Abstract
Induction of the toxic LamB-LacZ protein fusion, Hyb42-1, leads to a lethal generalized protein export defect. The prlF1 suppressor causes hyperactivation of the cytoplasmic Lon protease and relieves the inducer sensitivity of Hyb42-1. Since prlF1 does not cause a detectable change in the stability or level of the hybrid protein, we conducted a suppressor screen, seeking factors genetically downstream of lon with prlF1-like phenotypes. Two independent insertions in the ygdP open reading frame relieve the toxicity of the fusion protein and share two additional properties with prlF1: cold sensitivity and the ability to suppress the temperature sensitivity of a degP null mutation. Despite these similarities, ygdP does not appear to act in the same genetic pathway as prlF1 and lon, suggesting a fundamental link between the phenotypes. We speculate that the common properties of the suppressors relate to secretion defects. The ygdP gene (also known as nudH) has been shown to encode a Nudix protein that acts as a dinucleotide oligophosphate (alarmone) hydrolase. Our results suggest that loss of ygdP function leads to the induction of an alarmone-mediated response that affects secretion. Using an epitope-tagged ygdP construct, we present evidence that this response is sensitive to secretion-related stress and is regulated by differential proteolysis of YgdP in a self-limiting manner.
The general secretory (Sec) pathway is responsible for exporting the majority of proteins destined for extracytoplasmic locations in the gram-negative bacterium Escherichia coli. Export via this pathway in E. coli takes place largely posttranslationally, after most or all of the polypeptide has been synthesized. Sec substrates are targeted for export through the inner membrane translocation complex SecYEG, due to the presence of signal sequences in the amino-terminal region of the protein and are maintained in unfolded export-competent conformations by the actions of cytoplasmic chaperones. Hydrolysis of ATP by the cytoplasmic protein SecA pushes the protein through the translocator in an unfolded fashion (for recent reviews, see references 14, 29, and 31).
Heterologous cytoplasmic proteins can be directed to the Sec pathway by fusing a signal sequence to the amino terminus of the open reading frame (ORF) (39). The LamB-LacZ hybrid protein Hyb42-1 consists of the signal sequence and 173 residues of the mature portion of the outer membrane maltoporin LamB fused to the cytoplasmic protein LacZ (β-galactosidase) (4). The presence of the LacZ domain confers an uninduced Lac+ phenotype on lactose MacConkey agar and on Luria-Bertani (LB) agar containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). LamB production is under the control of the maltose regulon, and expression of the hybrid construct may be induced by the addition of maltose to the medium. Under inducing conditions, strains expressing Hyb42-1 exhibit maltose sensitivity (a Mals phenotype). This is due to the fact that the amino-terminal (LamB) region of the hybrid targets the protein for export but the cytoplasmically derived LacZ region folds more quickly than it can be translocated. Thus, the available SecYEG translocators are titrated out by the production of LamB-LacZ molecules that block the translocation pore. Induction of the hybrid ultimately prevents the export of other envelope proteins, resulting in a lethal secretion defect called hybrid jamming. This defect can be detected by a general accumulation of envelope precursor proteins in the cytoplasm (43).
A variety of mechanisms can suppress the maltose sensitivity conferred by the toxic hybrid. In the most trivial case, mutations that disrupt the gene fusion itself relieve jamming by preventing expression of the LacZ moiety. These can easily be distinguished from the parental strain on indicator media due to their Lac− Malr phenotype. A more interesting class of intragenic suppressors identifies the signal sequence. These mutant signal sequences fail to target the hybrid protein for export and are therefore resistant to maltose induction. The cytoplasmic retention of the hybrid protein in these mutants results in increased β-galactosidase activity, conferring a Lac++ Malr phenotype (15).
Extragenic suppressors of hybrid toxicity have also been isolated. Mutations that cause a decrease in the expression of the hybrid (because of an effect on the maltose regulon) allow compensatory mechanisms to alleviate the toxicity. Alternatively, increasing the activity of envelope stress response pathways allows cells to survive maltose induction (such as by a cpxA* allele). Suppression of hybrid jamming by the Cpx pathway results in the export of the hybrid protein to the periplasm by an unknown mechanism (42). The elevated Cpx response results in higher levels of the periplasmic protease DegP, which rapidly degrades the hybrid protein upon its entry into the periplasm. In fact, DegP is required for the suppression of hybrid toxicity in cpxA* strains. In the absence of a functional degP gene, these strains remain sensitive to induction of the hybrid, despite the fact that the hybrid-jamming phenotype has been relieved (12). While the secretion defect is relieved, the presence of the cysteine-rich LacZ domain in the oxidizing periplasmic environment results in the formation of high-molecular-weight disulfide-bonded aggregates that presumably interfere with normal periplasmic processes (43).
The prlF1 mutation was identified in a maltose-resistant mutant with a secretion-dependent Lac-down phenotype (Lac− Malr) (22). Spontaneous mutations in the prlF gene were independently isolated as cold-sensitive suppressors of the temperature-sensitive growth defect of a degP (also known as htrA) null strain (2). These alleles, sohA1 and sohA2 (for suppressor of htrA) are molecularly identical to the prlF1 allele (N. J. Hand, unpublished data). The degP suppression, cold sensitivity, and relief-of-jamming phenotypes conferred by the mutation in question (a 7-bp insertion in the prlF ORF that results in a truncated PrlF protein with a frameshifted C terminus) are all gain-of-function phenotypes (21). Null mutations in prlF have no detectable phenotype (21).
Selection for cold-resistant suppressors of prlF1 led to the identification of the cytoplasmic protease Lon as the only known effector of prlF1 function (44). In addition to suppressing prlF1 cold sensitivity, null mutations in lon revert the (Lac− Malr) prlF1 phenotype to wild type (Lac+ Mals). Furthermore, it has been shown that the prlF1 allele causes a posttranslational hyperactivation of Lon (44). However, unlike the activated Cpx strains, the prlF1 mutation affects neither the steady-state level nor the turnover of the hybrid protein (41). Weak sequence homology between PrlF and a chaperone-like domain of the VAT protein from Thermoplasma acidophilum hints at a direct interaction between PrlF and Lon (11). Both Lon and VAT are members of the AAA family of ATP-dependent proteases.
These observations present a conundrum. The suppression of hybrid jamming by prlF1 indicates that the hybrid protein is no longer interfering with translocator function, while the Lac− phenotype suggests that the hybrid protein is not in the cytoplasm (which, as in the case of the signal sequence mutants, would result in a Lac++ phenotype). Furthermore, the observation that the hybrid protein is toxic in the periplasm, unless it is degraded, means that the prlF1 mutation cannot simply relocate the hybrid to that compartment. Attempts to directly address the cellular localization of the hybrid protein in the prlF1 mutant by subcellular fractionation have been confounded by the anomalous behavior of the LamB-LacZ hybrid. Taken together, these observations indicate that the prlF1 allele links the function of PrlF to two proteases in distinct cellular compartments. The effect of the mutation is to cause an apparent relocalization of the LamB-LacZ hybrid protein to an unknown location, where it is both inactive and nontoxic. The only substantial clue to how this might take place was the involvement of Lon. Since we have shown that proteolysis of the hybrid itself is not responsible for the prlF1 phenotypes, we speculated that the degradation of an unknown substrate of Lon might instead be responsible for the maltose resistance. We postulated that by looking for prlF1-like suppressors of hybrid toxicity in a strain where the prlF1 phenotype is suppressed by a lon null mutation we might identify such a Lon substrate, or at least uncover clues to the mechanism of action of this elusive suppressor. In this paper, we present the surprising results of this screen. Rather than isolating a downstream effector of Lon, we have instead identified a novel suppressor, ygdP, which recapitulates the prlF1 phenotypes via a seemingly independent mechanism. This finding leads us to believe that there may be a fundamental link between cold sensitivity, the relief of hybrid jamming, and the temperature-sensitive defect caused by the degP null mutation.
MATERIALS AND METHODS
Media and chemicals.
