Abstract
Human immunodeficiency virus type 1 (HIV-1) can evade immunity shortly after transmission to a new host but the clinical significance of this early viral adaptation in HIV infection is not clear. We present an analysis of sequence variation from a longitudinal cohort study of HIV adaptation in 189 acute seroconverters followed for up to 3 years. We measured the rates of variation within well-defined epitopes to determine associations with the HLA-linked hazard of disease progression. We found early reversion across both the gag and pol genes, with a 10-fold faster rate of escape in gag (2.2 versus 0.27 forward mutations/1,000 amino acid sites). For most epitopes (23/34), variation in the HLA-matched and HLA-unmatched controls was similar. For a minority of epitopes (8/34, and generally associated with HLA class I alleles that confer clinical benefit), new variants appeared early and consistently over the first 3 years of infection. Reversion occurred early at a rate which was HLA-dependent and correlated with the HLA class 1-associated relative hazard of disease progression and death (P = 0.0008), reinforcing the association between strong cytotoxic T-lymphocyte responses, viral fitness, and disease status. These data provide a comprehensive overview of viral adaptation in the first 3 years of infection. Our findings of HLA-dependent reversion suggest that costs are borne by some escape variants which may benefit the host, a finding contrary to a simple immune evasion paradigm. These epitopes, which are both strongly and frequently recognized, and for which escape involves a high cost to the virus, have the potential to optimize vaccine design.
The dynamics of viral replication in acute and early human immunodeficiency virus (HIV) infection are not well understood as longitudinal data from large cohorts of seroconverters are hard to assemble. Recent studies have shown that new HIV infections may be the result of a single transmitted variant, that new env gene mutations can be detected within a few weeks (25), and that early immune escape can be detected at sites across the HIV genome (9). These data add to a body of work showing that cytotoxic T cells act early, contributing to the early reduction in viremia (8, 30).
Whether early cytotoxic T-lymphocyte (CTL) immune responses influence longer-term clinical outcome is not clear. Antigen-specific CTLs capable of producing gamma interferon and other cytokines are detectable at all stages of HIV infection (1, 3, 24, 41). Much weight is placed on the macaque/simian immunodeficiency virus model in which nearly total peripheral blood CD8+ T-cell elimination using monoclonal antibodies results in rising viremia (42). The role of other forms of host immunity (e.g., neutralizing antibodies, natural killer cells, and macrophages) has, to some extent, been pursued with less intensity in light of persuasive evidence that CTLs can control retrovirus infection (46). The extent to which the simian model mirrors HIV infection has been questioned (5) and, despite exhaustive cellular assays of T-cell function—from gamma interferon enzyme-linked immunospot assays(1, 27, 38) to polyfunctional cytokine matrices (2, 6)—no CTL function correlates robustly with HIV plasma viral load or viral dynamics. Moreover, analyses of evolutionary data suggest that CTLs are inefficient at killing HIV-infected cells (4).
However, statistical analysis of data from large cross-sectional studies link HLA class I alleles with specific genome-wide HIV polymorphisms, suggestive of a pervasive selection pressure enacted by CTLs (7, 10, 18, 36, 40). It is clear that associations between some HLA class I alleles and particular amino acid polymorphisms are robust although it is disputed whether immune escape influences disease progression. The viral fitness costs resulting from immune escape may even contribute to better clinical outcomes associated with the possession of HLA class I alleles such as B*27, B*57, and B*58 (18).
Evolutionary studies of HIV require longitudinal data from large cohorts of patients sampled since seroconversion to detect adaptation in new hosts as it accrues. HIV is one of the few pathogens where it is possible to do this within individuals because of the high viral turnover and rapid intrahost evolution. Here, we investigate a cohort of 189 acute seroconverters—the largest cohort reported to date—followed for up to 3 years to study the rates of viral mutation in individual epitopes within internal HIV proteins and to determine the association between HLA class I alleles and rates of immune escape and reversion.
MATERIALS AND METHODS
Patients.
From a cohort of 189 predominantly Caucasian male acute seroconverters recruited within a median of 60 days from the estimated date of seroconversion (interquartile range, 39 to 86 days), plasma samples were obtained at the time of first presentation or trial enrollment (“baseline”) and then 6 months, 1 year, 2 years, and 3 years later. Patients were recruited from a prospective study of acute HIV-1 infection at St. Mary's Hospital, London, United Kingdom (as fully described elsewhere [16]), and as part of randomized controlled trial of short course therapy in acute HIV infection (SPARTAC [http://www.ctu.mrc.ac.uk/studies/spartac.asp]). Criteria for acute HIV-1 infection were, as previously described (17), a documented seronegative HIV-1 antibody test within the previous 6 months, acute symptomatic seroconversion illness, an evolving HIV-specific antibody response as determined by enzyme-linked immunosorbent assay, and positive HIV-1 DNA PCR or positive HIV-1 RNA quantification (Chiron 3.0; Chiron Inc., Emeryville, CA) in the absence of an antibody response. In order to confirm primary HIV-1 infection, sequential serum samples were later tested blindly with a “detuned” enzyme-linked immunosorbent assay (data not shown).
We were unable to obtain baseline sequences on 15 pol and 16 gag genes. For the pol gene, plasma samples were obtained and sequenced at baseline (n = 174), 6 months (n = 66), 1 year (n = 111), 2 years (n = 69), and 3 years (n = 15) after seroconversion. For the gag gene, from the same cohorts, plasma samples were obtained and sequenced at baseline (n = 173), 6 months (n = 61), 1 year (n = 95), 2 years (n = 62), and 3 years (n = 30) after seroconversion. The majority of patients were subtype B (164/189), confirmed using the REGA subtyping tool (http://www.bioafrica.net/virus-genotype/html/index.html). The remainder, which were excluded from this analysis, comprised subtypes A, A/G, C, and F.
Most patients recruited from the nonrandomized cohort study (n = 88/101) received a short (0.5 to 6 months; median, 3.0 months) course of highly active antiretroviral therapy (HAART) at seroconversion but remained drug naïve until either viral load, CD4 cell count, or clinical parameters were met to require formal institution of HAART. For the SPARTAC trial, patients were randomized to receive no therapy or 12 weeks or 48 weeks of HAART and then to remain off therapy according to clinical need.
Viral sequencing.
Viral RNA was extracted from patient plasma (Qiagen Viral RNA Extraction Kit), and the HIV-1 pol and gag genes were amplified in three fragments using nested PCRs and primers described previously (19). The PCR products were purified and sequenced using Big Dye dideoxy terminator chemistry (ABI).
Data analysis.
Sequences were aligned and manually edited (Se-Al software [www.evolve.zoo.ox.ac.uk]). Mixed residues containing more than one amino acid were identified at a number of nucleotide positions within the raw sequence data. In these cases the nucleotides were encoded as mixed if the secondary peak reached 30% of the height of the primary peak. To determine the dynamics of the amino acid changes, if at the subsequent time point the mixed site had become homogeneous for one of the bases, this would be treated as the time at which fixation had been reached. Clonal analysis of a selection of longitudinal sequences was used to confirm the validity of using bulk sequences to identify the times at which mutations either occur or revert and showed that predominant homogeneity at the baseline time point was followed by accumulation of mutations in line with the bulk sequences (data not shown).
