion and entry in a single infection cycle assay (96, 97). Thus, we developed a novel HIV-2KR-based shuttle vector containing seven nucleotide substitutions, four of which were synonymous cloning site mutations (pHIV-2KR.X4), and three of which were nonsynonymous adaptive changes (pHIV-2KR.X7). The latter changes were identified in our sequence analysis of samples from the original HIV-2KR-MNV3 cell cultures (64), which we confirmed to enhance viral infectivity of the HIV-2KR.X7/HIV-1 V3 chimeras described in the current study (data not shown).
We initially created three HIV-2KR.X7/HIV-1 V3 chimeric viruses for our analyses of HIV-1 V3-specific NAbs: HIV-2KR.X7 YU2 V3, HIV-2KR.X7 Ccon V3, and HIV-2KR.X7 MN V3. The V3 sequence from HIV-1YU2 was selected because of the wealth of structural and biological data available for this primary virus strain (41), and HIV-2Ccon was selected because it was most representative of the most commonly circulating HIV-1 virus on a global scale. HIV-1MN was selected because it represented a common preclinical and clinical vaccine strain. Later in the study, a fourth chimera (HIV-2KR.X7 BORI V3) was generated to correspond to a primary transmitted/founder env (49). Assessment of the biological function and antigenicity of the HIV-2KR.X7/HIV-1 V3 chimeric viruses demonstrated that their Env glycoproteins maintained the essential functional and antigenic properties of a native HIV trimer that are requisite for use as a specific probe of V3-specific NAbs. HIV-2KR.X7 YU2 V3, HIV-2KR.X7 Ccon V3, and HIV-2KR.X7 MN V3 each conferred virus entry (Fig. 2A) to an extent that was proportional to the amount of processed gp120 present on the virion surface (Fig. 2B). HIV-2KR.X7 YU2 V3 and HIV-2KR.X7 Ccon V3 each maintained receptor engagement, coreceptor selection, and virus-cell membrane fusion kinetics typical of primary viruses, as assessed by inhibition studies with selective ligands (sCD4, TAK-779, AMD3100, T20, and T1249) (Table 1; Fig. 3A and B). Furthermore, each chimera, like the parental HIV-2KR.P1, HIV-2KR.X4, and HIV-2KR.X7 viruses, was resistant to neutralization by HIV-1 MAbs directed at CD4bs (b12), CD4i (17b, 19e, 21c, E51, 4.12D, ED47, and ED49), and MPER (2F5) epitopes (Fig. 4). The chimeric viruses were similar to HIV-2KR.X7 in sensitivity to 4E10, which is a property common to many primary HIV-2 viruses (H. Li et al., presented at the Keystone Symposium on HIV Vaccines, Keystone, CO, 2006). The fact that HIV-2KR.X7 was slightly more sensitive to 4E10 than were HIV-2KR.P1 and HIV-2KR.X4 suggests that accessibility of the 4E10 epitope is slightly greater in HIV-2KR.X7. Importantly, the HIV-2KR.P1, HIV-2KR.X4, and HIV-2KR.X7 viruses were each resistant to neutralization by plasma from HIV-1-infected subjects (Fig. 6A and Table 3). These findings are typical of primary HIV-2 strains (18) and indicate that the parental HIV-2 viruses and the HIV-2/HIV-1 V3 chimeras are not globally neutralization sensitive, a prerequisite for the HIV-2 Env to be a useful scaffold upon which to present HIV-1 epitopes for an HIV-1 epitope-specific NAb assay.
We tested the ability of the HIV-2KR.X7/HIV-1 V3 chimeras to detect V3-specific antibodies using two HIV-1 MAbs, 447-52D and F425 B4e8, whose V3 epitope specificities are well established (2, 72, 93, 108). HIV-2KR.X7 YU2 V3 was potently neutralized by both 447-52D and F425 B4e8 (IC50s of 0.001 μg/ml and 0.004 μg/ml, respectively), and HIV-2KR.X7 Ccon V3 was also effectively neutralized by the same antibodies but at higher concentrations (IC50s of 0.03 μg/ml and 0.48 μg/ml, respectively) (Table 1 and Fig. 4). The neutralization potencies of 447-52D against the HIV-2KR.X7/HIV-1 V3 chimeras is in agreement with binding assay data showing that the affinity of MAb 447-52D for clade C V3 sequences is reduced by approximately 10-fold relative to that seen with clade B V3 loops (A. Pinter, unpublished data). It is also of note that the HIV-2KR.X7 Ccon V3 chimera is nearly 1,000-fold more sensitive to neutralization by 447-52D than is an HIV-1SF162 Ccon V3 chimera (54), a finding we attribute to better V3 exposure in the HIV-2 Env scaffold than in the neutralization-sensitive HIV-1 Env scaffold. These findings suggested that the HIV-2KR.X7/HIV-1 V3 chimeric viruses had especially favorable properties for detecting HIV-1 V3-specific antibodies with sensitivity and specificity, and this was confirmed by competition studies using HIV-1 V3-specific peptides and fusion proteins (Fig. 5). The finding that 447-52D and F425-B4e8 MAbs neutralized the B-subtype (YU2) chimera more potently than the C-subtype (Ccon) chimera can be explained by the origins of the MAbs, their known binding specificities, and the V3 sequences of the two target viruses. MAbs 447-52D and F425 B4e8 were each derived from HIV-1 subtype B-infected patients, and the epitopes that they recognize have in common an amino acid at position 315 of the V3 crown. In most subtype B viruses and in the HIV-2KR.X7 YU2 V3 chimera, a highly conserved arginine (Arg315) residue is found at this position (Los Alamos Sequence Database [http://www.hiv.lanl.gov/]). Crystallographic data of both MAbs bound to V3 peptides indicate that antigen binding of 447-52D and F425 B4e8 relies heavily on side chain interactions with Arg315, thus conferring selectivity to these MAbs (2, 93). In most subtype C HIV-1 and in the HIV-2KR.X7 Ccon V3 chimera, Arg315 is replaced by a glutamine, thus accounting for decreased neutralization potency of 447-52D and F425 B4e8 for the HIV-2 chimera containing the Ccon V3 loop. Analysis of the V3-specific reactivity in polyclonal clade B and clade C HIV-1-positive human plasmas demonstrated that high-titer V3 reactivity is characteristic of chronic HIV-1 infection. These same plasmas showed greatly reduced potency against HIV-1YU2 with neutralizing titers >1,000-fold lower than those measured for the V3-specific component (Fig. 7). Fusion protein competition experiments verified that the reactivity detected in polyclonal plasma by the HIV-2KR.X7 YU2 V3 chimera is V3 specific and, furthermore, that it is dependent upon residues overlapping the 447-52D and F425 B4e8 epitopes located within the V3 crown (Fig. 6C).