Standard culture media were used for bacterial growth (38). For growth in liquid minimal medium, M63 medium (38) was used and was supplemented with 0.4% sugars, 0.5% (vol/vol) LB broth, 1 mM MgSO4, and 100 μg of thiamine HCl/ml. Where required, minimal medium was supplemented with individual amino acids at a final concentration of 100 μg/ml. For expression from the pTrc99a-derived plasmids and their λInCh derivatives, the medium was supplemented with isopropylthio-β-d-galactoside (IPTG) at 2 mM. Antibiotics were purchased from Sigma (St. Louis, Mo.) and were used at the following concentrations: ampicillin, 125 μg/ml for maintenance of high-copy-number plasmids and 25 μg/ml for selection of single-copy Ampr λ lysogens; chloramphenicol, 20 μg/ml; kanamycin, 50 μg/ml; tetracycline, 25 μg/ml; amikacin, 3 μg/ml; and spectinomycin, 50 μg/ml.
MalE (MBP) antiserum was from the laboratory collection. X-Gal was purchased from Fisher Scientific (Toronto, Ontario, Canada). o-Nitrophenyl-β-d-galactopyranoside, FLAG mouse (M2) monoclonal antibodies, and FLAG (M2) affinity agarose were purchased from Sigma. IPTG was purchased from Ambion (Austin, Tex).
Anti-rabbit immunoglobulin G horseradish peroxidase-linked whole antibodies (from sheep) and anti-mouse immunoglobulin G horseradish peroxidase-linked whole antibodies (from goat) were purchased from Amersham Life Sciences (Piscataway, N.J.). ECL Western blotting reagents were purchased from Amersham Life Sciences.
Reagents for DNA cloning, including restriction enzymes, and T4 DNA ligase were purchased from New England Biolabs (Beverly, Mass.) and were used according to the manufacturer's directions. Pfu polymerase, PfuTurbo polymerase, and QuikChange mutagenesis kits were purchased from Stratagene (La Jolla, Calif.) and were used as directed. Deoxyribonucleotides were purchased from Amersham Pharmacia Biotech (Piscataway, N.J.). DNA purification kits were purchased from Qiagen (Valencia, Calif.). Oligodeoxynucleotide primers were obtained from the synthesis/sequencing facility at Princeton University or purchased from IDT (Coralville, Iowa) or Sigma Genosys (The Woodlands, Tex.).
Protran 0.2-μm-pore-size nitrocellulose membranes were purchased from Schleicher and Schuell (Keene, N.H.). Ponceau S was purchased from Sigma. Kodak X-AR film was purchased from the Eastman Kodak Company (New York, N.Y.).
Microbiological techniques.
Standard techniques were used for the construction of strains by P1 transduction (38). P1vir, used to generate P1 lysates for transductions, was from our laboratory collection. The strains used in this study are shown in Table 1. Plasmid transformations were performed using a Bio-Rad (Hercules, Calif.) E. coli Gene Pulser according to the manufacturer's recommendations.
TABLE 1.
Strains used in this study
| Name | Genotype | Reference |
|---|---|---|
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | 10 |
| NJH101 | MC4100 φ(lamB-lacZ) Hyb42-1 [λpl(209)] | This study |
| NJH102 | MC4100 φ(lamB-lacZ) Hyb42-1 [λpl(209)] prlF1 | This study |
| NJH138 | MC4100 φ(lamB-lacZ) Hyb42-1 [λpl(209)] prlF::kan (StuI) | This study |
| NJH234 | MC4100 φ(lamB-lacZ) Hyb42-1 [λpl(209)] prlF1 lon::Tn10 non | This study |
| NJH350 | MC4100 φ(lamB-lacZ) Hyb42-1 [λpl(209)] prlF1 lon::Tn10 non ygdP::cm18 | This study |
| NJH351 | MC4100 φ(lamB-lacZ) Hyb42-1 [λpl(209)] ygdP::cm18 | This study |
| NJH402 | MC4100 φ(lamB-lacZ) Hyb42-1 [λpl(209)] ygdP::cm18 lon::Tn10 | This study |
| NJH445 | MC4100 λInCh TrcFLAG-YgdP | This study |
| NJH445.1 | MC4100 λInCh TrcFLAG-YgdP lon::Tn10 | This study |
| NJH446 | MC4100 λInCh TrcFLAG-YgdP prlF1 | This study |
| NJH446.1 | MC4100 λInCh TrcFLAG-YgdP prlF1 lon::Tn10 | This study |
| NJH455 | MC4100 φ(lamB-lacZ) Hyb42-1 [λpl(209)] λInCh TrcFLAG-YgdP | This study |
| NJH456 | MC4100 φ(lamB-lacZ) Hyb42-1 [λpl(209)] λInCh TrcFLAG-YgdP prlF1 | This study |
| NJH463 | MC4100 φ(lamB-lacZ) Hyb42-1 [λpl(209)] λInCh TrcYgdP(E51AE52A) ygdP::cm18 | This study |
| NJH464 | MC4100 φ(lamB-lacZ) Hyb42-1 [λpl(209)] λInCh TrcFLAG-YgdP lon::Tn10 | This study |
| NJH465 | MC4100 φ(lamB-lacZ) Hyb42-1 [λpl(209)] λInCh TrcFLAG-YgdP prlF1 lon::Tn10 | This study |
| NJH466 | MC4100 φ(lamB-lacZ) Hyb42-1 [λpl(209)] λInCh TrcFLAG-YgdP ygdP::cm18 lon::Tn10 | This study |
| NJH495 | MC4100 φ(lamB-lacZ) Hyb42-1 [λpl(209)] ygdP(E51AE52A) | This study |
| NJH522 | MC4100 φ(lamB-lacZX90) Hyb10-1 [λpl(209)] λInCh TrcFLAG-YgdP | This study |
| NJH523 | MC4100 φ(lamB-lacZX90) Hyb10-1 [λpl(209)] prlF1 λInCh TrcFLAG-YgdP | This study |
| NJH524 | MC4100 φ(lamB-lacZ S60) Hyb42-1 [λpl(209)] prlF1 | This study |
| NJH525 | MC4100 φ(lamB-lacZ S60) Hyb42-1 [λpl(209)] λInCh TrcFLAG-YgdP | This study |
| NJH526 | MC4100 φ(lamB-lacZ S60) Hyb42-1 [λpl(209)] prlF1 λInCh TrcFLAG-YgdP | This study |
Maltose sensitivity disk assays.
For maltose sensitivity assays, strains were grown to saturation in 5 ml of liquid minimal medium at 30°C with aeration. The cells were pelleted at 4,000 rpm and resuspended in 2.5 ml of fresh M63 medium. Fifty microliters of the cell suspension was added to 3 ml of molten F-Top agar (38) (cooled to 50°C) and plated on prewarmed minimal-glycerol agar plates. Two sterile 7-mm-diameter filter paper disks were placed on each plate, 10 μl of 10% maltose was pipetted onto the first disk, and 10 μl of 2.5% maltose was pipetted onto the second disk. The plates were incubated at 30°C overnight. The width of the zone of clearing around each disk was determined by averaging three measurements of the outer diameter of the zone of clearing, subtracting the diameter of the disk, and dividing the resulting value by two.
Identification of transposon insertion locations.
The insertion points of the mini-Tncam transposon from λNK1324 (24) TnCam candidate suppressors were identified using the anchored random-PCR strategy described by Gibson and Silhavy (17).
Protein analysis.
Sample buffer was prepared either as described by Silhavy et al. (with β-mercaptoethanol as a reducing agent) (38) or as described by Sambrook et al. (with dithiothreitol as a reducing agent) (37).
Crude whole-cell lysates were prepared for steady-state protein analysis by subculturing overnight cultures 1:100 into fresh media, growing the cells to an appropriate density, and harvesting them by centrifugation. The cell pellet was resuspended in 1/10 volume of 1× sodium dodecyl sulfate (SDS) sample buffer and boiled for 5 min, and 5 to 10 μl was loaded on denaturing SDS-polyacrylamide gels (10 or 15% acrylamide, depending on the size of the protein of interest).