For each patient, the baseline sequence was compared with HXB2, the Los Alamos National Laboratory subtype B reference sequence, to identify any mutants which might represent transmitted mutations or very early adaptation (i.e., escape that had taken place prior to sampling). Subsequent sequences collected in the first 3 years of infection were then compared with the patient's baseline sequence to determine the extent of intrahost evolution. As a quality control measure, HXB2 and the population consensus sequence were compared. The two sequences differed at four sites in Gag (p17, R76K and I94V; p24, I6L and V83L) and five in Pol (protease, V3I, S37N, and L63P; reverse transcriptase [RT], E122K and L214F). The affected epitopes in our analysis were ELRSLYNTV (R76K; HLA B*0801) and VPLDEDFRKY (E122K; B*3501) (mutated residues are in boldface), the latter being of interest as E122K has been reported as a T-cell receptor escape mutation but was the consensus sequence in our population.
For each amino acid in every patient, the timing of the emergence of polymorphisms was identified over 3 years for all amino acid positions (amino acids 1 to 417 for pol and amino acids 1 to 379 for gag). We defined a forward mutation (FM) as a mutation away from the wild-type baseline (according to HXB2 sequence), whereas a backward mutation (BM), or reversion, was identified as a mutation present in the baseline sequence which subsequently changed to the HXB2 wild type. Although FM and BM are likely to represent immune escape and reversion, respectively, in the absence of documented cellular immune responses for each patient, other causes of variation cannot be excluded, and these generic terms are used. For FMs, the number of events at each time point was compared with the number of baseline wild-type sites—i.e., the number with potential to escape—and adjusted for 1,000 amino acid sites. For BM, the denominator was the number of variant sites at baseline, representing sites with the potential for reversion. Known optimal epitopes, defined from the LANL database (www.lanl.com) were mapped onto the pol and gag gene sequences (Table 1). Mutations within these epitopes were identified, and the rates of their appearance were calculated.
TABLE 1.
Optimal epitopes used in the sequence analysis of the seroconverter cohort
| Epitope no. | Referencea | Optimal epitope | HLA restriction | HXB2 position (aa)b |
|---|---|---|---|---|
| Pol epitopes | ||||
| 1 | GPKVKQWPL | B*0801 | RT 18-26 | |
| 2 | ALVEICTEMEK | A*0301 | RT 33-43 | |
| 3 | TVLDVGDAY | B*3501 | RT 107-115 | |
| 4 | 15 | VLDVGDAYFSV | A*0201 | RT 108-118 |
| 5 | 44 | VPLDKDFRKY | B*3501 | RT 118-127 |
| 6 | TAFTIPSI | B*5101 | RT 128-135 | |
| 7 | 35 | SPAIFQSSMT | B*0702 | RT 156-165 |
| 8 | 44 | SPAIFQSSM | B*3501 | RT 156-164 |
| 9 | AIFQSSMTK | A*0301 | RT 158-166 | |
| 10 | NPDIVIYQY | B*3501 | RT 175-183 | |
| 11 | VIYQYMDDL | A*0201 | RT 179-187 | |
| 12 | 35 | EELRQHLLRW | B*44 | RT 203-212 |
| 13 | 35 | PIVLPEKDSW | B*5701 | RT 243-252 |
| 14 | ILKEPVHGV | A*0201 | RT 309-317 | |
| Gag epitopes | ||||
| 1 | 21 | KIRLRPGGKK | B*27 | p17 18-27 |
| 2 | GGKKKYKLK | B*0801 | p17 24-32 | |
| 3 | KYKLKHIVW | A*2402 | p17 28-36 | |
| 4 | ELRSLYNTV | B*0801 | p17 74-82 | |
| 5 | SLYNTVATL | A*0201 | p17 77-85 | |
| 6 | TLYCVHQRI | A*1101 | p17 84-92 | |
| 7 | ISPRTLNAW | B*5701 | p24 15-23 | |
| 8 | SPRTLNAWV | B*0702 | p24 16-24 | |
| 9 | KAFSPEVIPMF | B*5701 | p24 30-40 | |
| 10 | TPQDLNTML | B*0702 | p24 48-56 | |
| 11 | 39 | GHQAAMQMLKE | A*02 | p24 61-71 |
| 12 | TSTLQEQIGW | B*5701 | p24 108-117 | |
| 13 | 47 | PPIPVGEIY | B*35 | p24 122-130 |
| 14 | 45 | GEIYKRWII | B*0801 | p24 127-135 |
| 15 | KRWIILGLNK | B*2705 | p24 131-140 | |
| 16 | QASQEVKNW | B*5701 | p24 176-184 | |
| 17 | 44 | NANPDCKTI | B*51 | p24 193-201 |
| 18 | DCKTILKAL | B*0801 | p24 197-205 | |
| 19 | ACQGVGGPGHK | A*1101 | p24 217-227 | |
| 20 | GPGHKARVL | B*0702 | p24 223-231 |
All epitopes are derived from the Los Alamos National Laboratory A list of optimal epitopes (http://www.hiv.lanl.gov/content/immunology/pdf/2008/optimal_ctl_article.pdf), except for those indicated and referenced separately.
aa, amino acids.
HLA typing.
Patients’ HLA types were determined to the oligo-allelic level using Dynal RELITM reverse sequence-specific oligonucleotide kits for the HLA-A, HLA-B, and HLA-C loci (Dynal Biotech). To obtain four-digit typing, Dynal Biotech sequence-specific priming kits were used, in conjunction with the Sequence-Specific Oligonucleotide type.
Relative hazards for disease progression.
Hazards for disease progression were obtained from previous data using Cox model analyses based on the Caucasian samples from four cohorts for which patients had known dates of seroconversion: the Multicenter AIDS Cohort Study, the Multicenter Hemophilia Cohort Study, the San Francisco City Clinic Cohort, and AIDS Linked to Intravenous Experience (11, 21, 22, 37).
Statistics.
Fisher's exact tests, unpaired t tests, and linear regression were used in statistical analyses with Prism, version 4.0 (GraphPad Software).
RESULTS
Gene-specific rates of viral evolution following acute infection.
The rates of FMs and BMs were compared in the gag and pol genes. For FMs, which represent possible immune escape, there was a difference between the two genes (P < 0.001) up to 12 months (Fig. 1A). For pol, over the first 6 months the prevalence was 0.27 FM/1,000 amino acid sites, and there was a 10-fold greater prevalence of 2.2 FM/1,000 amino acid sites in gag. Only after the first 12 months did the rate of escape in pol approximate that in gag.
FIG. 1.
Prevalence of mutations in the HIV gag and pol genes in the first 3 years of infection. The prevalence of nonsynonymous mutations was measured at baseline, 6 months, 1 year, 2 years, and 3 years following seroconversion in the gag and pol genes. The prevalence of FMs (a) and BMs (b) were determined by coding when mutations at every amino acid either arose or reverted in each patient and normalizing these values per 1,000 amino acids (aa). Each data point is shown with 95% confidence intervals.