A primary aim of this study was to develop HIV-2/HIV-1 Env chimeric viruses that could identify HIV-1 V3-specific NAbs with sensitivity and specificity in complex polyclonal human or animal plasma. The sensitivity of the chimeric V3 Env assay was demonstrated to be quite extraordinary, as indicated by the IC50 titers of 447-52D and F425 B4e8 when tested against HIV-2KR.X7 YU2 V3 (0.001 μg/ml and 0.004 μg/ml, respectively) and compared with the same MAbs tested against the primary HIV-1YU2 virus strain containing the identical 35-amino-acid V3 sequence (4.15 μg/ml and > 10 μg/ml, respectively) (Table 1 and Fig. 4 and 8A). This 1,000-fold or greater enhancement in sensitivity of the HIV-2KR.X7 YU2 V3 chimera than of the corresponding HIV-1YU2 primary virus was corroborated by a similar analysis of 447-52D and F425 B4e8 neutralization of the HIV-1BORI V3 sequence when presented in the context of the primary HIV-1BORI Env trimer versus the HIV-2KR.X7 BORI V3 chimeric Env (Fig. 8B). Again, the latter showed a >1,000-fold enhancement in V3-mediated neutralization sensitivity. The exquisite specificity of the HIV-2/HIV-1 Env chimeric viruses was demonstrated by the ability of V3-containing fusion proteins (but not fusion proteins lacking V3 or the crown residues of V3) to remove >99% of the neutralizing activity of 447-52D, F425 B4e8, and antibodies from the clade B and C polyclonal plasmas against the HIV-2/HIV-1 V3 Env chimeras. HIV-1/HIV-1 V3 Env chimeras (53, 54, 79) and antibody depletion or enrichment strategies (19, 62, 68) have neither comparable sensitivity nor comparable specificity for detection of HIV-1 V3-specific NAbs.
Our findings provide new insights into questions raised at the outset of the study. First, the V3 region of primary clade B and C viruses is indeed highly immunogenic in naturally infected humans. Median within-clade V3-specific IC50 NAb titers of nearly 0.0001 were elicited by naturally replicating virus in both B clade and C clade infection. In comparison, median epitope-specific CD4i NAb titers (IC50) are approximately 0.003 (18), while median epitope-specific MPER NAb titers (IC50) are still lower (0.03) (F. Bibollet-Ruche et al., presented at the Keystone Symposium on HIV Vaccines, Keystone, CO, 2006). CD4i NAbs are found in >90% of HIV-1-infected subjects, whereas NAbs directed at the MPER are detectable in only one-third of subjects (18; F. Bibollet-Ruche et al., presented at the Keystone Symposium on HIV Vaccines, Keystone, CO, 2006); we show here that V3-specific NAbs were present in every subject tested. Our observation that a ∼10,000-fold dilution of HIV-1-positive patient plasma contains equivalent V3-specific NAb activity against the HIV-2KR.X7 YU2 V3 chimera as does ∼0.003 μg/ml of 447-52D or F425 B4e8 MAb suggests that neat HIV-1-positive human plasma contains the functional equivalent of ∼30 μg/ml of 447-52D or F425 B4e8 MAbs; this corresponds to approximately 0.3% of total IgG in normal humans (3). While different V3-specific NAbs may exhibit differences in affinity and NAb activity measured against functional Env trimers is not equivalent to binding titers measured against monomeric glycoprotein, we find notable the similarity of this estimate to values reported by Zolla-Pazner for total V3-specific antibody concentrations, which averaged 77 μg/ml in HIV-1-infected subjects (106). We also found that V3 antibodies exhibit broad within-clade and cross-clade reactivity. This was evident in the neutralizing activity of the two V3-specific MAbs and of the 21 HIV-1-positive clade B or C plasmas tested against viruses presenting representative clade B and C V3 sequences. Among the broadly neutralizing human MAbs, only 4E10 and the CD4i MAbs exhibit equivalent breadth (5, 18). Third, we found that the V3 loop in primary virus isolates such as HIV-1YU2 is heavily shielded in the intact gp120/gp41 trimer from 447-52D, F425 B4e8, and polyspecific V3 antibodies in human plasma. This was most evident in the >1,000-fold difference in sensitivity of primary HIV-1YU2 compared with the HIV-2KR.X7 YU2 V3 chimera to 447-52D, F425 B4e8, and to the 21 HIV-1-positive patient plasmas (Table 1 and Fig. 7A), a finding corroborated by HIV-1BORI and the corresponding HIV-2KR.X7 BORI V3 chimera (Fig. 7B and 8). Additionally, a comparative analysis of pseudotyped Env glycoproteins from 55 transmitted/early founder clade B HIV-1 viruses revealed that 93% of these viruses were resistant to 447-52D and F425 B4e8 (IC50 of >10 to 25 μg/ml), indicating that the V3 region is effectively shielded from NAb recognition. While there has been a growing awareness that the V3 region of primary virus strains is antigenically cross-reactive but sterically concealed (probably by V1, V2, and other regions of Env) (54, 79, 99), the results reported here of the differential sensitivity to neutralization of viruses with V3 sequences presented in the context of an HIV-2/HIV-1 Env chimera versus the native HIV-1 Env glycoprotein provide one of the clearest demonstrations yet of the profound effect that tertiary and quaternary HIV-1 Env structure has in shielding V3 loop sequences from antibody recognition. The data thus support a hypothesis that in vivo HIV-1 V3 in its many forms of presentation on gp120 (e.g., virion associated, cell surface CD4 bound, and shed glycoprotein) is highly immunogenic, but in the context of a functional Env trimer complex on the virion surface, V3 is generally shielded from antibody recognition and thus does not serve as an effective target of NAbs.
