Abstract
The mitochondrial antiviral signaling (MAVS) protein plays a central role in innate antiviral immunity. Upon recognition of a virus, intracellular receptors of the RIG-I-like helicase family interact with MAVS to trigger a signaling cascade. In this study, we investigate the requirement of the MAVS structure for enabling its signaling by structure-function analyses and resonance energy transfer approaches in live cells. We now report the essential role of the MAVS oligomer in signal transduction and map the transmembrane domain as the main determinant of dimerization. A combination of mutagenesis and computational methods identified a cluster of residues making favorable van der Waals interactions at the MAVS dimer interface. We also correlated the activation of IRF3 and NF-κB with MAVS oligomerization rather than its mitochondrial localization. Finally, we demonstrated that MAVS oligomerization is disrupted upon expression of HCV NS3/4A protease, suggesting a mechanism for the loss of antiviral signaling. Altogether, our data suggest that the MAVS oligomer is essential in the formation of a multiprotein membrane-associated signaling complex and enables downstream activation of IRF3 and NF-κB in antiviral innate immunity.
Viral nucleic acids, potent inducers of the antiviral innate immune response, are recognized at the extracellular level by a subset of endosomal Toll-like receptors and, upon permissive virus infection, at the cytoplasmic level by a family of DexD/H box RNA helicases including RIG-I (retinoic acid inducible gene I) and MDA5 (melanoma differentiation-associated gene 5) (26).
The sensor protein RIG-I is believed to be maintained in an auto-inhibited state in resting cells and to undergo a conformational change upon viral RNA binding. This conformational change exposes two N-terminal caspase activation and recruitment domains (CARDs) (36), induces cytoplasmic oligomerization (31), and promotes interaction with mitochondrial antiviral signaling (MAVS) protein (also known as IPS-1, Cardif, and VISA). The interaction between RIG-I and MAVS occurs through the protein-interacting CARDs of both proteins (17, 25, 32, 34) and initiates formation of an as-yet-undefined macromolecular signaling complex to the mitochondrial membrane. Formation of this complex involves the recruitment of multiple signaling components to activate interferon (IFN) regulatory factor 3 (IRF3) and nuclear factor-κB (NF-κB) transcription factors that are required for production of type-I IFNs (reviewed in reference 26). The RIG-I/MAVS pathway plays an important role in the antiviral host response to hepatitis C virus (HCV) infection, where the uncapped viral RNA of HCV triggers RIG-I signaling (33). However, HCV can counteract the antiviral response; its NS3/4A serine protease cleaves MAVS at cysteine 508, resulting in the loss of mitochondrial localization and the abrogation of signaling function (9, 19, 20, 25). This mechanism has been confirmed to occur in infected cells (22).
Here, we applied fluorescence and bioluminescence resonance energy transfer (FRET and BRET, respectively) to structure-function analysis of MAVS-mediated antiviral signaling in live cells. We demonstrated oligomerization of MAVS and identified the transmembrane (TM) domain as the main determinant of self-interaction. Site-directed mutagenesis identified four key residues for MAVS oligomerization, and computer modeling of the MAVS TM domain putative dimer supports an interacting surface, locating all of these four amino acids on the same side of the MAVS TM α-helix. Functional studies demonstrated that MAVS oligomerization, rather than localization at the mitochondria, best correlates with the activation of downstream effectors. Finally, we demonstrated that HCV NS3/4A protease-mediated cleavage of MAVS impaired oligomerization, suggesting a mechanism for the abrogation of antiviral signaling. Our results provide evidence of a crucial role for MAVS oligomerization in the formation of a multiprotein signaling complex that controls the antiviral innate immune response.
MATERIALS AND METHODS
Expression vectors.