For both genes, mutations present at baseline reverted early. For pol and gag, in the first 6 months, there were 26.2 reversions/1,000 amino acid sites and 29.9 reversions/1,000 amino acid sites, respectively. These rates were maintained for the first 2 years of infection, illustrating that transmitted mutations continued to revert (Fig. 1B). For a cohort of this size, it is not feasible to obtain donor sequences, and so it is possible that not all of these mutations were transmitted. Nevertheless, the changes observed are consistently toward wild type, with little evidence of recurrence of the mutations, suggesting that they were not originally selected, even if only temporarily, very early in infection.
As pol is also under selection pressure from antiretroviral therapy, we identified amino acid sites (Stanford University Drug Resistance Database [http://hivdb.stanford.edu/]) susceptible to drug resistance mutations. The proportion of both reversion and putative escape polymorphisms that arose at drug resistance residues was less than 5% of the new pol mutations observed although baseline variants were identified at M41 (four patients), T69 (five), A98 (three), K103 (five), V108 (four), V118 (six), V179 (two), Y181 (one), M184 (one), Y188 (one), L210 (three), and T215 (nine), the majority of which persisted over time. These were randomly distributed among HLA class I alleles, showing that the rates of observed mutation within CTL epitopes were not biased by drug resistance mutations. Although the median time between sampling and the estimated date of seroconversion was 60 days (interquartile range, 39 to 86), it is possible that the pol gene rate has been underestimated due to very early escape missed by our sampling. Although one might expect this bias to affect both genes equally, in order to answer this question robustly, one would need to analyze a cohort of acute and hyperacute patients identified within 4 weeks of infection.
Patterns of HLA-associated variation in HIV epitopes.
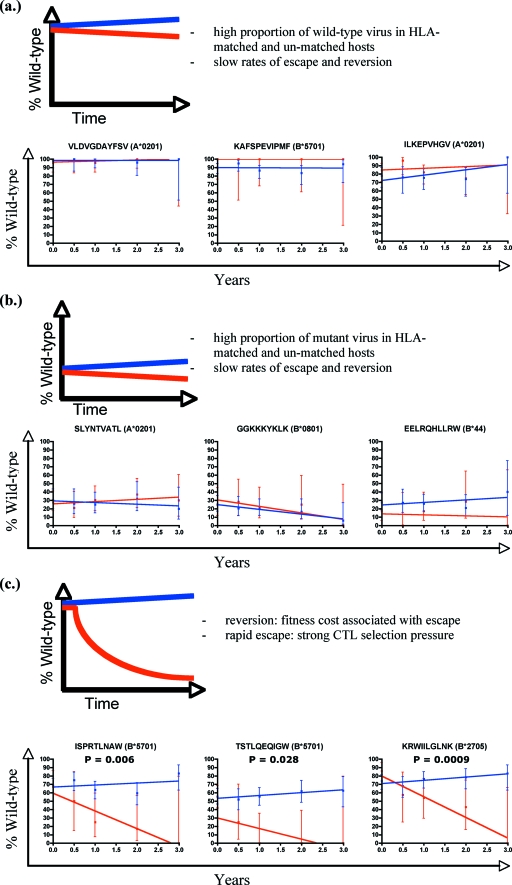
To determine how these patterns of forward mutation and reversion related to immune pressure, we studied 34 well-defined optimal epitopes restricted by HLA class I alleles (Table 1). For these epitopes we had previously documented HLA-restricted immune responses using gamma interferon enzyme-linked immunospot assays in a cohort of chronically infected patients (41). Epitope sequences were grouped according to whether patients carried the requisite restricting HLA class I allele. Any measure of variation needs to be controlled for sequence variation when the site is not acting as an antigen, i.e., in the absence of the restricting HLA class I allele. (Even then, confounding effects such as overlapping epitopes may occur). For each time point the proportion of wild-type epitopes (those with the same sequence as the index peptide from the Los Alamos National Laboratory HIV database [www.hiv.lanl.gov]) was plotted for both the HLA-matched and unmatched patients. A best-fit line was modeled to the data, the initial rate of change in percentage of wild-type amino acids (percentage points/year) was calculated, and the rates of change in the HLA-matched and unmatched patients were compared.
The rates of change were different among the 34 epitopes (Fig. 2). Three common patterns of epitope variation emerged, and these encompassed 31 of the 34 (91%) studied epitopes. For the first group (n = 16/34) the baseline sequences were conserved (usually greater than 80% wild type) regardless of whether the sequence was derived from the HLA-matched or HLA-unmatched group, followed by only minimal change (Fig. 2a). Such epitopes include KAFSPEVIPMF restricted by HLA B*5701, ILKEPVHGV (A*0201), and VLDGDAYFSV (A*0201). For the second group (n = 7/34), the epitopes are highly variable in both the HLA-matched and -unmatched patients (mostly <30% wild-type at baseline) but, again, with minimal change over time and no significant difference between the two (Fig. 2b). The examples shown are SLYNTVATL (A*0201), EERLQHLLRW (B44), and GGKKYKLK (B*0801). It is possible that there is so much background variation at these sites that we are not able to identify small HLA-attributable changes. However, since HLA-matched and HLA-unmatched patients are so similar, HLA class I is unlikely to be the major source of variation within these epitopes.
FIG. 2.
Prevalence of variation within defined optimal epitopes in HLA-matched and HLA-unmatched patients. For the epitopes shown, the percentage of those with wild-type sequences is shown for each time point. Each epitope is analyzed according to whether patients carried the restricting HLA class I allele (blue for HLA-unmatched and red for HLA-matched epitopes). Best-fit lines are drawn through the points using linear regression. Error bars show the 95% confidence intervals for each time point. Epitopes are divided into three groups and shown with an illustrative model: those that are conserved at baseline and over time regardless of HLA matching (a), those that are variant at baseline and over time (b), and those that are conserved at baseline but become more variable over time in the HLA-matched patients only (c). P values represent whether there is a statistically significant difference in the slopes of the two datasets.
The third group (n = 8/34) contained epitopes in which there was a divergence between the proportion of wild-type sequences in the HLA-matched and -unmatched patients over time (Fig. 2c). The HLA-matched epitopes became increasingly variant while baseline variation in unmatched epitopes reverted toward wild-type sequences. This third group of discordant epitopes included ISPRTLNAW (B*5701), KRWILGLNK (B*2705), TAFTIPSI (B*5101), TLYCVHQRI (A*1101), TSTLQEQIGW (B*5701), PIVLPEKDSW (B*5701), and QASKEVKNW (B*5701). This third group is dominated by epitopes restricted by HLA class I alleles which have previously been associated with lower viral loads (28), better clinical outcomes (37), and a high prevalence of variation in chronic infection (18). For some epitopes there were more mutant sequences in the HLA-matched patients than in the HLA-unmatched controls at baseline, which is suggestive of early escape prior to first sampling. For example, the HLA B*51-restricted epitope TAFTIPSI was mutant at the position 8 isoleucine in 10/13 patients at baseline compared with 68/151 B51-negative controls (P = 0.021), and the B*57/58-restricted TSTLQEQIGW was mutant at position 3 in 4/12 patients compared with 12/135 controls (P = 0.003). So, although the median interval between estimated seroconversion and first sampling is 60 days, at certain epitopes escape had clearly already taken place.
Epitopes restricted by beneficial HLA class I alleles show greater variability over time than less favorable epitopes.