Although not the central focus of our study, structural aspects of the transplantation itself are of interest. Within the middle 26 residues of V3, only 5 (20%) are conserved between HIV-1 and HIV-2. Two of these V3 residues relate to binding of sulfotyrosine at position 14 (Tys14) in the CCR5 N terminus (41); one is a site of glycosylation, and two include the Gly-Arg at the V3 loop tip, which is probably involved in binding to the second extracellular loop of CCR5 (42). This demonstrates how function forces conservation. It also is of interest that replication-competent HIV-2KR.X7/HIV-1 V3 chimeras expose HIV-1 V3 loop sequences on the unliganded Env trimer, as shown by monoclonal and polyclonal V3 antibody neutralization (Fig. 4), but do not simultaneously expose a preformed bridging sheet detectable by a panel of HIV-1 CD4i MAbs (18, 49). This finding suggests that the large spatial rearrangements in V3 and the bridging sheet that follow CD4 engagement and result in the exposure of antigenically conserved CD4i epitopes in the HIV-1 Env (42) may be disassociated in HIV-2KR.X7 HIV-1 V3 Env chimeras. This is not altogether surprising, given that we have shown that sCD4 binding to some HIV-1 Envs enhances 447-52D- and F425 B4e8-mediated neutralization (Fig. 8) but not neutralization by the CD4i MAb 17b (18, 49, 99). Also consistent with this interpretation is our finding (unpublished data) that HIV-2KR.X7 was sensitive to neutralization by nine HIV-2 patient plasmas with a median IC50 titer of 1:30,000 and that these titers were reduced to 1:1,000 when the HIV-1 V3 was substituted in the HIV-2KR.X7 Env scaffold. These results indicate both a spontaneous exposure of V3 in the HIV-2 scaffold and the existence of V3-specific NAbs in HIV-2-infected subjects just as in HIV-1-infected subjects.
Given the extraordinary titers and antigenic cross-reactivity of V3-specific antibodies elicited in human HIV-1 infection and also given the effective shielding of V3 within the functional Env trimer of primary viruses, what biological role might V3 antibodies play in vivo? Here, we suggest that V3 antibodies are like CD4i antibodies and that both have an important effect: V3 and CD4i antibodies are each elicited at high titers in natural infection or by vaccination (18, 24; also K. L. Davis, unpublished data). Each exhibits extensive within-clade and cross-clade binding specificity but fails to contribute to NAb breadth or potency against primary viruses because the target epitopes are concealed (18, 21, 56, 63, 91). Each has the potential to exert potent neutralizing activity against primary viruses if its epitopes are exposed on the native Env trimer as a result of spontaneous mutation or CD4 triggering (9, 18, 21, 39, 50, 51, 103). Studies of CD4-independent HIV-1 and SIV variants, in which Env assumes a conformation in which the coreceptor binding surface is constitutively formed, have shown that these viruses are globally sensitive to neutralization by antibodies targeting multiple regions of Env (18, 51, 103). Thus, we conclude that V3 and CD4i antibodies, as well as antibodies of other specificities, act to constrain the Env glycoprotein in the functional trimer to a tertiary and quaternary structure that, in the case of CD4i antibodies, precludes creation or exposure of the bridging sheet and associated coreceptor binding surface (18, 21, 50, 51) and, in the case of V3-specific antibodies, exposure of the V3 loop region. The presence of high-titer polyspecific CD4i and V3 antibodies in human plasma would be expected to exert strong selection pressure against the formation of CD4-independent variants in vivo and restrict the virus to a two-step entry process involving first the binding of the CD4 receptor at the cell surface and subsequently exposure of the bridging sheet and V3 epitopes to the coreceptor. At this point in the entry process, the CD4i (56) and V3 epitopes (42) are protected from neutralization by IgG due to steric restrictions at the virus-cell membrane interface. If V3 and CD4i NAbs were not present, HIV-1 Env could more readily assume a conformation wherein the V3 and bridging sheet were spontaneously exposed, bypass CD4 binding, and engage coreceptors directly and thus expand the tissue tropism of the virus to cells lacking surface CD4. There is evidence to support this hypothesis in the discovery of primary HIV-1 viruses within the human central nervous system (where antibody levels are scant) that exhibit a lower requirement for CD4 as a receptor (20, 32) and in the finding of spontaneous HIV-1 mutants that display CD4 independence and enhanced sensitivity to CD4i and V3 antibodies in the plasma of chronically infected individuals (18, 97, 103). An additional role for V3-specific antibodies in restricting CCR5-tropic viruses from undergoing coreceptor switching to use CXCR4 has been suggested by Lusso and coworkers (63). In regard to vaccine design, our data and those of others indicate that the HIV-1 V3 is inherently highly immunogenic and elicits antibodies having broad potential V3-neutralizing activity. However, the problem for V3 as a vaccine immunogen is that V3 antibodies are generally unable to access their target epitopes on the native Env trimer of primary viruses. It is not obvious how to overcome this obstacle.