A pcDNA3.1_MCS(MB) eukaryotic expression plasmid was generated by replacing the multicloning site of pcDNA3.1/Hygro(+) (Invitrogen) with an AflII-AgeI-BamHI-Pfl23II-NotI-BamHI-ClaI-XbaI multicloning site. Sequences of enhanced yellow fluorescent protein (eYFP), green fluorescent protein 2 (GFP2), and Renilla luciferase (Rluc) were amplified, without a stop codon, by PCR and inserted between the AflII and AgeI restriction sites, generating reporter plasmids pcDNA3.1_eYFP-MCS(MB), pcDNA3.1_GFP2-MCS(MB), and pcDNA3.1_Rluc-MCS(MB), respectively.
All MAVS, RIG-I, TOM22, and NS3/4A mutants were generated by PCR and inserted between the Pfl23II and NotI restriction sites of each pcDNA3.1_MCS(MB) reporter plasmid. The result is fusion proteins that are N-terminally linked to reporter genes by a flexible 6-amino-acid (aa) linker. All constructs were verified by nucleotide sequencing. To generate MAVS constructs that were resistant to degradation by short hairpin RNA (shRNA) 148945 (Sigma), the targeted sequence in the mRNA was mutated from CAAGTTGCCAACTAGCTCAAA to CAAACTTCCTACCTCCAGCAA (mutated nucleotides are underlined).
Cell culture and transfection.
293T (human embryonic kidney) and Huh7 (human hepatoma) cell lines were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM glutamine (all from Invitrogen) at 37°C in an atmosphere of 5% CO2. 293T cells were seeded at a density of 1 × 105 cells in 24-well plates (Ultident) for BRET and luciferase assays and at a density of 2 × 106 cells in 100-mm dishes (Sarstedt) for coimmunoprecipitation experiments and lentivirus production. Transient transfections were performed the following day with linear 25-kDa polyethylenimine (Polysciences, Inc.) at a ratio of 3 μg polyethylenimine to 1 μg DNA.
For fluorescence microscopy and FRET experiments, Huh7 cells were seeded at a density of 2 × 105 cells in 35-mm glass bottom dishes (thickness 0; MatTek). Transient transfections were performed the following day by using Lipofectamine 2000 (Invitrogen), according to the manufacturer's protocol.
Coimmunoprecipitation and Western blotting. (i) eYFP fusion proteins and endogenous MAVS.
293T cells were transfected with 5 μg of eYFP-MAVS or eYFP-TOM22. At 48 hours posttransfection, cells were washed with ice-cold phosphate-buffered saline (PBS), lysed in 1 ml of cold lysis buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 5 mM EDTA, 1% Triton X-100, 40 mM β-glycerophosphate, 50 mM NaF, and 1 mM Na-vanadate, supplemented with protease inhibitors [0.1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, and 1 μM pepstatin A]), and clarified by centrifugation at 13, 000 × g for 20 min at 4°C. Immunoprecipitations were performed using 20 μl of a 50% slurry of protein G-Sepharose 4 fast-flow medium (GE Healthcare) conjugated to 0.2 μg of a polyclonal goat anti-GFP antibody (US Biological), with constant agitation at 4°C for 4 h. Following incubation, immune complexes were washed four times with cold lysis buffer and resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer. Immunoprecipitated proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis, which was performed using a polyclonal goat anti-GFP antibody (US Biological) or a polyclonal rabbit anti-MAVS antibody (Alexis Biochemicals). Horseradish peroxidase-conjugated secondary antibodies were from Bio-Rad. The chemiluminescence reaction was performed using enhanced chemiluminescence reagent (GE Healthcare).
(ii) FLAG and Myc fusion proteins.
293T cells were transfected with 5 μg of FLAG-MAVS, FLAG-MAVSΔCARD, FLAG-MAVSΔ535-540, or FLAG-MAVSΔTM and 5 μg of myc-MAVS, myc-MAVSΔCARD, myc-MAVSΔ535-540, or myc-MAVSΔTM. Immunoprecipitation was performed as described for Rluc and eYFP, but with 10 μl ANTI-FLAG M2 affinity gel (Sigma), and Western blot analysis was performed using a mouse ANTI-FLAG M2 monoclonal antibody (1:1,000; Sigma) or a rabbit polyclonal c-Myc (A-14) antibody (1:200; Santa Cruz Biotechnology).