We plotted the initial rates of change for each epitope in the presence and absence of HLA class I restriction (Fig. 3). By intersecting the axes of the figure through (0, 0), four quadrants are defined. The inset figure shows the inferred behavior of epitopes located in each section of the plot. Most epitopes (20/34; 59%) are located on the left side of the y axis, suggesting that these epitopes become increasingly variant over time in the absence of HLA-associated pressure; i.e., there is ongoing nonsynonymous change over time not necessarily attributable to HLA class I-dictated immunity. We cannot rule out the possibility that overlapping or undefined epitopes might contribute to this. Twenty-three epitopes lie below the x axis, showing that variants accumulate over time in the HLA-matched patients. The less-variant epitopes lie close to the intersection of the x and y axes, suggestive of weaker selection pressure or structural constraint. Of interest are the epitopes located in the lower right quadrant. In this quadrant are epitopes that become more variable in the presence of HLA but that revert once this pressure is removed. They include KRWIILGLNK (restricted by HLA B*2705), ISPRTLNAW (HLA B*5701), TSTLQEQIGW (HLA B*5701), QASKEVKNW (HLA B*5701), and TLYCVHQRI (HLA A*1101). The epitopes in the other three quadrants show little evidence of HLA-specific selection.
FIG. 3.
Rates of mutation in the first 3 years of infection in HLA-matched and HLA-unmatched epitopes. Each epitope is plotted according to its rate of change (percent change in wild type/year) in HLA-matched and -unmatched patients. The insert shows the implications of a data point lying in each of the four quadrants. Each epitope can be identified using the key. HLA-ve, HLA negative; HLA+ve, HLA positive; WT, wild type; MT, mutation/mutant.
To demonstrate how epitope variation unfolds over time, we present key epitopes from Fig. 2 in a format that highlights the differences between the rates of mutation. On these plots the axes refer to the prevalence of variation within the cohort for each epitope at each time point. The figures reveal the patterns of epitope behavior through the movement of each epitope around the plot over 3 years. Figure 4a shows examples of neutral epitopes that remain close to the line x = y over time, with no strong evidence of HLA-driven adaptation, and that might represent the less effective “passenger” epitopes (29). Figure 4b shows the protective or dynamic “driver” epitopes (29) evolving over time, with specific adaptation to the HLA class I immune response. The figure includes the HLA B*5701-restricted ISPRTLNAW, the B*2705-restricted KRWIILGLNK, the B*5101-restricted TAFTIPSI, TSTLQEQIGW (B*5701), and QASKEVKNW (B*5701). As well as moving down the plot, these epitopes also move farther right over time. This reveals new mutations in the HLA-matched patients combined with reversion in the HLA-unmatched patients. This pattern is not seen in Fig. 4a. Of interest in Fig. 4b is the plot for the HLA B51-restricted TAFTIPSI. This is an epitope that is restricted by a favorable HLA class I allele (37) and mutates early (19) but is not associated with a fitness cost according to this in vivo analysis. One might expect, therefore, mutations in TAFTIPSI to accumulate in the population over time and to be prevalent in populations with a high frequency of HLA B*51-positive individuals.
FIG. 4.
The evolution of epitopes over time. The figure shows the transit of neutral (a) and dynamic (b) epitopes across the plot over time. Each epitope is plotted at each time point according to the relative prevalence of wild-type (WT) sequences in HLA-matched (y axis) and unmatched (x axis) patients. The epitopes shown in panel a are KYKLKHIVW, restricted by A*2402 (A24); SLYNTVATL, A*0201 (A2); ALVEICTEMEK A*0301 (A3); and GPKVKQWPL, B*0801 (B8). The epitopes shown in panel b are TAFTIPSI, restricted by B*5101 (B51); ISPRTLNAW, B*5701 (B57); KRWIILGLNK, B*2705 (B27); QASKEVKNW, B*5701 (B57); and TSTLQEQIGW, B*5701 (B57).
HLA-associated rates of reversion, but not escape, correlate with relative hazard of disease progression.
Recent data suggest that the frequencies with which epitopes are targeted by CTLs correlates with both the rate of escape in early infection (9) and the HLA-specific relative hazard of disease progression (41). The rate at which epitopes acquire new variation in early infection has not been directly compared with the risk of disease progression. In the absence of virological or clinical outcome data from the SPARTAC cohort, which remains blinded, the well-described relative hazards obtained from large Western seroconverter cohorts were used (11, 21, 22, 37). We correlated the rate of variation for each epitope in the absence and presence of the restricting HLA class I allele with the documented relative hazards for disease progression for specific HLA class I alleles for the time to reach a CD4 cell count of less than 200 cells/μl, time to AIDS using the CDC 1987 definition (time to AIDS 1987) (12), time to AIDS using the CDC 1993 definition (time to AIDS 1993) (13), and time to death. As previously described, the population HLA frequencies in these cohorts and our United Kingdom patients are strongly correlated (41).
There was no correlation between the rates of epitope mutation in the presence of their restricting HLA class I alleles and the associated relative hazards of disease progression (CD4 of <200, P = 0.08 and R2 = 0.09; time to AIDS 1987, P = 0.16 and R2 = 0.06; time to AIDS 1993, P = 0.09 and R2 = 0.09; death, P = 0.18 and R2 = 0.06) (data not shown). However, when a variant epitope is transmitted to an HLA-mismatched patient, the rate at which the epitope reverts (percent wild type/year) is strongly associated with the relative hazard for the restricting HLA class I allele (Fig. 5a to d). Measuring the epitope variation rate in an HLA-unmatched patient is effectively an in vivo fitness assay. The statistical correlation of reversion rates with the parameters for HLA-associated relative hazard is strong evidence that viral escape from beneficial HLA class I alleles results in strong fitness costs in the epitopes that they restrict.
FIG. 5.
Linear regression analysis of relative hazard of disease progression against rates of mutation within epitopes in HLA-unmatched subjects. Linear regression analysis of the rate of change of individual epitopes in HLA-unmatched patients (percent wild type/year) against the relative hazard of disease progression for the restricting HLA class I allele for time to a CD4 cell count of <200 cells/μl (a), time to AIDS 1987 (b), time to AIDS 1993 (c), and time to death (d). A negative mutation rate indicates mutation away from wild type and a positive mutation rate indicates change toward wild type (reversion).
DISCUSSION
We present a study of a large cohort of serially sampled HIV-infected seroconverters, with data analyzed for up to 3 years after infection. There are three main findings. First, the rates of FM in the two study genes vary, with escape mutations being more common in the more immunogenic gag gene although in both genes there is rapid and early reversion of transmitted mutants. Second, the patterns of variation are driven by changes within epitopes, with the most variant being restricted by beneficial HLA class I alleles. Third, we show a correlation between the HLA class I-associated hazard and the rate of reversion in HLA-unmatched epitopes, providing strong evidence for a role of viral fitness in the benefit conferred by certain HLA class I alleles.