Acknowledgments
We thank D. Burton, M. Zwick, J. Mascola, S. Zolla-Pazner, H. Katinger, L. Cavacini, J. Robinson, and the NIH AIDS Research and Reference Reagent Program for monoclonal antibodies; D. Looney for HIV-2 viruses and clones; S. Meleth for statistical analysis; David Edwards for technical assistance; J. White for manuscript preparation; and the clinical and sequencing cores of the University of Alabama at Birmingham Center for AIDS Research.
This work was supported by grants from the Bill and Melinda Gates Foundation (37874, 38619, and 38631) and the NIH (AI 67854, AI 27767, and AI 46238).
Footnotes
Published ahead of print on 19 November 2008.
REFERENCES
- 1.Albert, J., B. Abrahamsson, K. Nagy, E. Aurelius, H. Gaines, G. Nystrom, and E. M. Fenyo. 1990. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS 4107-112. [DOI] [PubMed] [Google Scholar]
- 2.Bell, C. H., R. Pantophlet, A. Schiefner, L. A. Cavacini, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2008. Structure of antibody F425-B4e8 in complex with a V3 peptide reveals a new binding mode for HIV-1 neutralization. J. Mol. Biol. 375969-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beutler, E., M. A. Lichtman, B. S. Coller, T. J. Kipps, and U. Seligsohn. 2000. Williams hematology, 6th ed. McGraw-Hill Professional, New York, NY.
- 4.Binley, J. M., E. A. Lybarger, E. T. Crooks, M. S. Seaman, E. Gray, K. L. Davis, J. M. Decker, D. Wycuff, L. Harris, N. Hawkins, B. Wood, C. Nathe, D. Richman, G. D. Tomaras, F. Bibollet-Ruche, J. E. Robinson, L. Morris, G. M. Shaw, D. C. Montefiori, and J. R. Mascola. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 8211651-11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 7813232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 686103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3205-211. [DOI] [PubMed] [Google Scholar]
- 8.Bottiger, B., A. Karlsson, P. A. Andreasson, A. Naucler, C. M. Costa, E. Norrby, and G. Biberfeld. 1990. Envelope cross-reactivity between human immunodeficiency virus types 1 and 2 detected by different serological methods: correlation between cross-neutralization and reactivity against the main neutralizing site. J. Virol. 643492-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bou-Habib, D. C., G. Roderiquez, T. Oravecz, P. W. Berman, P. Lusso, and M. A. Norcross. 1994. Cryptic nature of envelope V3 region epitopes protects primary monocytotropic human immunodeficiency virus type 1 from antibody neutralization. J. Virol. 686006-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burton, D. R., R. C. Desrosiers, R. W. Doms, W. C. Koff, P. D. Kwong, J. P. Moore, G. J. NAbel, J. Sodroski, I. A. Wilson, and R. T. Wyatt. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5233-236. [DOI] [PubMed] [Google Scholar]
- 11.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 3002065-2071. [DOI] [PubMed] [Google Scholar]
- 12.Cardoso, R. M., F. M. Brunel, S. Ferguson, M. Zwick, D. R. Burton, P. E. Dawson, and I. A. Wilson. 2007. Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10. J. Mol. Biol. 3651533-1544. [DOI] [PubMed] [Google Scholar]
- 13.Cavacini, L., M. Duval, L. Song, R. Sangster, S. H. Xiang, J. Sodroski, and M. Posner. 2003. Conformational changes in env oligomer induced by an antibody dependent on the V3 loop base. AIDS 17685-689. [DOI] [PubMed] [Google Scholar]
- 14.Centers for Disease Control and Prevention. 1989. Interpretation and use of the Western blot assay for serodiagnosis of human immunodeficiency virus type 1 infections. MMWR Morb. Mortal. Wkly. Rep. 381-7. [PubMed] [Google Scholar]
- 15.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433834-841. [DOI] [PubMed] [Google Scholar]
- 16.Cormier, E. G., M. Persuh, D. A. Thompson, S. W. Lin, T. P. Sakmar, W. C. Olson, and T. Dragic. 2000. Specific interaction of CCR5 amino-terminal domain peptides containing sulfotyrosines with HIV-1 envelope glycoprotein gp120. Proc. Natl. Acad. Sci. USA 975762-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cormier, E. G., D. N. Tran, L. Yukhayeva, W. C. Olson, and T. Dragic. 2001. Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J. Virol. 755541-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Decker, J. M., F. Bibollet-Ruche, X. Wei, S. Wang, D. N. Levy, W. Wang, E. Delaporte, M. Peeters, C. A. Derdeyn, S. Allen, E. Hunter, M. S. Saag, J. A. Hoxie, B. H. Hahn, P. D. Kwong, J. E. Robinson, and G. M. Shaw. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 2011407-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhillon, A. K., H. Donners, R. Pantophlet, W. E. Johnson, J. M. Decker, G. M. Shaw, F. H. Lee, D. D. Richman, R. W. Doms, G. Vanham, and D. R. Burton. 2007. Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J. Virol. 816548-6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunfee, R. L., E. R. Thomas, P. R. Gorry, J. Wang, J. Taylor, K. Kunstman, S. M. Wolinsky, and D. Gabuzda. 2006. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc. Natl. Acad. Sci. USA 10315160-15165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards, T. G., T. L. Hoffman, F. Baribaud, S. Wyss, C. C. LaBranche, J. Romano, J. Adkinson, M. Sharron, J. A. Hoxie, and R. W. Doms. 2001. Relationships between CD4 independence, neutralization sensitivity, and exposure of a CD4-induced epitope in a human immunodeficiency virus type 1 envelope protein. J. Virol. 755230-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eggink, D., C. E. Baldwin, Y. Deng, J. P. Langedijk, M. Lu, R. W. Sanders, and B. Berkhout. 2008. Selection of T1249-resistant human immunodeficiency virus type 1 variants. J. Virol. 826678-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiebig, E. W., D. J. Wright, B. D. Rawal, P. E. Garrett, R. T. Schumacher, L. Peddada, C. Heldebrant, R. Smith, A. Conrad, S. H. Kleinman, and M. P. Busch. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 171871-1879. [DOI] [PubMed] [Google Scholar]