BRET assay.
293T cells were transfected in 24-well plates with 5 ng/well of the DNA construct coding for the BRET donor (Rluc-x) and increasing amounts (0 to 200 ng/well) of the DNA construct coding for the BRET acceptor (eYFP-x), and total DNA was completed to 1 μg using salmon sperm DNA (Invitrogen). At 48 hours posttransfection, cells were harvested in Dulbecco's modified Eagle's medium and washed twice with PBS. For each sample, two aliquots of 1 × 105 cells were distributed to wells of a 96-well microplate (Costar 3912; Corning). Total eYFP expression was measured using a Flex Station II (Molecular Devices), with an excitation wavelength at 485 nm, an emission filter at 535 nm, and a cutoff of 530 nm. The luciferase substrate coelenterazine H (Lucigen Corporation) was then added to give a final concentration of 5 μM, and emissions of luminescence and fluorescence were measured simultaneously using a Mithras LB940 (Berthold Technologies). Filters were set to 485 ± 10 nm for luciferase emission and 530 ± 12.5 nm for eYFP emission. BRET ratios were calculated as described by Angers et al. (2).
Fluorescence microscopy and FRET.
At 48 hours posttransfection, Huh7 cells were infected with Sendai virus for 4 h before being examined under a LSM 510 laser scanning confocal microscope (Zeiss). To label mitochondria, cells were incubated for 30 min at 37°C with cell-permeable MitoTracker deep red (Molecular Probes) to give a final concentration of 200 nM. Cells stained with Mitotracker deep red were scanned at a 633-nm excitation wavelength (HeNe laser at 50% intensity), with a 650-nm long-pass filter. Nuclei were labeled with Hoechst 33342 (Invitrogen) at a final concentration of 1 μg/ml, and cells were scanned at a 405-nm excitation wavelength (diode laser at 10% intensity), with a 420- to 480-nm band pass filter. GFP2 and eYFP scans used 405-nm and 488-nm excitation wavelengths (argon laser at 10% intensity), with 470- to 500-nm band pass and 560-nm long-pass filters, respectively. FRET scans used a 405-nm excitation wavelength with a 530- to 560-nm band pass filter. eYFP was photobleached with 200 iterations of the 488-nm argon laser at 80% intensity to produce ∼90% bleaching. Images (1,024 by 1,024 pixels) were acquired in multitrack mode to minimize cross talk between fluorophores, processed using Northern Eclipse software, and optimized for contrast using Adobe Photoshop.
Luciferase assay.
293T cells were transfected with 100 ng of an IFN-β promoter (−280/+20 IFN-β promoter) (21), an NF-κB-dependent promoter (human immunodeficiency virus type 1 long terminal repeats) (4), or an IRF3-dependent promoter (pGL3_ISG56) (10) driving firefly luciferase expression; 1 ng of an Rluc-expressing plasmid (or 100 ng of β-galactosidase expression plasmids, when Rluc fusion proteins were used), used to normalize transfection efficiency; and 100 ng (see Fig. 1, 5, and 7) or 0.1 ng (see Fig. 8) of a protein expression plasmid. Cells were harvested 48 h later, washed twice with PBS, distributed in two wells of a 96-well microplate (Costar 3912; Corning), and lysed, and luciferase activities were determined by a dual-luciferase reporter assay system (Promega).
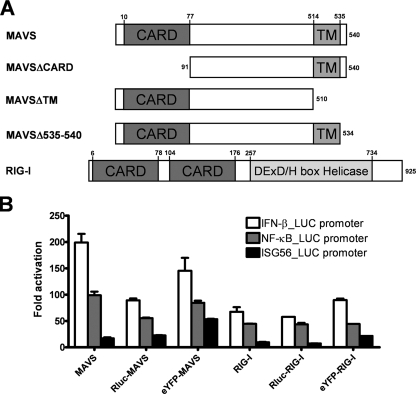
FIG. 1.