In both the gag and pol genes, reversion occurred early—generally within the first year of infection—whereas FMs arose faster in the gag gene. There may be a number of explanations for this. In new hosts, the controlling effect of the previous donor's immune response on HIV variability has been relaxed before the new recipient has mounted his own T-cell response. Accordingly, an initial limiting factor to viral replication will be the fitness cost of the transmitted mutations. If one assumes that HIV can produce extensive de novo variants early in infection, then one might expect the most costly escape mutations to revert quickly and early. Our data agree with the findings of Li et al., who reported early reversion on clonal analysis of seven acute seroconverters, also predominantly within 6 months (32). These data are in contrast to the transmission of drug resistance mutations to drug-naïve hosts. Here, it would appear that transmitted variants—mostly in pol—are maintained, often for a number of years (20). Why should there be a different dynamic for the transmission of immune escape and drug resistance mutations? Possibilities include the different potencies of the selection pressures driving variation, the role of compensatory mutations, and the pathways for reversion. For certain antiretroviral agents including nucleoside reverse transcriptase inhibitors and protease inhibitors, a number of mutations cluster together to confer the reduction in drug susceptibility, some of which require two nucleotide mutations within the same codon. It is feasible, therefore, that the reversion pathways for these drug resistance mutations require much greater kinetics on behalf of the virus and are more likely to persist once transmitted.
We found that escape occurs more quickly in the gag gene. This is not surprising as Gag has consistently been reported as one of the main targets of the immune response in acute infection, and the early detection of escape reflects this. In addition, the enzymatic function of Pol may impose greater sequence conservation than the structural Gag protein. It is possible that we have underestimated the rate of escape in pol as in some patients escape mutations may have occurred prior to sampling; however, data from Brumme et al. (9) also reveals a similar discrepancy in gag and pol.
We find that, in early infection, the variation appears most rapidly within epitopes restricted by HLA class I alleles that have been associated with lower viral loads and delayed progression. In particular HLA B*2705, B*5101, and B*5701 were associated with faster rates of mutation, but this was not the case for all the epitopes restricted by these alleles. A key finding from this study is the correlation between the rates of reversion of transmitted mutations and the relative hazard of disease progression reported for the alleles that select them. The rate of reversion for a transmitted mutation, when assayed across a large number of patients, is an in vivo measure of its associated fitness cost. Accordingly, our data reflect the correlation between HLA class I alleles that are linked with a delayed progression to AIDS and the fitness costs incurred by the virus due to the strong selection pressures these alleles impose. We cannot comment on whether transmission of an escape variant to an HLA-mismatched recipient is of clinical advantage or disadvantage. This is a separate analysis that will be undertaken once the SPARTAC trial has been unblinded.
Recently, the clinical significance of reverting sites and their association with lower viral loads have been inferred from cross-sectional data from chronically infected patients (34). Although we agree with the findings of that study, to comment on reversion requires longitudinal analysis of acutely infected patients, ideally, although not necessarily, with viral sequence data from the donor. Cross-sectional analyses from chronic infection make assumptions on the nature and frequency of mutation transmission. Although our study is a longitudinal analysis of acutely infected patients, we do not know the sequence of the donor virus for the members of our cohort. How might this bias our results? In the analysis, we identified any variant epitope present in the baseline sample, i.e., the sample taken at first clinical presentation within a median of 60 days of the estimated date of seroconversion. Variation was defined against the sequence of the wild-type optimal epitope in the Los Alamos National Laboratory HIV database (www.lanl.gov). If the variant remained unchanged over time, this was classified as a nonrevertant. There are two potential sources of error that arise in the absence of the donor sequence. First, rather than being transmitted the variant may have risen anew in the recipient, reflecting adaptation to the new selective environment. If such a variant changed to a wild-type genotype at a later time point, then this would suggest a transient escape, a process which we encountered rarely in our cohort (0.15% of all amino acids) (data not shown). Accordingly, a change from a variant to wild-type sequence is likely to reflect true reversion. Alternatively, if the variant persisted, this could reflect a nonreverting transmitted mutation or a stable new mutation in the new host. Without the donor sequence it is impossible to distinguish between these two outcomes. However, when the reverting mutation is a well-documented escape mutation in a patient who does not carry the restricting HLA class I allele, this additional information supports the argument that this is true reversion. We also cannot exclude the influence of overlapping epitopes with escape mutations at the same amino acid sites.
Our data are intriguing since some escape is evident, but for most epitopes (23/34) there is no significant difference in the rates of variation in the HLA-matched and HLA-unmatched patients. If one assumes that HIV can respond to selection pressure by accepting mutations with fitness costs (26), the lack of variation in so many epitopes suggests weak selection rather than structural constraint and, by inference, a weak CTL immune response. Alternatively, these epitopes may simply be critical sites of conservation within the HIV genome. If they are conserved, then CTLs should have a “good” antigen to target. If no escape is detected yet variation is tolerated, it follows that the CTLs are weak, which is compatible with recent mathematical models (4, 5). Cellular assays of the CTL immune response are currently being undertaken on the seroconverter cohort to resolve this issue. The results of these studies in combination with the sequence variation data will inform vaccine design by highlighting epitopes that are both strongly and frequently recognized and for which escape involves high cost to the virus. However, the observation that variation and immune escape are limited to a few epitopes associated with slower disease progression is compatible with data from African and European cohorts of chronically infected patients (18).
The evidence that immune escape is the precursor to a rise in viral load and clinical progression is limited (14, 23). We have previously argued that the fitness cost imposed by certain immune escape mutations may be one of a number of factors which benefit the host (18). These hypotheses have been borne out by in vitro fitness assays (33). Since escape polymorphisms occur in patients with protective HLA class I alleles, one has to conclude that the selection pressure imposed by CTLs dictated by these alleles is so strong that it is still to the virus's advantage to escape, despite the fitness cost. Hence, there is rapid reversion of these mutations once they are transmitted to an environment where they are no longer beneficial. Some mutations do not appear to carry a fitness cost (e.g., the HLA B51-restricted TAFTIPSI), and mutants are therefore likely to accrue over time, possibly rising to fixation and so becoming the consensus sequence (31). Should our analysis include epitopes that already contain an escape variant as part of the consensus sequence (“negatopes”), this would present as apparent reversions in HLA-matched hosts and a slow rate of “escape” in HLA-mismatched hosts. Such events were extremely rare in this cohort (data not shown), and although negatopes have been described within the genome (31), we do not feel that they bias this analysis.
Our cohort is part of a clinical trial, so a majority of patients received a short course of antiretroviral therapy (ART) at seroconversion. Could our findings have been influenced by a short course of drug therapy? The reduction in viral load, replication, antigenic load, and the predictable decline in CTL immune responses that might be expected from a short course of ART would be expected to slow down viral evolution and immune escape and decrease genetic diversity. Our data show that mutations that could reflect both escape and reversion are present within 6 months, and in a number of epitopes (e.g., the pol epitope TAFTIPSI), the escape variants had already reached fixation on clonal analysis (data not shown). In addition, as therapy will be distributed randomly across the different HLA class I alleles, any difference in viral variation attributed to different HLA class I alleles should not be significantly biased. Evidence from unpublished data from a similar seroconverter cohort (Stephane Hue, personal communication) suggests that the impact on genetic diversity of a short course of HAART at seroconversion is limited although this needs to be formally assessed in a controlled trial. The epitopes that we have shown to mutate, do so quickly although it is possible that, if anything, we have underestimated this. Most other epitopes do very little; if there is a differential impact of a short course of ART on epitope variability, this warrants further analysis once the SPARTAC trial is unblinded.