Samples were separated on SDS-polyacrylamide gels according to the technique of Laemmli (26). A Bio-Rad Mini Trans-Blot apparatus was used for SDS-polyacrylamide gel electrophoresis and for electrotransfer to Protran 0.2-μm-pore-size nitrocellulose membranes, according to the manufacturer's recommendations. The transferred proteins were visualized by staining them with 0.5% (wt/vol in 5% acetic acid) Ponceau S and destained by washing them several times with deionized water and then Western Wash Solution (10 mM Tris · HCl [pH 7.5], 0.9% NaCl, 0.2% Tween 20). The membranes were blocked by shaking them in blocking solution (10 mM Tris · HCl [pH 7.5], 0.9% NaCl, 0.2% Tween 20, 2.5% powdered milk, 0.5% bovine serum albumin) either at 4°C overnight or at room temperature for 1 h. The membranes were then incubated with primary antibodies in fresh blocking solution (1:8,000 for LacZ, LamB, and MalE antisera and 1:1,000 for FLAG mouse monoclonal antiserum) with shaking, either at 4°C overnight or at room temperature for 2 h. The membranes were washed for at least 1 h at room temperature, with shaking, in Western Wash Solution. The membranes were incubated with secondary antibody (1:8,000 in blocking solution) for 45 min at room temperature and then washed as described above. ECL reagents and Kodak X-AR film were used for chemiluminescence detection of peroxidase activity according to the manufacturer's instructions. The relative intensities of the bands were determined using the ImageQuant program (Molecular Dynamics). For the FLAG Western blots, expression of the chromosomal Trc-FLAG-YgdP construct was induced by the addition of 2 mM IPTG to the culture medium.
Plasmid construction.
Plasmid pCRScript-YgdP was constructed using DNA amplified from MC4100 by PCR with primers NH4GDP5F (5′-AACTCTGCACATAACTGTG-3′) and NH4GDP3R (5′-ATTGGCGTGACTTAACCTC-3′). The resulting plasmid contained DNA sequence corresponding to nucleotides (nt) 9494 to 10162 of GenBank entry AE000366. The cloned DNA is inserted at nt 728 of pPCRScript-Amp such that the orientation of the YgdP ORF is the same as that of the LacZα fragment (nt 9494 of the insert abuts nt 728 of the vector, and nt 10162 of the insert abuts nt 729 of the vector).
Plasmid pTrc-YgdP+ was constructed as follows. DNA was amplified from MC4100 by PCR, using primers NH4GDP11 (5′-TGAGGTGAATTCGTGATTGATGACGATGGCT-AC-3′) and NH4GDP12 (5′-CGCAGGAAGCTTAGCATAATTGGCGTGACTTAAC-3′). Recognition sequences for restriction enzymes used in the plasmid construction are underlined.
The purified PCR product was cut with EcoRI and HindIII and cloned into EcoRI-HindIII-cut pTrc99a. The resulting plasmid, pTrc-YgdP+, carries the DNA sequence corresponding to nt 10035 to 9489 of GenBank entry AE000366, replacing nt 276 to 320 of pTrc99a (GenBank entry U13872).
Plasmid pTrc-FLAGYgdP was constructed using primers NHFL5YGD (5′-AACAGACCATGGACTACAAGGACGATGATGACAAGATTGATGACGATGGCTAC-3′) and NH4GDP12 (5′-CGCAGGAAGCTTAGCATAATTGGCGTGACTTAAC-3′). The resulting plasmid, pTrc-FLAGYgdP, attaches the DNA sequence encoding the FLAG epitope, with a start codon, to nt 10032 to 9489 of GenBank entry AE000366 (the ygdP ORF minus the start codon). The resulting DNA sequence replaces nt 271 to 320 of pTrc99a (GenBank entry U13872).
Generation of a chromosomal ygdPE56AE57A mutation by allelic exchange.
The mutagenic primers NHFMUTRA (5′-CAGGCGATGTACCGTGAATTGTTTGCTGCAGTAGGATTAAGCCGCAAAGACGTTC-3′) and NHRMUTRA (5′-GAACGTCTTTGCGGCTTAATCCTACTGCAGCAAACAATTCACGGTACATCGCCTG-3′) were used to generate mutations in the conserved Nudix motif of ygdP. The mutated derivatives of pCRScript-YgdP+ and pTrc-YgdP+ contain the double mutation E56AE57A and were designated pCRScript-YgdP− and pTrc-YgdP−, respectively. Mutation of the native ygdP chromosomal locus was accomplished by subcloning the insert from pCRScript-YgdP− into pGS284 (40) in strain S17λpir. The parental plasmid carries the counterselectable sacB gene from Bacillus amyloliquefaciens. The mutated ygdP was introduced into strain NJH351 by conjugal transfer, and exconjugants were selected on LB agar containing low levels (25 μg/ml) of ampicillin and chloramphenicol. Since pGS284 has an R6K origin of replication, the plasmid cannot replicate in the recipient strain (which lacks λpir). Selection for ampicillin resistance results in the formation of a plasmid cointegrate at the chromosomal ygdP locus. The resolution of the cointegrate was selected by plating it on LB agar without salt, with 5% sucrose, to counterselect the plasmid-borne sacB gene. The sucrose-resistant colonies were screened for chloramphenicol sensitivity (indicating loss of the ygdP::cm18 allele). The presence of the desired ygdPE56AE57A allele was screened by colony PCR followed by PstI restriction digestion and was confirmed by DNA sequencing.
Construction of single-copy gene expression strains.
The plasmids pTrc-YgdP+, pTrc-YgdP−, and pTrc-FLAGYgdP were targeted to the chromosome as stable single-copy lysogens using the λInCh procedure as described by Boyd et al. (8).
Targeted disruption of the ygdP and ptsP genes.
The chromosomal disruptions of the ygdP and ptsP ORFs were made according to the procedure of Datsenko and Wanner (13).
The linear DNA used for the disruption of the ORFs was produced by amplifying the kanamycin resistance cassette from the pUC4K plasmid with primers NHYGDPKF (5′-TATCCACCCCTTCCTCTGTTTATAACTCTGCACATAACTGTGAGTTATAAGAATTCTGATTAGAAAAACTC-3′) and NHYGDPKR (5′-AACCAGTTACGCGTTGAAGCAAGGATTCGAACGTCTTTGCGGCTTAATCCGGATTCTTCAACTCAGCAAAAG-3′) (in the case of ygdP) and with primers NHPTSPKF (5′-CCACAAAACGCATCTGCTTATCGACGTAAAAGAGGTTAAGTCACAGAATTCTGATTAGAAAAACTCATCGAG-3′)and NHPTSPKR (5′-TAGTAACAACGTCGATCATGATCGGCCAGGTAGACCGAACAGACCTCGGTTAGGATTCTTCAACTCAGCAAAAG-3′) (in the case of ptsP). Disruption of the ORFs was confirmed by PCR.
RESULTS
Rationale.
The prlF1 mutation confers a Lac-down, maltose-resistant (Lac− Malr) phenotype in a strain carrying the toxic, maltose-inducible LamB-LacZ hybrid protein (Hyb42-1). In an effort to further understand the mechanism by which prlF1 suppresses hybrid jamming, we sought mutants with prlF1-like phenotypes. Since we knew that lon null mutations suppress all known phenotypes of prlF1, we used as a parent strain NJH234 (Hyb42-1 prlF1 lon::Tn10 non), hoping to identify genes functioning downstream or independently of lon.
Null mutations in lon produce a mucoid phenotype due to the overproduction of capsular polysaccharide (18, 45). The non (for “nonmucoid”) mutation maps to the cps (for “capsular polysaccharide synthesis”) gene cluster at 45.7 min and suppresses mucoidy (33). We therefore used a strain with an uncharacterized non mutation for practical reasons: it is difficult to pick individual colonies, and to score their phenotypes, from crowded plates of mucoid strains.