- 24.Forsell, M. N., B. Dey, A. Morner, K. Svehla, S. O'Dell, C. M. Hogerkorp, G. Voss, R. Thorstensson, G. M. Shaw, J. R. Mascola, G. B. Karlsson Hedestam, and R. T. Wyatt. 2008. B cell recognition of the conserved HIV-1 co-receptor binding site is altered by endogenous primate CD4. PLoS Pathog. 4e1000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frost, S. D., T. Wrin, D. M. Smith, S. L. Kosakovsky Pond, Y. Liu, E. Paxinos, C. Chappey, J. Galovich, J. Beauchaine, C. J. Petropoulos, S. J. Little, and D. D. Richman. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc. Natl. Acad. Sci. USA 10218514-18519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorny, M. K., A. J. Conley, S. Karwowska, A. Buchbinder, J. Y. Xu, E. A. Emini, S. Koenig, and S. Zolla-Pazner. 1992. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 667538-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorny, M. K., T. C. VanCott, C. Hioe, Z. R. Israel, N. L. Michael, A. J. Conley, C. Williams, J. A. Kessler, 2nd, P. Chigurupati, S. Burda, and S. Zolla-Pazner. 1997. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and interclade cross-reactivity. J. Immunol. 1595114-5122. [PubMed] [Google Scholar]
- 28.Gorny, M. K., C. Williams, B. Volsky, K. Revesz, S. Cohen, V. R. Polonis, W. J. Honnen, S. C. Kayman, C. Krachmarov, A. Pinter, and S. Zolla-Pazner. 2002. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J. Virol. 769035-9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorny, M. K., C. Williams, B. Volsky, K. Revesz, X. H. Wang, S. Burda, T. Kimura, F. A. Konings, A. Nadas, C. A. Anyangwe, P. Nyambi, C. Krachmarov, A. Pinter, and S. Zolla-Pazner. 2006. Cross-clade neutralizing activity of human anti-V3 monoclonal antibodies derived from the cells of individuals infected with non-B clades of human immunodeficiency virus type 1. J. Virol. 806865-6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorny, M. K., J. Y. Xu, V. Gianakakos, S. Karwowska, C. Williams, H. W. Sheppard, C. V. Hanson, and S. Zolla-Pazner. 1991. Production of site-selected neutralizing human monoclonal antibodies against the third variable domain of the human immunodeficiency virus type 1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 883238-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorny, M. K., J. Y. Xu, S. Karwowska, A. Buchbinder, and S. Zolla-Pazner. 1993. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J. Immunol. 150635-643. [PubMed] [Google Scholar]
- 32.Gorry, P. R., J. Taylor, G. H. Holm, A. Mehle, T. Morgan, M. Cayabyab, M. Farzan, H. Wang, J. E. Bell, K. Kunstman, J. P. Moore, S. M. Wolinsky, and D. Gabuzda. 2002. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J. Virol. 766277-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goudsmit, J., C. Debouck, R. H. Meloen, L. Smit, M. Bakker, D. M. Asher, A. V. Wolff, C. J. Gibbs, Jr., and D. C. Gajdusek. 1988. Human immunodeficiency virus type 1 neutralization epitope with conserved architecture elicits early type-specific antibodies in experimentally infected chimpanzees. Proc. Natl. Acad. Sci. USA 854478-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray, E. S., P. L. Moore, F. Bibollet-Ruche, H. Li, J. M. Decker, T. Meyers, G. M. Shaw, and L. Morris. 2008. 4E10-resistant variants in a human immunodeficiency virus type 1 subtype C-infected individual with an anti-membrane-proximal external region-neutralizing antibody response. J. Virol. 822367-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gray, E. S., P. L. Moore, I. A. Choge, J. M. Decker, F. Bibollet-Ruche, H. Li, N. Leseka, F. Treurnicht, K. Mlisana, G. M. Shaw, S. S. Karim, C. Williamson, and L. Morris. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 816187-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gurgo, C., H. G. Guo, G. Franchini, A. Aldovini, E. Collalti, K. Farrell, F. Wong-Staal, R. C. Gallo, and M. S. Reitz, Jr. 1988. Envelope sequences of two new United States HIV-1 isolates. Virology 164531-536. [DOI] [PubMed] [Google Scholar]
- 37.Hartley, O., P. J. Klasse, Q. J. Sattentau, and J. P. Moore. 2005. V3: HIV's switch-hitter. AIDS Res. Hum. Retrovir. 21171-189. [DOI] [PubMed] [Google Scholar]
- 38.Haynes, B. F., J. Fleming, E. W. St Clair, H. Katinger, G. Stiegler, R. Kunert, J. Robinson, R. M. Scearce, K. Plonk, H. F. Staats, T. L. Ortel, H. X. Liao, and S. M. Alam. 2005. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 3081906-1908. [DOI] [PubMed] [Google Scholar]
- 39.Hoffman, T. L., C. C. LaBranche, W. Zhang, G. Canziani, J. Robinson, I. Chaiken, J. A. Hoxie, and R. W. Doms. 1999. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc. Natl. Acad. Sci. USA 966359-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Honnen, W. J., C. Krachmarov, S. C. Kayman, M. K. Gorny, S. Zolla-Pazner, and A. Pinter. 2007. Type-specific epitopes targeted by monoclonal antibodies with exceptionally potent neutralizing activities for selected strains of human immunodeficiency virus type 1 map to a common region of the V2 domain of gp120 and differ only at single positions from the clade B consensus sequence. J. Virol. 811424-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang, C. C., S. N. Lam, P. Acharya, M. Tang, S. H. Xiang, S. S. Hussan, R. L. Stanfield, J. Robinson, J. Sodroski, I. A. Wilson, R. Wyatt, C. A. Bewley, and P. D. Kwong. 2007. Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4. Science 3171930-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 3101025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hung, C. S., N. Vander Heyden, and L. Ratner. 1999. Analysis of the critical domain in the V3 loop of human immunodeficiency virus type 1 gp120 involved in CCR5 utilization. J. Virol. 738216-8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hwang, S. S., T. J. Boyle, H. K. Lyerly, and B. R. Cullen. 1991. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science 25371-74. [DOI] [PubMed] [Google Scholar]