Our results support the notion that HIV adaptation to new hosts begins quickly following transmission but also show that this process continues for at least 36 months, predominantly at a few key immunogenic epitopes. In addition, the differential rates of reversion according to associated clinical hazard is strong evidence that immune escape from “protective” HLA class I alleles comes at a cost to the virus. Further work is needed to determine why many epitope sequences are conserved. If fitness cost, rather than a weak selection pressure, is the key, then these will be epitopes worth targeting for vaccine design.
Acknowledgments
The following is a list of the SPARTAC Investigators. The Trial Steering Committee members were A. Breckenridge (Chair), C. Conlon, D. Cooper, F. Conradie, J. Kaldor, M. Schechter, P. Claydon, P. Kaleebu, G. Ramjee, F. Ssali, G. Tambussi, and J. Weber; the Trial Physician was Sarah Fidler; and the Trial Statistician was Abdel Babiker. Members of the Data and Safety Monitoring Committee were A. McLaren (in memoriam), V. Beral, G. Chene, and J. Hakim. Members of the Central Virology Laboratories and Repositories Jefferiss Trust Laboratories, Imperial College, London, United Kingdom, were M. McClure, D. Muir, I. Blain, A. Helander, O. Erlwien, and S. Kaye. The Clinical Endpoint Review Committee members were N. Paton and S. Fidler. The following were coordinating trial centers: in Australia, the National Centre in HIV Epidemiology and Clinical Research, University of New South Wales, Sydney (P. Gray, D. Cooper, T. Kelleher, and M. Law), and in the United Kingdom and Ireland, the MRC Clinical Trials Unit, London (A. Babiker, K. Porter, P. Kelleher, K. Boyd, D. Johnson, and D. Nock). The investigators and staff at participating sites in Australia were as follows: St. Vincent's Hospital, Sydney (D. Cooper); Carlton Clinic, Melbourne (J. Anderson); 407 Doctors, Sydney (R. McFarlane); Prahran Market Clinic, Melbourne (N. Roth); Taylor Square Private Clinic, Sydney (R. Finlayson); The Centre Clinic, Melbourne (B. Kiem Tee); Sexual Health Centre, Melbourne (T. Read); AIDS Medical Unit, Brisbane (M. Kelly); and Centre for Immunology, Sydney (P. Cunningham). In Brazil, the participating site (investigators) was the Projeto Praça Onze, Hospital Escola São Francisco de Assis, Universidade federal do Rio de Janeiro, Rio de Janeiro (M. Schechter, R. Zajdenverg, and M. Merçon). Participating sites (investigators) in Italy were the Ospedale San Raffaele, Milan (G. Tambussi, C. Tassan Din, C. Ronchetti, G. Travi, V. Rusconi, and G. de Bartolo), and the Ospedale Lazzaro Spallanzani, Roma (G. D'Offizi, C. Vlassi, and A. Corpolongo). Participating sites (investigators) in South Africa were the Desmond Tutu HIV Centre, Institute of Infectious Diseases, Capetown (R. Wood, J. Pitt, L.-G. Bekker, J. Aploon, L. Fielder, N. Killa, and T. Buhler); the Reproductive Health and HIV Research Unit, Bara Clinic, Chris Hani Baragwanath Hospital, Johannesburg (H. Rees, J. Moyes, S. Walaza, and K. Moitse); Contract Laboratory Services, Johannesburg Hospital, Johannesburg (W. Stevens, C. Wallis, C. Ingram, and M. Majam); and in KwaZulu-Natal, the HIV Prevention Unit, Medical Research Council, Durban (G. Ramjee, D. Singh, T. Mtambo, S. Gappoo, H. Somaroo, J. Moodley, M. Mills, A. Premrajh, N. Nozulu, and K. Naidoo). In Uganda, the participating site (investigators) was the MRC/Uganda Virus Research Institute, Entebbe (H. Grosskurth, A. Kamali, P. Kaleebu, J. Mugisha, U. Bahemuka, F. Lyagoba, and P. Tabuga). In Spain, the participating site (investigators) was the Hospital Clinic-IDIBAPS, University of Barcelona, Barcelona (J. M. Miro, M. López-Dieguez, F. Agüero, J. A. Arnaiz, T. Pumarola, M. Plana, M. Tuset, M. C. Ligero, C. Gil, T. Gallart, and J. M. Gatell). In the United Kingdom and Ireland, the participating sites (investigators) were the Royal Sussex County Hospital, Brighton (M. Fisher, L. Heald, N. Perry, D. Pao, and D. Maitland); St. James's Hospital, Dublin (F. Mulcahy, G. Courtney, and D. Reidy); Regional Infectious Diseases Unit, Western General Hospital and Genitourinary Department, Royal Infirmary of Edinburgh, Edinburgh (C. Leen, G. Scott, L. Ellis, S. Morris, P. Simmonds, and T. Shaw); Chelsea and Westminster Hospital, London (B. Gazzard, D. Hawkins, C. Higgs, and C. Mahuma); Homerton Hospital, London (J. Anderson and L. Muromba); Mortimer Market Centre, London (I. Williams, J. Turner, D. Mullan, and D. Aldam); North Middlesex Hospital (J. Ainsworth and A. Waters); Royal Free Hospital (M. Johnson, S. Kinloch, A. Carroll, P. Byrne, and Z. Cuthbertson); St. Bartholomew's Hospital, London (C. Orkin, J. Hand, and C. De Souza); and St. Mary's Hospital, London (J. Weber, S. Fidler, E. Thomson, J. Fox, K. Legg, S. Mullaney, A. Winston, N. Poulter, and S. Wilson). Trial Secretariat members were D. Winogron and S. Keeling.
We thank the patients and staff of the St. Mary's Hospital Jefferiss Wing clinic and the patients and staff involved in the SPARTAC trial. We thank Paul Klenerman for his advice on the manuscript.
A.D. and J.F. are supported by the MRC. J.W. and R.E.P. are supported by the Wellcome Trust (United Kingdom).
Footnotes
Published ahead of print on 19 November 2008.