Isolation of lon suppressors.
A pooled P1vir lysate representing ∼40,000 independent mini-TnCam insertions in an MC4100 strain was prepared as described previously (24). Random insertions from this pool were introduced into strain NJH234 (Hyb42-1 prlF1 lon::Tn10 non) by P1 transduction, and the transductants were plated on LB agar containing X-Gal and chloramphenicol. Approximately 30,000 independent transductants were screened, representing a theoretical coverage of the genome at an average density of one insertion every 133 bp. The parent strain, NJH234, produces dark-blue colonies on LB agar containing X-Gal. Transductants with decreased Lac activity, which produced light-blue colonies, were colony purified for further study. This initial screen yielded 67 mutants with stable Lac-down phenotypes.
In addition to identifying transposon insertions with novel effects on the hybrid phenotypes, we expected to obtain Lac-down transductants from this due to the cotransduction of the wild-type lon+ allele from the MC4100 strain used to generate the TnCam pooled P1 lysate. In the latter case, the Lac phenotype would simply be due to the (unsuppressed) prlF1 mutation. In order to distinguish the two classes, each of the candidate Lac suppressors was tested for the presence of the lon::Tn10 allele by screening for the ability to grow on LB agar containing tetracycline. Of the 67 Lac-down TnCam insertions with stable phenotypes, 44 proved to be sensitive to tetracycline and were thus identified as lon+ transductants. These transductants were not subjected to further study.
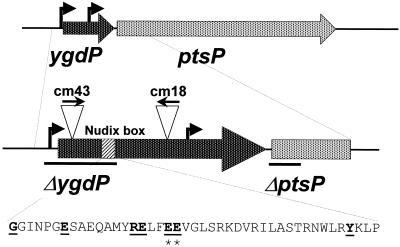
The insertion points of the remaining 23 candidate suppressors were identified using the degenerate PCR method described by Gibson and Silhavy (17). In most cases, the reduced β-galactosidase activity of the LamB-LacZ gene fusion caused by the transposon insertions could be explained by decreased expression of the maltose regulon. The phenotypes of these strains were likely due either to nonspecific effects on protein synthesis (spoT tnaA) or on carbohydrate metabolism (gatZ uhpC cya) or to specific effects on the maltose regulon (malI). However, two strains, cm18 and cm43, contained independent TnCam insertions in the ygdP ORF (Fig. 1), the effects of which could not easily be explained in terms of a known phenotype of the gene.
FIG. 1.
Positions of the TnCam transposon insertions in the ygdP ORF. The arrows above the insertions indicate the direction of the chloramphenicol acetyltransferase promoter. The bars indicate the regions deleted in the ygdP(Δ5′)::kan and ptsP(Δ5′)::kan alleles. The amino acid sequence corresponding to the Nudix box is shown, with the invariant signature residues underlined. The asterisks identify the residues mutated in the ygdPE56AE57A allele.
The decrease in Lac activity of the hybrid protein is secretion dependent.
The ygdP insertions do not decrease the activity of a derivative of the lamB-lacZ Hyb42-1 gene fusion (BZR60) in which the signal sequence is disrupted (data not shown). In contrast, mutations that decrease the expression of the maltose regulon produce a Lac-down phenotype in strains with this cytoplasmically localized LamB-LacZ hybrid. This indicates that, like the prlF1 mutation, the decrease in the Lac activity of LamB-LacZ conferred by the ygdP mutation depends on proper targeting of the hybrid to the translocator.
Two independent insertions in ygdP act as antisuppressors of lon.
The ygdP insertions in an NJH234 (Hyb42-1 prlF1 lon::Tn10 non) background produce a maltose-resistant phenotype that is indistinguishable from that of a prlF1 lon+ strain (Table 2).
TABLE 2.
Hybrid protein phenotypes
| Name | Relevant genotype | Phenotype
|
|
|---|---|---|---|
| Laca | Malb | ||
| NJH101 | Hyb42-1 | + | S |
| NJH102 | Hyb42-1 prlF1 | − | R |
| NJH234 | Hyb42-1 prlF1 lon::Tn10 | + | S |
| NJH350 | Hyb42-1 prlF1 lon::Tn10 ygdP::cm18 | − | R |
| NJH351 | Hyb42-1 ygdP::cm18 | +/− | R/S |
| NJH402 | Hyb42-1 ygdP::cm18 lon::Tn10 | − | R |
| NJH495 | Hyb42-1 ygdPE51AE52A | +/− | R/S |
| NJH463 | Hyb42-1 ydsP::cm18 {λInCh Trc YgdP(E51AE52A)} | +/− | R/S |
+, positive; −, negative; +/−, intermediate.
S, sensitive; R, resistant; R/S, partial resistance.
In addition to the effects on the Lac and Mal phenotypes of the Hyb42-1 protein, the prlF1 mutation confers a cold-sensitive phenotype that is suppressed in prlF1 lon double-mutant strains. Both of the ygdP insertions resulted in a cold-sensitive growth defect at 18°C that was more severe than that of a prlF1 single-mutant strain. Furthermore, when the ygdP::cm insertions were introduced into a degP::Tn10 strain, they increased the efficiency of plating of the strain at 42°C by 2 to 3 log units relative to an isogenic ygdP+ strain. As noted previously, the prlF1 mutation was isolated independently as a suppressor of the temperature-sensitive phenotype of a null mutant of htrA (degP). Taken together, these results showed that the ygdP::cm insertions affected all of the phenotypes known to be affected by prlF1 and initially suggested to us that ygdP acts in the same genetic pathway as prlF1, downstream of lon. Both insertions in ygdP behaved identically with respect to all of the phenotypes assayed.
A 5′ deletion of the ygdP gene phenocopies the ygdP::cm insertions.
To eliminate the possibility that the effects of ygdP::cm might be due to an effect on the expression of the downstream gene ptsP, we independently constructed defined null alleles of ygdP and ptsP using the single-step PCR strategy of Datsenko and Wanner (13). In the case of the ygdP(Δ5′)::kan allele, the promoter region and approximately one-third of the ygdP ORF were replaced by a kanamycin resistance cassette. The deleted region of ptsP(Δ5′)::kan replaces 150 bp of the 5′ end of the ptsP gene with a kanamycin resistance cassette, thus removing the start codon of ptsP, and several potential alternative start codons in the 5′ region of the gene, without affecting the ygdP ORF (Fig. 1). When transduced into the NJH234 (Hyb42-1 prlF1 lon::Tn10 non) strain, the ygdP::kan allele was indistinguishable from the ygdP::cm insertions while the ptsP::kan allele had no effect on either the Lac or Mal phenotype of the parent strain (data not shown). This indicated that the original ygdP::cm insertions were nulls and that the phenotypes associated with them were due to loss of ygdP function and not to an effect on ptsP.
The ygdP gene product is a member of the conserved family of Nudix proteins.
Sequence homology identifies the ygdP gene product as a member of the Nudix family of proteins (5). Members of the Nudix family are cytoplasmic proteins that possess the highly conserved signature sequence GX5EX7REUXEEXGU, where X is any amino acid and U represents a bulky hydrophobic amino acid (isoleucine, leucine, or valine). The acronym Nudix is derived from the observation that the substrates of these proteins are nucleoside diphosphates linked to an additional component, X. To test if mutation of the Nudix motif could produce an antisuppression phenotype, we generated a site-directed ygdPE56AE57A mutant allele on a plasmid. This allele was transferred to the native locus in the chromosome by allelic exchange. In the strain thus formed, NJH495 (Hyb42-1 ygdPE51AE52A), the mutation of the Nudix motif resulted in a Lac-down, maltose-resistant phenotype indistinguishable from that of the ygdP null alleles. Furthermore, the expression of a single copy of ygdPE51AE52A (at the λ attachment site) driven by the IPTG-inducible pTrc promoter fails to complement a chromosomal ygdP null in strain NJH463 [Hyb42-1 λInCh TrcYgdP(E51AE52A) ygdP::cm18].