Schematic presentation of Rluc and eYFP constructs and their activation of the IFN-β promoter and the NF-κB- and IRF3-dependent promoters. (A) Domain structure of the MAVS and RIG-I constructs showing the position of the CARD and the TM and DexD/H box helicase domains. (B) Expression vectors for MAVS and RIG-I, either alone or in fusion with Rluc or eYFP, were cotransfected with an IFN-β-, NF-κB-, or ISG56-firefly luciferase reporter in 293T cells. Results are expressed as activation levels of the promoter compared to those in cells transfected with an empty expression vector. The error bars represent the standard deviations from the mean values obtained from three independent experiments performed in duplicate.
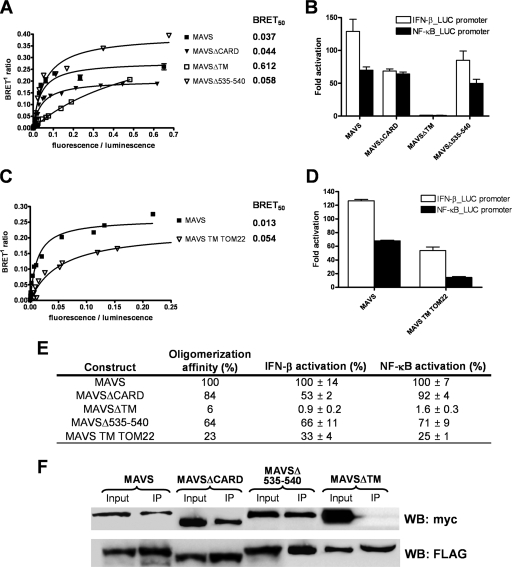
FIG. 5.
The ability of MAVS to form oligomers correlates with its activation of IFN-β and NF-κB. (A) BRET saturation assays were performed as described for Fig. 4A. BRET50, which is a reflection of the relative affinity of the acceptor fusion for the donor fusion, is indicated to the right of the curve for every deletion mutant of MAVS. The curves shown represent the means ± standard deviations of results from one representative experiment carried out in duplicate. (B) Expression vectors for MAVS deletion mutants in fusion with the Rluc or eYFP were cotransfected with an IFN-β- or NF-κB-firefly luciferase reporter in 293T cells. The Rluc or eYFP activity of the fusion protein was used to normalize firefly luciferase activity. Results are expressed as average activation levels of the promoter in cells transfected with Rluc- or eYFP fusion proteins, compared to those in cells transfected with an empty expression vector. The error bars represent the standard deviations from the mean values obtained from three independent experiments performed in duplicate. (C) BRET saturation assays were performed as described for panel A. (D) The activations of the IFN-β promoter and the NF-κB-dependent promoter were determined as described for panel B. (E) Summary table of the relative oligomerization affinities and activations of IFN-β and NF-κB promoters for each MAVS mutant, compared to the levels for wild-type MAVS. (F) MAVS, MAVSΔCARD, MAVSΔ535-540, or MAVSΔTM tagged in N-terminal with FLAG or myc epitopes were cotransfected in 293T cells. After 48 h, cell lysates were immunoprecipitated (IP) with anti-FLAG antibodies, followed by immunoblot analysis (WB) with anti-FLAG (lower panel) and anti-myc (upper panel) antibodies.
FIG. 7.