REFERENCES
- 1.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 772081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almeida, J. R., D. A. Price, L. Papagno, Z. A. Arkoub, D. Sauce, E. Bornstein, T. E. Asher, A. Samri, A. Schnuriger, I. Theodorou, D. Costagliola, C. Rouzioux, H. Agut, A. G. Marcelin, D. Douek, B. Autran, and V. Appay. 2007. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J. Exp. Med. 2042473-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altfeld, M., E. T. Kalife, Y. Qi, H. Streeck, M. Lichterfeld, M. N. Johnston, N. Burgett, M. E. Swartz, A. Yang, G. Alter, X. G. Yu, A. Meier, J. K. Rockstroh, T. M. Allen, H. Jessen, E. S. Rosenberg, M. Carrington, and B. D. Walker. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8+ T cell response against HIV-1. PLoS Med. 3e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asquith, B., C. T. Edwards, M. Lipsitch, and A. R. McLean. 2006. Inefficient cytotoxic T lymphocyte-mediated killing of HIV-1-infected cells in vivo. PLoS Biol. 4e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asquith, B., and A. R. McLean. 2007. In vivo CD8+ T cell control of immunodeficiency virus infection in humans and macaques. Proc. Natl. Acad. Sci. USA 1046365-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betts, M. R., M. C. Nason, S. M. West, S. C. De Rosa, S. A. Migueles, J. Abraham, M. M. Lederman, J. M. Benito, P. A. Goepfert, M. Connors, M. Roederer, and R. A. Koup. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 1074781-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharya, T., M. Daniels, D. Heckerman, B. Foley, N. Frahm, C. Kadie, J. Carlson, K. Yusim, B. McMahon, B. Gaschen, S. Mallal, J. I. Mullins, D. C. Nickle, J. Herbeck, C. Rousseau, G. H. Learn, T. Miura, C. Brander, B. Walker, and B. Korber. 2007. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science 3151583-1586. [DOI] [PubMed] [Google Scholar]
- 8.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 686103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brumme, Z. L., C. J. Brumme, J. Carlson, H. Streeck, M. John, Q. Eichbaum, B. L. Block, B. Baker, C. Kadie, M. Markowitz, H. Jessen, A. D. Kelleher, E. Rosenberg, J. Kaldor, Y. Yuki, M. Carrington, T. M. Allen, S. Mallal, M. Altfeld, D. Heckerman, and B. D. Walker. 2008. Marked epitope and allele-specific differences in rates of mutation in human immunodeficiency virus type 1 (HIV-1) Gag, Pol, and Nef cytotoxic T-lymphocyte epitopes in acute/early HIV-1 infection. J. Virol. 829216-9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brumme, Z. L., C. J. Brumme, D. Heckerman, B. T. Korber, M. Daniels, J. Carlson, C. Kadie, T. Bhattacharya, C. Chui, J. Szinger, T. Mo, R. S. Hogg, J. S. Montaner, N. Frahm, C. Brander, B. D. Walker, and P. R. Harrigan. 2007. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 3e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hoots, and S. J. O'Brien. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 2831748-1752. [DOI] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention. 1987. Revision of the CDC surveillance case definition for acquired immunodeficiency syndrome. MMWR Morb. Mortal Wkly. Rep. 361-15S. [Google Scholar]
- 13.Centers for Disease Control and Prevention. 1992. 1993 revised classification system for HIV infection and expanded surveillance case definition for AIDS among adolescents and adults. MMWR Recommend. Rep. 41(RR-17)1-19. [PubMed] [Google Scholar]
- 14.Feeney, M. E., Y. Tang, K. A. Roosevelt, A. J. Leslie, K. McIntosh, N. Karthas, B. D. Walker, and P. J. Goulder. 2004. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J. Virol. 788927-8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrari, G., D. D. Kostyu, J. Cox, D. V. Dawson, J. Flores, K. J. Weinhold, and S. Osmanov. 2000. Identification of highly conserved and broadly cross-reactive HIV type 1 cytotoxic T lymphocyte epitopes as candidate immunogens for inclusion in Mycobacterium bovis BCG-vectored HIV vaccines. AIDS Res. Hum. Retrovir. 161433-1443. [DOI] [PubMed] [Google Scholar]
- 16.Fidler, S., J. Fox, G. Touloumi, N. Pantazis, K. Porter, A. Babiker, and J. Weber. 2007. Slower CD4 cell decline following cessation of a 3 month course of HAART in primary HIV infection: findings from an observational cohort. AIDS 211283-1291. [DOI] [PubMed] [Google Scholar]
- 17.Fidler, S., A. Oxenius, M. Brady, J. Clarke, I. Cropley, A. Babiker, H. T. Zhang, D. Price, R. Phillips, and J. Weber. 2002. Virological and immunological effects of short-course antiretroviral therapy in primary HIV infection. AIDS 162049-2054. [DOI] [PubMed] [Google Scholar]
- 18.Frater, A. J., H. Brown, A. Oxenius, H. F. Gunthard, B. Hirschel, N. Robinson, A. J. Leslie, R. Payne, H. Crawford, A. Prendergast, C. Brander, P. Kiepiela, B. D. Walker, P. J. Goulder, A. McLean, and R. E. Phillips. 2007. Effective T-cell responses select human immunodeficiency virus mutants and slow disease progression. J. Virol. 816742-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frater, A. J., C. T. Edwards, N. McCarthy, J. Fox, H. Brown, A. Milicic, N. Mackie, T. Pillay, J. W. Drijfhout, S. Dustan, J. R. Clarke, E. C. Holmes, H. T. Zhang, K. Pfafferott, P. J. Goulder, M. O. McClure, J. Weber, R. E. Phillips, and S. Fidler. 2006. Passive sexual transmission of human immunodeficiency virus type 1 variants and adaptation in new hosts. J. Virol. 807226-7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gandhi, R. T., A. Wurcel, E. S. Rosenberg, M. N. Johnston, N. Hellmann, M. Bates, M. S. Hirsch, and B. D. Walker. 2003. Progressive reversion of human immunodeficiency virus type 1 resistance mutations in vivo after transmission of a multiply drug-resistant virus. Clin. Infect. Dis. 371693-1698. [DOI] [PubMed] [Google Scholar]
- 21.Gao, X., A. Bashirova, A. K. Iversen, J. Phair, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, M. Altfeld, S. J. O'Brien, and M. Carrington. 2005. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nat. Med. 111290-1292. [DOI] [PubMed] [Google Scholar]
- 22.Gao, X., G. W. Nelson, P. Karacki, M. P. Martin, J. Phair, R. Kaslow, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, S. J. O'Brien, and M. Carrington. 2001. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 3441668-1675. [DOI] [PubMed] [Google Scholar]
- 23.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3212-217. [DOI] [PubMed] [Google Scholar]