The ygdP single mutants do not fully phenocopy prlF1.
In the simplest case, if ygdP were in a genetic pathway downstream of lon, then disruptions of the gene should produce prlF1-like phenotypes in a prlF+ lon+ background. We therefore transduced the insertions into a strain containing the lamB-lacZ Hyb42-1 fusion in an otherwise wild-type background and tested both the maltose resistance and the Lac phenotypes on lactose MacConkey agar. The results of the tests are shown in Table 2.
The maltose sensitivity and Lac phenotypes of the two ygdP transposon insertions are indistinguishable from one another but clearly intermediate between the wild type and prlF1. Both insertions produce a more severe cold-sensitive growth defect at 18°C than prlF1. Since protein export is an inherently cold-sensitive process (32), a tantalizing possibility is that the cold sensitivities of both the prlF1 allele and the ygdP::cm mutants reflect a perturbation of the secretion machinery. Consistent with this notion, the presence of the LamB-LacZ hybrid protein enhances the cold sensitivity of the ygdP::cm strains, even in the absence of maltose induction, as does growth on (lactose) MacConkey agar. These growth conditions place additional stress on the cell by the presence of the slowly exported hybrid protein or due to the bile salts in the MacConkey agar.
A lon null allele enhances the effects of ygdP::cm on Hyb42-1.
The strains NJH350 (Hyb42-1 prlF1 lon::Tn10 non ygdP::cm18) and NJH102 (Hyb42-1 prlF1) behave identically with respect to the Lac and Mal phenotypes associated with the hybrid protein. In contrast, NJH351 (Hyb42-1 ygdP::cm18) has higher Lac activity and is less maltose resistant (exhibiting partial sensitivity in a maltose disk assay) than either NJH350 or NJH102. Remarkably, when the lon::Tn10 allele was introduced into NJH351, the Lac activity was decreased and the maltose resistance was increased (Table 2). Thus, while the ygdP mutations behaved as antisuppressors of lon when introduced into NJH234, the lon null mutant enhanced the partial suppression due to the ygdP mutation.
Insertions in ygdP relieve hybrid jamming.
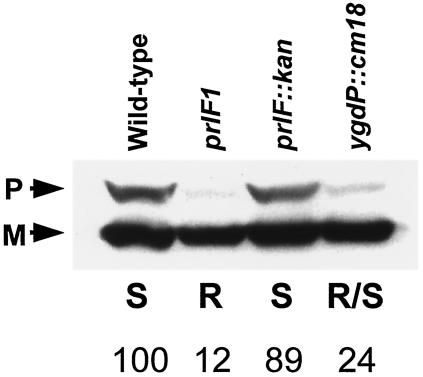
Maltose induction of the hybrid protein in a wild-type strain leads to jamming of the secretion apparatus, which results in a general cytoplasmic accumulation of unprocessed precursors of envelope proteins. The prlF1 mutation relieves hybrid jamming. To test whether the ygdP::cm insertions could also relieve hybrid jamming, we assayed the accumulation of unprocessed MalE in strains containing the LamB-LacZ hybrid, Hyb42-1, with a prlF+, prlF1, prlF::kan, or ygdP::cm18 mutation (Fig. 2).
FIG. 2.
Disruption of ygdP relieves hybrid jamming. Western analysis of whole-cell lysates of strains containing the LamB-LacZ hybrid protein Hyb42-1 was performed after 30 min of maltose induction. The analysis was performed using antibody against the maltose-binding protein, MalE. The presence of unprocessed precursor MalE (P) is a consequence of hybrid jamming. The position of the mature MalE protein (M) is indicated. No accumulation of precursor is observed in the prlF1 strain, while the jamming phenotype is partially relieved by the ygdP::cm18 null mutation. The strains are (from left to right) NJH101, NJH102, NJH138, and NJH351. The maltose phenotypes of the strains are indicated: S, sensitive; R, resistant; R/S, partial resistance. The numbers indicate the intensity of the pMalE band (corrected for loading using the lower, mature MalE band) expressed relative to the level seen in the wild-type strain.
No suppression of jamming occurred in the wild-type and prlF::kan (null) strains. In contrast, jamming was relieved in the prlF1 strain. A dramatic reduction in jamming occurred in the ygdP::cm strain; however, there is still detectable precursor MalE present. This intermediate amount of precursor MalE observed in the ygdP::cm strain is consistent with the intermediate effects of the mutation on both the Lac activity and the maltose sensitivity of the hybrid protein in an otherwise wild-type strain. Furthermore, as with the prlF1 mutation, Western analysis indicated that the levels of the Hyb42-1 hybrid protein were not significantly altered by the ygdP mutations. Thus, the relief of jamming is not due to the degradation of the hybrid protein.
Levels of epitope-tagged YgdP do not correlate with the allele of prlF.
Given that prlF1 causes a hyperactivation of Lon (44), a major cytoplasmic protease, and since degradation of the hybrid is clearly not responsible for the suppression of hybrid jamming, we had originally reasoned that YgdP might be the relevant Lon substrate. If this were true, then we would expect significant degradation of YgdP in prlF1 lon+ strains. To test this, we generated a FLAG-tagged derivative of ygdP by PCR and cloned the epitope-tagged gene into pTrc99a. To eliminate the possibility of artifactual effects of the presence of the epitope-tagged protein in high copy numbers, we transferred the construct to the λ attachment site in single copy, using the λInCh procedure (8). We compared the properties of pTrc-Flg-YgdP in single copy to a corresponding untagged pTrc-YgdP+ construct at the λ attachment site to address the possibility that the presence of the epitope tag might alter the properties of the YgdP protein. Our results show that in trans to a ygdP::cm allele at the chromosomal locus, the epitope-tagged YgdP behaves identically to the wild-type construct. Both constructs complement the ygdP null mutant, restoring the phenotypes associated with the LamB-LacZ hybrid protein to wild type (Lac+ Mals). Based on this observation, we concluded that the Flg-YgdP protein was functional.
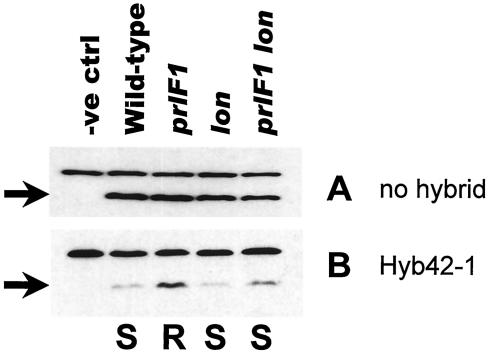
Whole-cell protein extracts were prepared from wild-type, prlF1, lon::Tn10, and prlF1 lon::Tn10 strains carrying the tagged construct on the chromosome, and steady-state levels of the Flg-YgdP protein were examined by Western analysis. The results are shown in Fig. 3A. The inclusion of the MC4100 control sample (in which there is no Flg-YgdP construct) identified the upper band as a cross-reacting band which serves as an internal loading control. No significant difference was found among the levels of Flg-YgdP in the strains tested. We repeated this experiment using isogenic strains containing the toxic lamB-lacZ gene fusion under conditions of maltose induction. Surprisingly, while there were differences in the levels of Flg-YgdP, the alteration in the levels of the tagged protein was opposite to that anticipated. Rather than seeing enhanced degradation of Flg-YgdP in the activated Lon (prlF1) strain, we saw an increased steady-state level of the protein compared to the corresponding prlF+ strain (Fig. 3B). Contrary to our expectation, the decrease in the Flg-YgdP levels correlated, not with the allele of prlF present, but with the Mal phenotype, and by extension, with the state of cellular stress. The level of Flg-YgdP was lower in the maltose-sensitive strains than in the maltose-resistant prlF1 strain. Similarly, strains carrying the LamB-LacZ hybrid protein have low levels of Flg-YgdP relative to the corresponding strain lacking the hybrid (compare Fig. 3B to A), except in the case of the prlF1 mutants.
FIG. 3.