Mutagenesis analysis and theoretical three-dimensional model of the monomer and dimer forms of MAVS TM. (A) Sequence of the MAVS TM domain (boxed). Asterisks indicate positions that were mutated to alanine (W517A and Y535A) or tryptophan (Q519W, V520W, A521W, V522W, T523W, G524W, V525W, L526W, V527W, and V528W). (B) Summary table of the relative oligomerization affinities and activations of IFN-β and NF-κB promoters for each MAVS TM mutant, compared to the levels for wild-type MAVS. Affinities were calculated with the BRET50 value obtained from BRET saturation assays for all TM mutants. Important results are colored magenta. The error bars represent the standard deviations from the mean values obtained from three independent experiments performed in duplicate. (C) Similarity of the MAVS TM sequence to the TM α-helix of protein J of the photosystem II reaction center (Protein Data Bank entry 1S5L) (8). The sequences display 25% identity and 85% similarity, with a 20-aa overlap (aa 516 to 535 and 2010 to 2029, respectively). “Structure” represents the secondary structure of the protein J TM segment, deduced from its three-dimensional structure: h, helix; s, bend; t, hydrogen-bonded turn. #, prediction of MAVS TM segment, deduced from various prediction methods. (D) Molecular model of MAVS TM. A ribbon representation including residue side chains and an amino acid van der Waals surface representation are shown on the left and the right, respectively. Residues identified by mutagenesis analysis as essential for MAVS dimer formation are located on one side of the TM α-helix (W517, A521, G524, and V528; colored magenta). Other hydrophobic residues are colored in gray, except Tyr, which is colored in dark yellow. Hydrophilic residues Thr and Gln are colored in green, and Arg is colored in blue. (E) Representative dimer model of MAVS TM. Ribbon representations including the side chains of interfacial residues (W517, V520, A521, G524, V528, and L531) as sticks and the van der Waals surface are shown on the left and the right, respectively. The orientation of the left structure highlights the crossing angle (31°) of the symmetric dimer, while the right structure is rotated 90° to highlight the contacts between interfacial residues. The dimer model was manually positioned within a phospholipid bilayer membrane represented as a simulated model of a 1-palmitoyl-2-oleoyl-3-sn-glycero-3-phospholcholine (POPC) bilayer (obtained from Peter Tieleman; http://moose.bio.ucalgary.ca/). Polar heads and hydrophobic tails of phospholipids (surface structures) are light yellow and gray, respectively. Figures were generated from structure coordinates by using VMD (15) (http://www.ks.uiuc.edu/Research/vmd/) and rendered with POV-Ray (http://www.povray.org/).
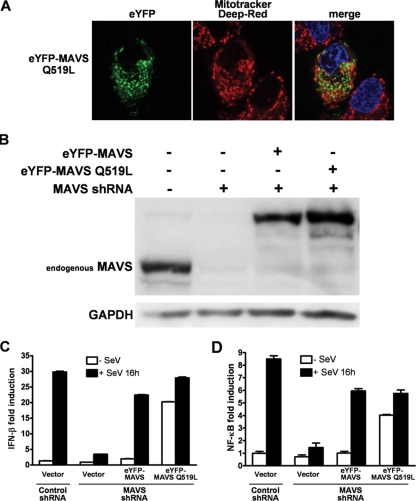
FIG. 8.
Mitochondrial localization is required for negative regulation of MAVS. (A) Fluorescence confocal microscopy was used to visualize an optical cross-section of live Huh7 cells. Forty-eight hours after transfection with eYFP-MAVS Q519L (green), mitochondria were stained with Mitotracker deep red (red) and nuclei were labeled with Hoechst 33342 (blue). (B) Cell lysates used for panels C and D were immunoblotted with anti-MAVS and anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (loading control) antibodies. (C) 293T cells stably expressing nontarget or MAVS shRNAs were cotransfected with an IFN-β-firefly luciferase reporter and either an empty vector or a vector encoding eYFP-MAVS or eYFP-MAVS Q519L. Sixteen hours after infection with Sendai virus, IFN-β-firefly luciferase reporter activity was measured as described for Fig. 5B. (D) Experiments were performed as described for panel C, but with an NF-κB-firefly luciferase reporter.
Lentiviral vector production and transduction.
293T cells were transfected with 6 μg pLKO.1-puro carrying shRNA 148945 or nontarget shRNA (Sigma), 1.5 μg pMDLg/pRRE, 1.5 μg pRSV-REV, and 3 μg pVSVg (as described in reference 7). On the next day, medium was replaced with fresh medium. On the third day, medium containing lentiviral vector was harvested and cellular debris were removed by filtration before being transferred on 293T cells for 6 h of infection with 4 μg/ml polybrene (Sigma). On the fourth day, 1 μg/ml puromycin (Sigma) was added to infected 293T cells, and these stable cell lines were used 72 h later.