- 45.Javaherian, K., A. J. Langlois, C. McDanal, K. L. Ross, L. I. Eckler, C. L. Jellis, A. T. Profy, J. R. Rusche, D. P. Bolognesi, S. D. Putney, et al. 1989. Principal neutralizing domain of the human immunodeficiency virus type 1 envelope protein. Proc. Natl. Acad. Sci. USA 866768-6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones, N. A., X. Wei, D. R. Flower, M. Wong, F. Michor, M. S. Saag, B. H. Hahn, M. A. Nowak, G. M. Shaw, and P. Borrow. 2004. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J. Exp. Med. 2001243-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karlsson Hedestam, G. B., R. A. Fouchier, S. Phogat, D. R. Burton, J. Sodroski, and R. T. Wyatt. 2008. The challenges of eliciting neutralizing antibodies to HIV-1 and to influenza virus. Nat. Rev. Microbiol. 6143-155. [DOI] [PubMed] [Google Scholar]
- 48.Kayman, S. C., Z. Wu, K. Revesz, H. Chen, R. Kopelman, and A. Pinter. 1994. Presentation of native epitopes in the V1/V2 and V3 regions of human immunodeficiency virus type 1 gp120 by fusion glycoproteins containing isolated gp120 domains. J. Virol. 68400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keele, B. F., E. E. Giorgi, J. F. Salazar-Gonzalez, J. M. Decker, K. T. Pham, M. G. Salazar, C. Sun, T. Grayson, S. Wang, H. Li, X. Wei, C. Jiang, J. L. Kirchherr, F. Gao, J. A. Anderson, L. H. Ping, R. Swanstrom, G. D. Tomaras, W. A. Blattner, P. A. Goepfert, J. M. Kilby, M. S. Saag, E. L. Delwart, M. P. Busch, M. S. Cohen, D. C. Montefiori, B. F. Haynes, B. Gaschen, G. S. Athreya, H. Y. Lee, N. Wood, C. Seoighe, A. S. Perelson, T. Bhattacharya, B. T. Korber, B. H. Hahn, and G. M. Shaw. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA 1057552-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kolchinsky, P., E. Kiprilov, P. Bartley, R. Rubinstein, and J. Sodroski. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J. Virol. 753435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kolchinsky, P., E. Kiprilov, and J. Sodroski. 2001. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J. Virol. 752041-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 684650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krachmarov, C., A. Pinter, W. J. Honnen, M. K. Gorny, P. N. Nyambi, S. Zolla-Pazner, and S. C. Kayman. 2005. Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade a and clade B v3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J. Virol. 79780-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krachmarov, C. P., W. J. Honnen, S. C. Kayman, M. K. Gorny, S. Zolla-Pazner, and A. Pinter. 2006. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J. Virol. 807127-7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laakso, M. M., F. H. Lee, B. Haggarty, C. Agrawal, K. M. Nolan, M. Biscone, J. Romano, A. P. Jordan, G. J. Leslie, E. G. Meissner, L. Su, J. A. Hoxie, and R. W. Doms. 2007. V3 loop truncations in HIV-1 envelope impart resistance to coreceptor inhibitors and enhanced sensitivity to neutralizing antibodies. PLoS Pathog. 3e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Labrijn, A. F., P. Poignard, A. Raja, M. B. Zwick, K. Delgado, M. Franti, J. Binley, V. Vivona, C. Grundner, C. C. Huang, M. Venturi, C. J. Petropoulos, T. Wrin, D. S. Dimitrov, J. Robinson, P. D. Kwong, R. T. Wyatt, J. Sodroski, and D. R. Burton. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 7710557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li, B., J. M. Decker, R. W. Johnson, F. Bibollet-Ruche, X. Wei, J. Mulenga, S. Allen, E. Hunter, B. H. Hahn, G. M. Shaw, J. L. Blackwell, and C. A. Derdeyn. 2006. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J. Virol. 805211-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 7910108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li, Y., H. Hui, C. J. Burgess, R. W. Price, P. M. Sharp, B. H. Hahn, and G. M. Shaw. 1992. Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J. Virol. 666587-6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li, Y., J. C. Kappes, J. A. Conway, R. W. Price, G. M. Shaw, and B. H. Hahn. 1991. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J. Virol. 653973-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li, Y., S. A. Migueles, B. Welcher, K. Svehla, A. Phogat, M. K. Louder, X. Wu, G. M. Shaw, M. Connors, R. T. Wyatt, and J. R. Mascola. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 131032-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li, Y., K. Svehla, M. K. Louder, D. Wycuff, S. Phogat, M. Tang, S. A. Migueles, X. Wu, A. Phogat, G. M. Shaw, M. Conners, J. Hoxie, J. R. Mascola, and R. T. Wyatt. 12 November 2008. Analysis of the neutralization specificities in polyclonal sera derived from human immunodeficiency virus type-1 infected individuals. J. Virol. doi: 10.1128/JVI.01992-08. [DOI] [PMC free article] [PubMed]