Levels of Flg-YgdP are lower in the presence of the LamB-LacZ hybrid protein. Western analysis was performed using a monoclonal antibody against the FLAG epitope. The arrows indicate Flg-YgdP protein; the upper cross-reacting band serves as an internal loading control. The strains in panels A and B are isogenic, except that the strains in panel B carry the lamb-lacZ Hyb42-1 fusion. (A) Strains (from left to right): MC4100, NJH445, NJH446, NJH445.1, and NJH446.1. (B) Strains (from left to right): NJH101, NJH455, NJH456, NJH464, and NJH465. The maltose sensitivity phenotypes (S, sensitive; R, resistant), where relevant, are indicated below panel B. MC4100 and NJH101 do not carry Flg-YgdP and serve as negative controls (−ve ctrl).
Decreased levels of epitope-tagged YgdP correlate with cellular stress.
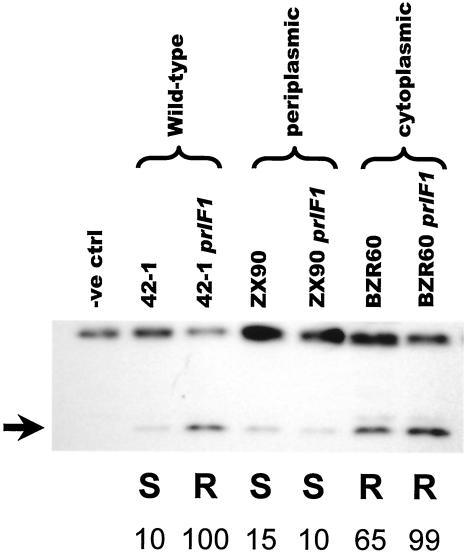
To test if the higher level of Flg-YgdP in strain NJH456 relative to that in NJH455 might reflect lower cellular stress in the prlF1 strain relative to its prlF+ counterpart, we took advantage of two alleles of the lamB-lacZ gene fusion. The lamBS60 mutation deletes part of the signal sequence of the LamB domain. Thus, the BZR60 derivative of the hybrid (carrying the S60 mutation in the LamB domain) is not targeted for export and therefore causes neither hybrid jamming nor maltose sensitivity. The prlF1 allele has no effect on the Lac phenotype of BZR60. The X90 allele of lamB-lacZ results in an eight-residue truncation of the carboxy terminus of the LacZ domain of the hybrid protein. The carboxy terminus of LacZ is apparently important for efficient protein folding, since this small alteration relieves jamming of the ZX90 derivative of the hybrid protein. Instead, the mutant hybrid protein is secreted into the periplasm. As noted previously, the presence of the cysteine-rich LacZ domain in the periplasm is toxic. Thus, the ZX90 hybrid has a Mals phenotype that is not suppressed by prlF1.
Figure 4 shows Western analysis of strains containing the chromosomal flg-ygdP construct and either wild-type LamB-LacZ, cytoplasmic LamB-LacZ (nontoxic), or periplasmic LamB-LacZ (toxic). A comparison of the levels of Flg-YgdP relative to the cross-reacting higher-molecular-weight band again suggests a correlation with the Mal phenotype rather than with the prlF allele. Thus, levels of Flg-YgdP are higher in the Malr BZR60 strains than in the Mals ZX90 strains. We consistently see a subtle difference between the levels of Flg-YgdP in the prlF+ and prlF1 BZR60 strains (NJH525 and NJH526). This difference likely reflects some leakiness of the S60 signal sequence mutation such that in strain NJH525 a small proportion of the mutant hybrid protein causes a minor perturbation of the export machinery.
FIG. 4.
Flg-YgdP levels correlate with cellular stress, not with the location of the hybrid protein. We used three alleles of the hybrid protein to test if the location of the LamB-LacZ hybrid affected the levels of Flg-YgdP. The wild-type 42-1 fusion allele is toxic but is suppressed by the prlF1 mutation. The ZX90 allele of the lamB-lacZ fusion is efficiently secreted into the periplasm, where the hybrid protein is toxic. In contrast, the BZR60 allele is not efficiently targeted for export due to a signal sequence mutation. Western analysis was performed using the anti-FLAG antibody. Low Flg-YgdP levels in the jammed 42-1 strain, and in both ZX90 strains, indicate that the smaller amounts of Flg-YgdP reflect cellular stress rather than localization of the hybrid protein. Strains (from left to right): NJH101, NJH455, NJH456, NJH522, NJH523, NJH525, and NJH526. The maltose sensitivity phenotypes (S, sensitive; R, resistant) are indicated. The arrow indicates the Flg-YgdP band. Strain NJH101 does not carry Flg-YgdP and serves as a negative control (−ve ctrl). The numbers indicate the intensities of the Flg-YgdP band (corrected for loading using the upper cross-reacting band) expressed relative to the level seen in the Hyb42-1 prlF1 strain.
DISCUSSION
We present here the identification of a novel suppressor of LamB-LacZ hybrid jamming, ygdP. The screen that identified this gene was intended to elucidate the mechanism by which the prlF1 mutation suppresses hybrid jamming. Since prlF1 increases the activity of the Lon protease, we postulated that the degradation of some unknown protein might be responsible for the effects of the prlF1 allele. We hoped to identify this hypothetical Lon substrate and thus gain insight into prlF1 suppression. We have instead discovered a prlF-independent mode of suppression of hybrid jamming. The phenotypes associated with this new gene are the same as those of prlF1, namely, cold-sensitive growth, relief of hybrid jamming, and suppression of the temperature sensitivity of the degP null strain.
The fact that two independent suppressors, the prlF1 allele and ygdP null mutations, affect three phenotypes in common suggests that these phenotypes may be fundamentally linked. Although the nature of the link among the phenotypes is unclear, it is tempting to speculate that they all relate to aspects of translocator function. While it is known that secretion is a fundamentally cold-sensitive process (32) and that induction of the LamB-LacZ hybrid causes a severe general protein export defect, the connection between degP and secretion is less clear. Previous models explaining the temperature sensitivity of the degP null strain suggested that the DegP requirement is for the proteolysis of unfolded periplasmic proteins. This conclusion is based on studies implicating DegP in the degradation of unchaperoned pilin subunits (20) and of heterologous proteins targeted to the periplasm (12), as well as on the synthetic phenotypes of degP null mutants with some periplasmic protein folding factors (36). We favor the view that the lethal stress caused by the absence of DegP at 42°C is due to the misfolding of newly synthesized proteins rather than the unfolding of folded periplasmic proteins. If this is the case, at higher temperatures misfolded translocating proteins may remain associated with the translocator in the absence of DegP, impairing secretion in a manner that can be relieved either by prlF1 or by ygdP null mutations.
The connection among prlF1, ygdP, and degP hints at a general role for DegP in secretion. It may be that DegP assists in the clearance of exported proteins from the translocator. This could reflect an involvement of the DegP chaperone function in the translocation of many substrates, analogous to that of Kar2p (9, 25, 46) in yeast, or a more specific secretion-related stress function, in which the protease activity degrades misfolded, partially translocated polypeptides that might otherwise impair secretion.
Although for technical reasons we cannot satisfactorily address the question of the subcellular location of the hybrid protein, our data suggest that in the prlF1 and ygdP mutant strains the hybrid is most likely released from jammed transloctors into the inner membrane. It may be that these suppressors cause an increase in an innate property of the SecYEG pore, namely, lateral release. Lateral opening of the translocator is required for the integration of transmembrane segments into the inner membrane. Altering the properties of the translocator itself might result in cold sensitivity, and increasing lateral release rates would be likely to have the profoundest effect on slowly translocating or stalled translocation substrates.
The E. coli gene encoding the Nudix protein YgdP has recently been cloned, overexpressed, and purified (6). Analysis has shown that this protein catalyzes the hydrolysis of diadenosine oligophosphates in vitro (tetra-, penta-, and hexaphosphates), with a preference for diadenosine pentaphosphate. YgdP catalyzes the hydrolysis of Ap5A into ATP and ADP. YgdP has no detectable activity for other typical substrates of the Nudix family of hydrolases: NADH, modified sugars (ADP-, GDP-, and UDP-sugars), and nucleotide triphosphates and their derivatives (6). Collectively, the Nudix hydrolases share the property of recycling of small, undesirable molecules, and for this reason they are sometimes referred to as “housecleaning” proteins (5).