Sendai virus infection.
Cells were infected with 100 hemagglutinating units/ml of Sendai virus (Cantell strain, from Charles River Laboratories) for 4 h before coimmunoprecipitation or confocal microscopy experiments and for 16 h before measurement of activation of the IFN-β promoter and the NF-κB-dependent promoter when required.
Sequence analyses and molecular modeling.
Various methods were combined for the prediction of the MAVS TM domain, as follows: PHDhtm (http://www.predictprotein.org/), TMHMM (http://www.cbs.dtu.dk/services/TMHMM), DAS (http://www.sbc.su.se/∼miklos/DAS/), TopPred2 (http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py?form=toppred), Tmpred (http://www.ch.embnet.org/software/TMPRED_form.html), HMMTOP (http://www.enzim.hu/hmmtop/), and SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/). Sequences similar to that of the MAVS TM segment were searched for in the Protein Data Bank of three-dimensional structures by using the SSEARCH program in the IBCP website facilities (http://npsa-pbil.ibcp.fr/). The molecular model of the MAVS TM domain (aa 514 to 536) was constructed by using the α-helix TM segment comprising aa 2008 to 2030 of the J protein of the photosystem II reaction center as a template and the Swiss-PdbViewer program (12) (http://spdbv.vital-it.ch/). The model of the MAVS TM domain dimer was calculated using the protein-protein docking program HADDOCK (high-ambiguity driven protein-protein docking) (6) (http://www.nmr.chem.uu.nl/haddock/) with the standard protocol. The docking was performed with the assumption that residues W517, A521, G525, and V528, observed to be essentials for MAVS dimer formation, are located at the interface of each monomer, and a C2 symmetry was set. A set of 50 models was generated, and the symmetric, lowest-energy structures compatible with a TM insertion (i.e., with a crossing angle between helix monomers lower than 60°) were analyzed for the numbers and types of contacts of interfacial residues between monomers as well as for the molecular surface of the interface. The molecular surface was calculated using the Swiss-PdbViewer program. The crossing angle between the two-helix axis was performed with MolMol (http://www.mol.biol.ethz.ch/groups/wuthrich_group/software).
RESULTS
Tagged and nontagged MAVS proteins behave similarly in live-cell cultures.
To investigate the biochemical mechanism of MAVS, we created a panel of proteins (Fig. 1A) tagged with N-terminal eYFP, GFP2, or Rluc for use in protein-protein interaction studies by FRET and BRET in live cells. All fusion proteins were assessed for signaling activity in 293T cells by cotransfection alongside firefly luciferase controlled by the promoter region of IFN-β or the NF-κB- and IRF3-dependent promoters (4, 10, 21). With the exception of Rluc-MAVS, all N-terminally tagged MAVS and RIG-I proteins induced the activation of the IFN-β promoter and the NF-κB- and IRF3-dependent promoters to levels that were comparable to levels obtained with the nontagged protein (Fig. 1B). The twofold reductions in IFN-β and NF-κB induction observed with Rluc-MAVS were attributed to the expression level being lower than that of the MAVS protein (data not shown). It is noteworthy that overexpression of MAVS and RIG-I without stimulation was sufficient to induce strong promoter activation and that addition of Sendai virus increased this activation by only about 20% (data not shown).