- 63.Lusso, P., P. L. Earl, F. Sironi, F. Santoro, C. Ripamonti, G. Scarlatti, R. Longhi, E. A. Berger, and S. E. Burastero. 2005. Cryptic nature of a conserved, CD4-inducible V3 loop neutralization epitope in the native envelope glycoprotein oligomer of CCR5-restricted, but not CXCR4-using, primary human immunodeficiency virus type 1 strains. J. Virol. 796957-6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mamounas, M., D. J. Looney, R. Talbott, and F. Wong-Staal. 1995. An infectious chimeric human immunodeficiency virus type 2 (HIV-2) expressing the HIV-1 principal neutralizing determinant. J. Virol. 696424-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McKeating, J. A., C. Shotton, J. Cordell, S. Graham, P. Balfe, N. Sullivan, M. Charles, M. Page, A. Bolmstedt, S. Olofsson, et al. 1993. Characterization of neutralizing monoclonal antibodies to linear and conformation-dependent epitopes within the first and second variable domains of human immunodeficiency virus type 1 gp120. J. Virol. 674932-4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Montefiori, D. C., T. S. Hill, H. T. Vo, B. D. Walker, and E. S. Rosenberg. 2001. Neutralizing antibodies associated with viremia control in a subset of individuals after treatment of acute human immunodeficiency virus type 1 infection. J. Virol. 7510200-10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moog, C., H. J. Fleury, I. Pellegrin, A. Kirn, and A. M. Aubertin. 1997. Autologous and heterologous neutralizing antibody responses following initial seroconversion in human immunodeficiency virus type 1-infected individuals. J. Virol. 713734-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moore, P. L., E. S. Gray, I. A. Choge, N. Ranchobe, K. Mlisana, S. S. Abdool Karim, C. Williamson, and L. Morris. 2008. The c3-v4 region is a major target of autologous neutralizing antibodies in human immunodeficiency virus type 1 subtype C infection. J. Virol. 821860-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nara, P. L., L. Smit, N. Dunlop, W. Hatch, M. Merges, D. Waters, J. Kelliher, R. C. Gallo, P. J. Fischinger, and J. Goudsmit. 1990. Emergence of viruses resistant to neutralization by V3-specific antibodies in experimental human immunodeficiency virus type 1 IIIB infection of chimpanzees. J. Virol. 643779-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ofek, G., M. Tang, A. Sambor, H. Katinger, J. R. Mascola, R. Wyatt, and P. D. Kwong. 2004. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J. Virol. 7810724-10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Palker, T. J., M. E. Clark, A. J. Langlois, T. J. Matthews, K. J. Weinhold, R. R. Randall, D. P. Bolognesi, and B. F. Haynes. 1988. Type-specific neutralization of the human immunodeficiency virus with antibodies to env-encoded synthetic peptides. Proc. Natl. Acad. Sci. USA 851932-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pantophlet, R., R. O. Aguilar-Sino, T. Wrin, L. A. Cavacini, and D. R. Burton. 2007. Analysis of the neutralization breadth of the anti-V3 antibody F425-B4e8 and re-assessment of its epitope fine specificity by scanning mutagenesis. Virology 364441-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pantophlet, R., and D. R. Burton. 2006. GP120: target for neutralizing HIV-1 antibodies. Annu. Rev. Immunol. 24739-769. [DOI] [PubMed] [Google Scholar]
- 74.Pantophlet, R., T. Wrin, L. A. Cavacini, J. E. Robinson, and D. R. Burton. 2008. Neutralizing activity of antibodies to the V3 loop region of HIV-1 gp120 relative to their epitope fine specificity. Virology 381251-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parren, P. W., J. P. Moore, D. R. Burton, and Q. J. Sattentau. 1999. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13(Suppl. A)S137-162. [PubMed] [Google Scholar]
- 76.Patel, M. B., N. G. Hoffman, and R. Swanstrom. 2008. Subtype-specific conformational differences within the V3 region of subtype B and subtype C human immunodeficiency virus type 1 Env proteins. J. Virol. 82903-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pilgrim, A. K., G. Pantaleo, O. J. Cohen, L. M. Fink, J. Y. Zhou, J. T. Zhou, D. P. Bolognesi, A. S. Fauci, and D. C. Montefiori. 1997. Neutralizing antibody responses to human immunodeficiency virus type 1 in primary infection and long-term-nonprogressive infection. J. Infect. Dis. 176924-932. [DOI] [PubMed] [Google Scholar]
- 78.Pinter, A. 2007. Roles of HIV-1 Env variable regions in viral neutralization and vaccine development. Curr. HIV Res. 5542-553. [DOI] [PubMed] [Google Scholar]
- 79.Pinter, A., W. J. Honnen, Y. He, M. K. Gorny, S. Zolla-Pazner, and S. C. Kayman. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 785205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Poignard, P., R. Sabbe, G. R. Picchio, M. Wang, R. J. Gulizia, H. Katinger, P. W. Parren, D. E. Mosier, and D. R. Burton. 1999. Neutralizing antibodies have limited effects on the control of established HIV-1 infection in vivo. Immunity 10431-438. [DOI] [PubMed] [Google Scholar]
- 81.Profy, A. T., P. A. Salinas, L. I. Eckler, N. M. Dunlop, P. L. Nara, and S. D. Putney. 1990. Epitopes recognized by the neutralizing antibodies of an HIV-1-infected individual. J. Immunol. 1444641-4647. [PubMed] [Google Scholar]
- 82.Ratner, L., A. Fisher, L. L. Jagodzinski, H. Mitsuya, R. S. Liou, R. C. Gallo, and F. Wong-Staal. 1987. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res. Hum. Retrovir. 357-69. [DOI] [PubMed] [Google Scholar]
- 83.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 1004144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rizzuto, C., and J. Sodroski. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum. Retrovir. 16741-749. [DOI] [PubMed] [Google Scholar]
- 85.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 2801949-1953. [DOI] [PubMed] [Google Scholar]