Dinucleoside oligophosphates were first detected as by-products of tRNA charging aminoacyl tRNA synthetase reactions (47). These compounds have since been found to be involved in a wide variety of cellular responses in both prokaryotes and eukaryotes (reviewed in references 3 and 23) and are believed to constitute a novel class of signaling molecules. In bacteria, concentrations of diadenosine oligophosphates have been shown to increase >100-fold over the endogenous level in response to heat or oxidative stress, and for this reason these molecules are collectively called alarmones (7, 19, 27, 28).
We have shown that mutagenesis of the signature Nudix box in ygdP results in phenotypes similar to those caused by the prlF1 mutation. Our results suggest that YgdP mediates an alarmone response with an effect on protein secretion. The likely role of YgdP is to purge the cell of an alarmone, allowing it to exit a stress response state. In the absence of YgdP function, a stress-adapted state would exist, enabling cells to tolerate conditions such as hybrid jamming and growth at 42°C in the absence of DegP. We propose that the normal function of YgdP is to turn over the alarmone and dampen this response. Thus, in a wild-type strain containing the LamB-LacZ fusion, levels of the alarmone are low and jamming occurs. However, in a ygdP null mutant strain, the higher endogenous level of the alarmone partially phenocopies the prlF1 mutation.
In a ygdP mutant strain, the higher endogenous levels of alarmone may lead to increased activity of a protease. This might reflect an increase in either the expression or the activity of the protease. Overlapping substrate specificities between this putative protease and Lon would explain the similar phenotypes of ygdP and prlF1. Support for this hypothesis comes from the work of Fuge and Farr (16), who characterized a disruption of the apaH gene. While ApaH is unrelated to YgdP at the primary sequence level, it is also a hydrolase and has been shown to have overlapping substrate specificity with YgdP in vitro. The disrupted copy of apaH was found to enhance a heat shock-stimulated proteolytic activity in the absence of Lon. Specifically, degradation of abnormal proteins (produced by puromycin treatment) following heat shock was 40% higher in an apaH lon mutant strain than in a lon single mutant. If a similar effect results from the disruption of ygdP, then the ygdP suppression phenotypes may be due to increased activity by a protease other than Lon. Since the presence of the LamB-LacZ hybrid protein induces the heat shock response (although not to levels sufficient to suppress its toxicity), disruption of ygdP may cause an alarmone-mediated increase in the activity of a heat shock protease. Thus, the effect of the ygdP null mutations would be convergent with that of the prlF1 mutation, with both circumstances leading to the activation of proteases. Since neither the prlF1 allele nor the ygdP null mutations result in detectable degradation of the hybrid protein, it seems likely that the substrate(s) of the proteases has yet to be identified.
Homology between the PrlF protein sequence and that of VAT-Nn, a domain of the T. acidophilum thermosome (11) that assists in the unfolding of proteins prior to degradation, hints that PrlF itself may have a chaperone function. This, coupled with the genetic evidence implicating the Lon protease in the phenotypes associated with the prlF1 mutation (44), suggests that PrlF may act as a Lon-associated chaperone. If this is the case, presumably the mutant PrlF1 protein has higher-than-wild-type chaperone activity, resulting in the observed posttranslational hyperactivation of Lon. Interestingly, while the mechanisms by which alarmones exert their pleiotropic effects in bacteria are unclear, it has been shown that the alarmone AppppA (diadenosine tetraphosphate) binds to a number of proteins, including the cytoplasmic chaperones, DnaK, GroEL, and ClpB (16, 19). Thus, it may be that alarmones modulate the activities of protease-associated chaperones, thus increasing the activity of the proteases.
Rendering the localization of the hybrid protein independent of the prlF allele by using mutations in the lamB-lacZ gene fusion suppresses the allele-specific stabilization of the tagged Flg-YgdP protein. This indicates that it is the suppression of stress that is responsible for the higher level of Flg-YgdP in the Hyb42-1 prlF1 strain and not the localization of the hybrid protein per se. In the experiments involving Flg-YgdP, all of the sequence elements necessary for the transcription and translation of the tagged protein originate from the vector sequences in the parent construct. It seems clear, therefore, that the differences in the steady-state level of the protein are achieved by the differential regulation of the proteolysis of the tagged protein. Either the proteolysis of Flg-YgdP is enhanced under stressed conditions or proteolysis is decreased in the absence of cellular stress. Whichever is the case, the identity of the protease responsible, the mechanism by which the difference between the strains is sensed, and how this leads to differences in the levels of Flg-YgdP all remain issues to be resolved. Regulation of the levels of an alarmone hydrolase would allow negative feedback on an alarmone-mediated response, thus making the response self-limiting. Such a feedback mechanism is consistent with other secretion-related stress-responsive pathways (34, 35).
Consistent with the notion that the prlF1 and ygdP suppression phenotypes occur via convergent pathways, Phenotypic MicroArray (Biolog, Inc., Hayward, Calif.) analysis of the effects of prlF1 and ygdP::cm in strain MG1655 indicate that while the effects of prlF1 are very subtle (in the absence of the hybrid protein), the ygdP mutant has more pleiotropic effects (X. Lei and B. Bochner, personal communication). This indicates that while the response mediated by ygdP overlaps with the effects of the prlF1 mutation, its effects are likely not limited to secretion.
Homologues of YgdP appear to be associated with invasion of cellular tissues. For example, the Bartonella bacilliformis homologue, IalA, is required for the invasion of human erythrocytes in vitro, and transformation of a minimally invasive E. coli strain with the Bartonella ialA operon increases the invasiveness of the strain (30). Moreover, in E. coli K1 (the primary gram-negative bacterium responsible for neonatal meningitis), the expression of ygdP was found to be elevated in bacterial cells isolated from brain microvascular endothelial cells (1), and introduction of a disrupted copy of ygdP rendered the resulting strain noninvasive. Finally, of the 16 closest homologues of E. coli YgdP, 13 are from known invasive pathogens. Thus, the alarmone-mediated response regulated by ygdP appears to play a physiologically significant role in the survival of bacteria invading host tissue. Interestingly, in the context of the role in pathogenesis, wild-type ygdP strains have an advantage. While this may reflect a general selective disadvantage of the stress-adapted state, based on our results, it is tempting to speculate that the loss of invasiveness of the ygdP mutants reflects an alarmone-mediated perturbation of secretion in these strains.
Acknowledgments
We thank Greg Smith for providing the pGS284 plasmid and the S17-λpir E. coli strain and for helpful discussions concerning the allelic-exchange procedure. We thank M. Bessman, B. Bochner, and S. Gottesman for helpful suggestions. Technical assistance was provided by L. Shi and V. Register. We thank members of the Pohlschröder and Silhavy laboratories for helpful discussions, especially P. DiGiuseppe, K. Dilks, M. Pohlschröder, R. W. Rose, and N. Ruiz for critical reading of the manuscript. We also thank S. DiRenzo for invaluable assistance in the preparation of the manuscript. Finally, we thank B. Bochner, R. Dalbey, and X. Lei for discussions of unpublished results.
This work was supported by an NIGMS MERIT Award (GM34821) to T.J.S.