- 24.Hay, C. M., D. J. Ruhl, N. O. Basgoz, C. C. Wilson, J. M. Billingsley, M. P. DePasquale, R. T. D'Aquila, S. M. Wolinsky, J. M. Crawford, D. C. Montefiori, and B. D. Walker. 1999. Lack of viral escape and defective in vivo activation of human immunodeficiency virus type 1-specific cytotoxic T lymphocytes in rapidly progressive infection. J. Virol. 735509-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keele, B. F., E. E. Giorgi, J. F. Salazar-Gonzalez, J. M. Decker, K. T. Pham, M. G. Salazar, C. Sun, T. Grayson, S. Wang, H. Li, X. Wei, C. Jiang, J. L. Kirchherr, F. Gao, J. A. Anderson, L. H. Ping, R. Swanstrom, G. D. Tomaras, W. A. Blattner, P. A. Goepfert, J. M. Kilby, M. S. Saag, E. L. Delwart, M. P. Busch, M. S. Cohen, D. C. Montefiori, B. F. Haynes, B. Gaschen, G. S. Athreya, H. Y. Lee, N. Wood, C. Seoighe, A. S. Perelson, T. Bhattacharya, B. T. Korber, B. H. Hahn, and G. M. Shaw. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. USA 1057552-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiepiela, P., A. J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, P. Rathnavalu, C. B. Moore, K. J. Pfafferott, L. Hilton, P. Zimbwa, S. Moore, T. Allen, C. Brander, M. M. Addo, M. Altfeld, I. R. James, S. A. Mallal, M. Bunce, L. D. Barber, J. Szinger, C. Day, P. Klenerman, J. Mullins, B. T. Korber, H. M. Coovadia, B. D. Walker, and P. J. Goulder. 2004. Dominant influence of HLA-B In mediating the potential co-evolution of HIV and HLA. Nature 432769-774. [DOI] [PubMed] [Google Scholar]
- 28.Kiepiela, P., K. Ngumbela, C. Thobakgale, D. Ramduth, I. Honeyborne, E. Moodley, S. Reddy, C. de Pierres, Z. Mncube, N. Mkhwanazi, K. Bishop, M. van der Stok, K. Nair, N. Khan, H. Crawford, R. Payne, A. Leslie, J. Prado, A. Prendergast, J. Frater, N. McCarthy, C. Brander, G. H. Learn, D. Nickle, C. Rousseau, H. Coovadia, J. I. Mullins, D. Heckerman, B. D. Walker, and P. Goulder. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 1346-53. [DOI] [PubMed] [Google Scholar]
- 29.Klenerman, P., Y. Wu, and R. Phillips. 2002. HIV: current opinion in escapology. Curr. Opin. Microbiol. 5408-413. [DOI] [PubMed] [Google Scholar]
- 30.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 684650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leslie, A., D. Kavanagh, I. Honeyborne, K. Pfafferott, C. Edwards, T. Pillay, L. Hilton, C. Thobakgale, D. Ramduth, R. Draenert, S. Le Gall, G. Luzzi, A. Edwards, C. Brander, A. K. Sewell, S. Moore, J. Mullins, C. Moore, S. Mallal, N. Bhardwaj, K. Yusim, R. Phillips, P. Klenerman, B. Korber, P. Kiepiela, B. Walker, and P. Goulder. 2005. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J. Exp. Med. 201891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, B., A. D. Gladden, M. Altfeld, J. M. Kaldor, D. A. Cooper, A. D. Kelleher, and T. M. Allen. 2007. Rapid reversion of sequence polymorphisms dominates early human immunodeficiency virus type 1 evolution. J. Virol. 81193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Picado, J., J. G. Prado, E. E. Fry, K. Pfafferott, A. Leslie, S. Chetty, C. Thobakgale, I. Honeyborne, H. Crawford, P. Matthews, T. Pillay, C. Rousseau, J. I. Mullins, C. Brander, B. D. Walker, D. I. Stuart, P. Kiepiela, and P. Goulder. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 803617-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthews, P. C., A. Prendergast, A. Leslie, H. Crawford, R. Payne, C. Rousseau, M. Rolland, I. Honeyborne, J. Carlson, C. Kadie, C. Brander, K. Bishop, N. Mlotshwa, J. I. Mullins, H. Coovadia, T. Ndung'u, B. D. Walker, D. Heckerman, and P. J. Goulder. 2008. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J. Virol. 828548-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menendez-Arias, L., A. Mas, and E. Domingo. 1998. Cytotoxic T-lymphocyte responses to HIV-1 reverse transcriptase (review). Viral Immunol. 11167-181. [DOI] [PubMed] [Google Scholar]
- 36.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 2961439-1443. [DOI] [PubMed] [Google Scholar]
- 37.O'Brien, S. J., X. Gao, and M. Carrington. 2001. HLA and AIDS: a cautionary tale. Trends. Mol. Med. 7379-381. [DOI] [PubMed] [Google Scholar]
- 38.Oxenius, A., D. A. Price, H. F. Gunthard, S. J. Dawson, C. Fagard, L. Perrin, M. Fischer, R. Weber, M. Plana, F. Garcia, B. Hirschel, A. McLean, and R. E. Phillips. 2002. Stimulation of HIV-specific cellular immunity by structured treatment interruption fails to enhance viral control in chronic HIV infection. Proc. Natl. Acad. Sci. USA 9913747-13752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plana, M., F. Garcia, A. Oxenius, G. M. Ortiz, A. Lopez, A. Cruceta, G. Mestre, E. Fumero, C. Fagard, M. A. Sambeat, F. Segura, J. M. Miro, M. Arnedo, L. Lopalcos, T. Pumarola, B. Hirschel, R. E. Phillips, D. F. Nixon, T. Gallart, and J. M. Gatell. 2004. Relevance of HIV-1-specific CD4+ helper T-cell responses during structured treatment interruptions in patients with CD4+ T-cell nadir above 400/mm3. J. Acquir. Immune Defic. Syndr. 36791-799. [DOI] [PubMed] [Google Scholar]
- 40.Rousseau, C. M., M. G. Daniels, J. M. Carlson, C. Kadie, H. Crawford, A. Prendergast, P. Matthews, R. Payne, M. Rolland, D. N. Raugi, B. S. Maust, G. H. Learn, D. C. Nickle, H. Coovadia, T. Ndung'u, N. Frahm, C. Brander, B. D. Walker, P. J. Goulder, T. Bhattacharya, D. E. Heckerman, B. T. Korber, and J. I. Mullins. 2008. HLA class I-driven evolution of human immunodeficiency virus type 1 subtype c proteome: immune escape and viral load. J. Virol. 826434-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scherer, A., J. Frater, A. Oxenius, J. Agudelo, D. A. Price, H. F. Gunthard, M. Barnardo, L. Perrin, B. Hirschel, R. E. Phillips, and A. R. McLean. 2004. Quantifiable cytotoxic T lymphocyte responses and HLA-related risk of progression to AIDS. Proc. Natl. Acad. Sci. USA 10112266-12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283857-860. [DOI] [PubMed] [Google Scholar]
- 43.Tomiyama, H., K. Miwa, H. Shiga, Y. I. Moore, S. Oka, A. Iwamoto, Y. Kaneko, and M. Takiguchi. 1997. Evidence of presentation of multiple HIV-1 cytotoxic T lymphocyte epitopes by HLA-B*3501 molecules that are associated with the accelerated progression of AIDS. J. Immunol. 1585026-5034. [PubMed] [Google Scholar]
- 44.Tomiyama, H., T. Sakaguchi, K. Miwa, S. Oka, A. Iwamoto, Y. Kaneko, and M. Takiguchi. 1999. Identification of multiple HIV-1 CTL epitopes presented by HLA-B*5101 molecules. Hum. Immunol. 60177-186. [DOI] [PubMed] [Google Scholar]
- 45.Turnbull, E. L., A. R. Lopes, N. A. Jones, D. Cornforth, P. Newton, D. Aldam, P. Pellegrino, J. Turner, I. Williams, C. M. Wilson, P. A. Goepfert, M. K. Maini, and P. Borrow. 2006. HIV-1 epitope-specific CD8+ T cell responses strongly associated with delayed disease progression cross-recognize epitope variants efficiently. J. Immunol. 1766130-6146. [DOI] [PubMed] [Google Scholar]
- 46.Weiss, R. A. 2008. Special anniversary review: twenty-five years of human immunodeficiency virus research: successes and challenges. Clin. Exp. Immunol. 152201-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson, J. D., G. S. Ogg, R. L. Allen, C. Davis, S. Shaunak, J. Downie, W. Dyer, C. Workman, S. Sullivan, A. J. McMichael, and S. L. Rowland-Jones. 2000. Direct visualization of HIV-1-specific cytotoxic T lymphocytes during primary infection. AIDS 14225-233. [DOI] [PubMed] [Google Scholar]