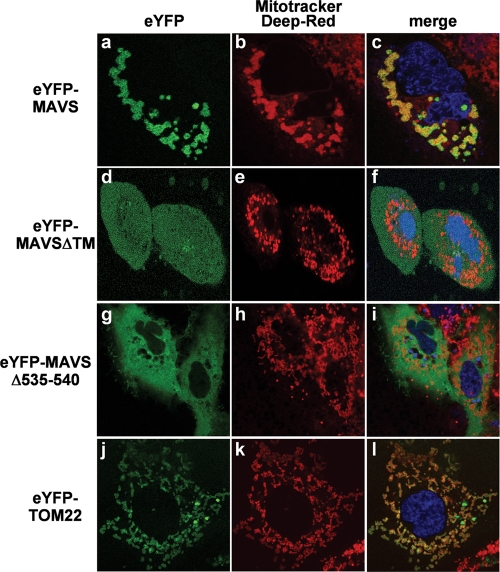
Subcellular localization of MAVS was assessed by confocal microscopy with live Huh7 cells expressing eYFP fusion proteins. Prior to visualization, mitochondria and nuclei were labeled with Mitotracker deep red and Hoechst, respectively. Figure 2 shows eYFP-MAVS localized to the mitochondrial membrane (panels a to c), in contrast to the TM domain-deleted MAVS mutant (eYFP-MAVSΔTM) that localized within the cytoplasm (panels d to f), as previously reported for fixed cells (13, 20, 32). Interestingly, the deletion of the last six residues of MAVS (YRRRLH), predicted to serve as the mitochondrial targeting sequence, led to a predominant staining of all intracellular membranes as well as the cytosol (eYFP-MAVSΔ535-540) (Fig. 2g to i), with minimal colocalization with Mitotracker deep red. As a control protein, we used eYFP-TOM22, an import receptor of translocase located at the outer mitochondrial membrane (35) that colocalized with the Mitotracker deep red staining (Fig. 2j to l). Altogether, these data demonstrated that the tagged proteins behave similarly to the nontagged MAVS protein and confirmed their value in structure-function studies of MAVS protein interactions.
FIG. 2.
Live-cell confocal fluorescence imaging of eYFP fusion proteins and mitochondria. Fluorescence confocal microscopy was used to visualize an optical cross-section of live Huh7 cells. Forty-eight hours after transfection with various eYFP fusion proteins (green), mitochondria were stained with Mitotracker deep red (red) and nuclei were labeled with Hoechst 33342 (blue). Yellow labeling in the merge image indicates colocalization of eYFP fusion constructs with mitochondria.
MAVS interaction with RIG-I and MAVS oligomerization are visible with FRET in live cells.
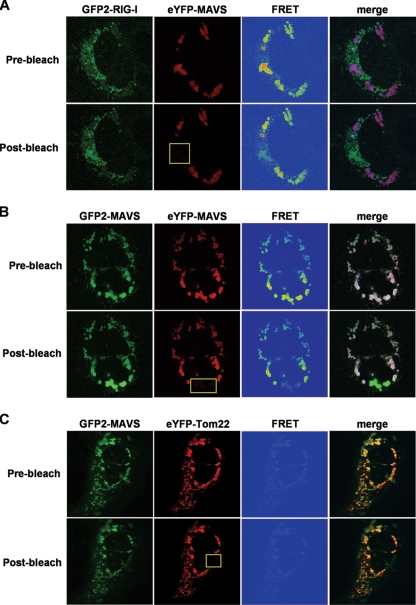
To investigate the protein interaction between MAVS and RIG-I and MAVS self-interaction, GFP2-RIG-I or GFP2-MAVS fusion proteins were coexpressed with eYFP-MAVS in Huh7 cells, which were then stimulated for an innate antiviral response by infection with Sendai virus. The cells were visualized by confocal microscopy and analyzed for FRET, allowing visualization of steady-state protein-protein interactions in organelles of live cells. Interactions are detected when a nonradiative transfer of energy from the energy donor, GFP2, excites the energy acceptor, eYFP (28). Upon coexpression of GFP2-RIG-I and eYFP-MAVS, we observed no apparent colocalization of these two proteins (Fig. 3A, prebleach). However, a FRET signal indicative of a direct interaction between RIG-I and MAVS was detected. To confirm the specificity of the FRET signal, we performed an acceptor photobleaching experiment, where an increase in donor fluorescence is expected following the release of its quenching (3). Here, photobleaching of eYFP-MAVS decreased both eYFP-MAVS fluorescence and FRET signal by more than 90%, while increasing GFP2-RIG-I fluorescence (Fig. 3A, postbleach). These FRET experiments demonstrate the direct interaction of a GFP2-RIG-I cellular subset with eYFP-MAVS that localizes at the mitochondrial membrane in live Huh7 cells (Fig. 2a to c).