- 86.Rong, R., F. Bibollet-Ruche, J. Mulenga, S. Allen, J. L. Blackwell, and C. A. Derdeyn. 2007. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J. Virol. 811350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rong, R., S. Gnanakaran, J. M. Decker, F. Bibollet-Ruche, J. Taylor, J. N. Sfakianos, J. L. Mokili, M. Muldoon, J. Mulenga, S. Allen, B. H. Hahn, G. M. Shaw, J. L. Blackwell, B. T. Korber, E. Hunter, and C. A. Derdeyn. 2007. Unique mutational patterns in the envelope alpha 2 amphipathic helix and acquisition of length in gp120 hypervariable domains are associated with resistance to autologous neutralization of subtype C human immunodeficiency virus type 1. J. Virol. 815658-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rusche, J. R., K. Javaherian, C. McDanal, J. Petro, D. L. Lynn, R. Grimaila, A. Langlois, R. C. Gallo, L. O. Arthur, P. J. Fischinger, et al. 1988. Antibodies that inhibit fusion of human immunodeficiency virus-infected cells bind a 24-amino acid sequence of the viral envelope, gp120. Proc. Natl. Acad. Sci. USA 853198-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sagar, M., X. Wu, S. Lee, and J. Overbaugh. 2006. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J. Virol. 809586-9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Salazar-Gonzalez, J. F., E. Bailes, K. T. Pham, M. G. Salazar, M. B. Guffey, B. F. Keele, C. A. Derdeyn, P. Farmer, E. Hunter, S. Allen, O. Manigart, J. Mulenga, J. A. Anderson, R. Swanstrom, B. F. Haynes, G. S. Athreya, B. T. Korber, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 823952-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Salzwedel, K., E. D. Smith, B. Dey, and E. A. Berger. 2000. Sequential CD4-coreceptor interactions in human immunodeficiency virus type 1 Env function: soluble CD4 activates Env for coreceptor-dependent fusion and reveals blocking activities of antibodies against cryptic conserved epitopes on gp120. J. Virol. 74326-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Saphire, E. O., P. W. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 2931155-1159. [DOI] [PubMed] [Google Scholar]
- 93.Stanfield, R. L., M. K. Gorny, C. Williams, S. Zolla-Pazner, and I. A. Wilson. 2004. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure 12193-204. [DOI] [PubMed] [Google Scholar]
- 94.Sullivan, N., Y. Sun, J. Binley, J. Lee, C. F. Barbas, 3rd, P. W. Parren, D. R. Burton, and J. Sodroski. 1998. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J. Virol. 726332-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.von Brunn, A., M. Brand, C. Reichhuber, C. Morys-Wortmann, F. Deinhardt, and F. Schodel. 1993. Principal neutralizing domain of HIV-1 is highly immunogenic when expressed on the surface of hepatitis B core particles. Vaccine 11817-824. [DOI] [PubMed] [Google Scholar]
- 96.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 461896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422307-312. [DOI] [PubMed] [Google Scholar]
- 98.Weiss, R. A., P. R. Clapham, J. N. Weber, D. Whitby, R. S. Tedder, T. O'Connor, S. Chamaret, and L. Montagnier. 1988. HIV-2 antisera cross-neutralize HIV-1. AIDS 295-100. [DOI] [PubMed] [Google Scholar]
- 99.Wu, X., A. Sambor, M. C. Nason, Z. Yang, L. Wu, S. Zolla-Pazner, G. J. NAbel, and J. R. Mascola. 2008. Soluble CD4 broadens neutralization of V3-directed monoclonal antibodies and guinea pig vaccine sera against HIV-1 subtype B and C reference viruses. Virology 380285-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393705-711. [DOI] [PubMed] [Google Scholar]
- 101.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 2801884-1888. [DOI] [PubMed] [Google Scholar]
- 102.Yuste, E., H. B. Sanford, J. Carmody, J. Bixby, S. Little, M. B. Zwick, T. Greenough, D. R. Burton, D. D. Richman, R. C. Desrosiers, and W. E. Johnson. 2006. Simian immunodeficiency virus engrafted with human immunodeficiency virus type 1 (HIV-1)-specific epitopes: replication, neutralization, and survey of HIV-1-positive plasma. J. Virol. 803030-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang, P. F., P. Bouma, E. J. Park, J. B. Margolick, J. E. Robinson, S. Zolla-Pazner, M. N. Flora, and G. V. Quinnan, Jr. 2002. A variable region 3 (V3) mutation determines a global neutralization phenotype and CD4-independent infectivity of a human immunodeficiency virus type 1 envelope associated with a broadly cross-reactive, primary virus-neutralizing antibody response. J. Virol. 76644-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang, Y., B. Lou, R. B. Lal, A. Gettie, P. A. Marx, and J. P. Moore. 2000. Use of inhibitors to evaluate coreceptor usage by simian and simian/human immunodeficiency viruses and human immunodeficiency virus type 2 in primary cells. J. Virol. 746893-6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhou, T., L. Xu, B. Dey, A. J. Hessell, D. Van Ryk, S. H. Xiang, X. Yang, M. Y. Zhang, M. B. Zwick, J. Arthos, D. R. Burton, D. S. Dimitrov, J. Sodroski, R. Wyatt, G. J. NAbel, and P. D. Kwong. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zolla-Pazner, S. 2005. Improving on nature: focusing the immune response on the V3 loop. Hum. Antibodies 1469-72. [PubMed] [Google Scholar]
- 107.Zolla-Pazner, S., S. S. Cohen, C. Krachmarov, S. Wang, A. Pinter, and S. Lu. 2008. Focusing the immune response on the V3 loop, a neutralizing epitope of the HIV-1 gp120 envelope. Virology 372233-246. [DOI] [PubMed] [Google Scholar]
- 108.Zolla-Pazner, S., P. Zhong, K. Revesz, B. Volsky, C. Williams, P. Nyambi, and M. K. Gorny. 2004. The cross-clade neutralizing activity of a human monoclonal antibody is determined by the GPGR V3 motif of HIV type 1. AIDS Res. Hum. Retrovir. 201254-1258. [DOI] [PubMed] [Google Scholar]