REFERENCES
- 1.Badger, J. L., C. A. Wass, and K. S. Kim. 2000. Identification of Escherichia coli K1 genes contributing to human brain microvascular endothelial cell invasion by differential fluorescence induction. Mol. Microbiol. 36:174-182. [DOI] [PubMed] [Google Scholar]
- 2.Baird, L., and C. Georgopoulos. 1990. Identification, cloning, and characterization of the Escherichia coli sohA gene, a suppressor of the htrA (degP) null phenotype. J. Bacteriol. 172:1587-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxi, M. D., and J. K. Vishwanatha. 1995. Diadenosine polyphosphates: their biological and pharmacological significance. J. Pharmacol. Toxicol. Methods 33:121-128. [DOI] [PubMed] [Google Scholar]
- 4.Benson, S. A., E. Bremer, and T. J. Silhavy. 1984. Intragenic regions required for LamB export. Proc. Natl. Acad. Sci. USA 81:3830-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bessman, M. J., D. N. Frick, and S. F. O'Handley. 1996. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 271:25059-25062. [DOI] [PubMed] [Google Scholar]
- 6.Bessman, M. J., J. D. Walsh, C. A. Dunn, J. Swaminathan, J. E. Weldon, and J. Shen. 2001. The gene ygdP, associated with the invasiveness of Escherichia coli K1, designates a Nudix hydrolase, Orf176, active on adenosine (5′)-pentaphospho-(5′)-adenosine (Ap5A). J. Biol. Chem. 276:37834-37838. [DOI] [PubMed] [Google Scholar]
- 7.Bochner, B. R., P. C. Lee, S. W. Wilson, C. W. Cutler, and B. N. Ames. 1984. AppppA and related adenylylated nucleotides are synthesized as a consequence of oxidation stress. Cell 37:225-232. [DOI] [PubMed] [Google Scholar]
- 8.Boyd, D., D. S. Weiss, J. C. Chen, and J. Beckwith. 2000. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J. Bacteriol. 182:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brodsky, J. L., J. Goeckeler, and R. Schekman. 1995. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 92:9643-9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 11.Coles, M., T. Diercks, J. Liermann, A. Groger, B. Rockel, W. Baumeister, K. K. Koretke, A. Lupas, J. Peters, and H. Kessler. 1999. The solution structure of VAT-N reveals a ′missing link' in the evolution of complex enzymes from a simple βαββ element. Curr. Biol. 9:1158-1168. [DOI] [PubMed] [Google Scholar]
- 12.Cosma, C. L., P. N. Danese, J. H. Carlson, T. J. Silhavy, and W. B. Snyder. 1995. Mutational activation of the Cpx signal transduction pathway of Escherichia coli suppresses the toxicity conferred by certain envelope-associated stresses. Mol. Microbiol. 18:491-505. [DOI] [PubMed] [Google Scholar]
- 13.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Driessen, A. J., P. Fekkes, and J. P. van der Wolk. 1998. The Sec system. Curr. Opin. Microbiol. 1:216-222. [DOI] [PubMed] [Google Scholar]
- 15.Emr, S. D., J. Hedgpeth, J. M. Clement, T. J. Silhavy, and M. Hofnung. 1980. Sequence analysis of mutations that prevent export of lambda receptor, an Escherichia coli outer membrane protein. Nature 285:82-85. [DOI] [PubMed] [Google Scholar]
- 16.Fuge, E. K., and S. B. Farr. 1993. AppppA-binding protein E89 is the Escherichia coli heat shock protein ClpB. J. Bacteriol. 175:2321-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibson, K. E., and T. J. Silhavy. 1999. The LysR homolog LrhA promotes RpoS degradation by modulating activity of the response regulator sprE. J. Bacteriol. 181:563-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottesman, S., and V. Stout. 1991. Regulation of capsular polysaccharide synthesis in Escherichia coli K12. Mol. Microbiol. 5:1599-1606. [DOI] [PubMed] [Google Scholar]
- 19.Johnstone, D. B., and S. B. Farr. 1991. AppppA binds to several proteins in Escherichia coli, including the heat shock and oxidative stress proteins DnaK, GroEL, E89, C45 and C40. EMBO J. 10:3897-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, C. H., P. N. Danese, J. S. Pinkner, T. J. Silhavy, and S. J. Hultgren. 1997. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 16:6394-6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiino, D. R., G. J. Phillips, and T. J. Silhavy. 1990. Increased expression of the bifunctional protein PrlF suppresses overproduction lethality associated with exported beta-galactosidase hybrid proteins in Escherichia coli. J. Bacteriol. 172:185-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiino, D. R., and T. J. Silhavy. 1984. Mutation prlF1 relieves the lethality associated with export of beta-galactosidase hybrid proteins in Escherichia coli. J. Bacteriol. 158:878-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kisselev, L. L., J. Justesen, A. D. Wolfson, and L. Y. Frolova. 1998. Diadenosine oligophosphates (Ap(n)A), a novel class of signalling molecules? FEBS Lett. 427:157-163. [DOI] [PubMed] [Google Scholar]
- 24.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139-180. [DOI] [PubMed] [Google Scholar]
- 25.Kurihara, T., and P. Silver. 1993. Suppression of a sec63 mutation identifies a novel component of the yeast endoplasmic reticulum translocation apparatus. Mol. Biol. Cell 4:919-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Lee, P. C., B. R. Bochner, and B. N. Ames. 1983. AppppA, heat-shock stress, and cell oxidation. Proc. Natl. Acad. Sci. USA 80:7496-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, P. C., B. R. Bochner, and B. N. Ames. 1983. Diadenosine 5′,5′′′-P1,P4-tetraphosphate and related adenylylated nucleotides in Salmonella typhimurium. J. Biol. Chem. 258:6827-6834. [PubMed] [Google Scholar]
- 29.Manting, E. H., and A. J. Driessen. 2000. Escherichia coli translocase: the unravelling of a molecular machine. Mol. Microbiol. 37:226-238. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell, S. J., and M. F. Minnick. 1995. Characterization of a two-gene locus from Bartonella bacilliformis associated with the ability to invade human erythrocytes. Infect. Immun. 63:1552-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori, H., and K. Ito. 2001. The Sec protein-translocation pathway. Trends Microbiol. 9:494-500. [DOI] [PubMed] [Google Scholar]
- 32.Pogliano, K. J., and J. Beckwith. 1993. The Cs sec mutants of Escherichia coli reflect the cold sensitivity of protein export itself. Genetics 133:763-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radke, K. L., and E. C. Siegel. 1971. Mutation preventing capsular polysaccharide synthesis in Escherichia coli K-12 and its effect on bacteriophage resistance. J. Bacteriol. 106:432-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ecf sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 35.Raivio, T. L., and T. J. Silhavy. 1999. The sigmaE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr. Opin. Microbiol. 2:159-165. [DOI] [PubMed] [Google Scholar]
- 36.Rizzitello, A. E., J. R. Harper, and T. J. Silhavy. 2001. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J. Bacteriol. 183:6794-6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Silhavy, T. J., H. A. Shuman, J. Beckwith, and M. Schwartz. 1977. Use of gene fusions to study outer membrane protein localization in Escherichia coli. Proc. Natl. Acad. Sci. USA 74:5411-5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snyder, W. 1995. Ph.D. thesis. Princeton University, Princeton, N.J.
- 42.Snyder, W. B., L. J. Davis, P. N. Danese, C. L. Cosma, and T. J. Silhavy. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J. Bacteriol. 177:4216-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snyder, W. B., and T. J. Silhavy. 1995. Beta-galactosidase is inactivated by intermolecular disulfide bonds and is toxic when secreted to the periplasm of Escherichia coli. J. Bacteriol. 177:953-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Snyder, W. B., and T. J. Silhavy. 1992. Enhanced export of β-galactosidase fusion proteins in prlF mutants is Lon dependent. J. Bacteriol. 174:5661-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stout, V., A. Torres-Cabassa, M. R. Maurizi, D. Gutnick, and S. Gottesman. 1991. RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J. Bacteriol. 173:1738-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogel, J. P., L. M. Misra, and M. D. Rose. 1990. Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J. Cell Biol. 110:1885-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zamecnik, P. C., M. L. Stephenson, C. M. Janeway, and K. Randerath. 1966. Enzymatic synthesis of diadenosine tetraphosphate and diadenosine triphosphate with a purified lysyl-sRNA synthetase. Biochem. Biophys. Res. Commun. 24:91-97. [DOI] [PubMed] [Google Scholar]