FIG. 3.
RIG-I-MAVS and MAVS-MAVS interactions visualized by FRET and acceptor photobleaching. Four hours after infection with Sendai virus, fluorescence confocal microscopy was used to visualize an optical cross-section of live Huh7 cells coexpressing either GFP2-RIG-I (donor, green) and eYFP-MAVS (acceptor, red) (A), GFP2-MAVS and eYFP-MAVS (B), or GFP2-MAVS and eYFP-TOM22 (C). FRET images are represented with pixel-by-pixel FRET intensities, using a color code (from blue to red). Acceptor photobleaching was performed at 488 nm (yellow rectangle).
Upon cotransfection of GFP2-MAVS and eYFP-MAVS fusion constructs, we observed colocalization of both proteins to the mitochondrial membrane and detected a strong FRET signal (Fig. 3B, prebleach). Acceptor photobleaching of eYFP-MAVS decreased eYFP-MAVS fluorescence and FRET signal by more than 90%, while leading to a significant increase of GFP2-MAVS fluorescence, confirming the energy transfer between the two MAVS fusion proteins upon interaction (Fig. 3B, postbleach).
To ensure that colocalization of the two fusion proteins to the mitochondria is not sufficient for detection of a FRET signal, we expressed TOM22 in fusion to eYFP (eYFP-TOM22). TOM22 is a component of a translocase located at the outer mitochondrial membrane (35) (Fig. 2j to l). MAVS localization to the outer mitochondrial membrane has already been demonstrated by subcellular fractionation experiments (20, 32). Upon coexpression of GFP2-MAVS and eYFP-TOM22 fusion constructs in Huh7 cells, we observed a colocalization of the two proteins to the mitochondrial membrane; however, no FRET signal was detected (Fig. 3C, prebleach). Photobleaching of the eYFP-TOM22 acceptor protein did not increase GFP2-MAVS fluorescence but rather decreased it due to partial bleaching of GFP2 upon exposure to high laser intensity at 488 nm (Fig. 3C, postbleach). These experiments demonstrate the absence of a direct interaction between MAVS and TOM22, regardless of the fact that both are located at the outer mitochondrial membrane.
Overall, our data provide strong evidence of a direct interaction between MAVS and a subset of RIG-I as well as MAVS oligomerization at the mitochondrial membrane in live Huh7 cells following Sendai virus infection.
MAVS oligomerization is confirmed by BRET saturation assays and semiendogenous coimmunoprecipitation.
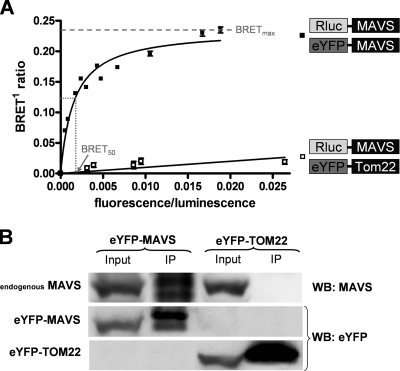
MAVS oligomerization, earlier visualized by FRET images of the mitochondrial membrane in live Huh7 cells, was further investigated by BRET analyses and coimmunoprecipitation. BRET is detected when energy emitted by Rluc of a donor fusion protein is transferred to an acceptor eYFP fusion partner following catalytic degradation of the cell-permeable Rluc substrate coelenterazine H. Figure 4A shows a nonlinear increase in BRET intensity and rapid saturation of signal occurring when a fixed amount of the Rluc-MAVS construct is cotransfected with increasing quantities of the eYFP-MAVS construct. This characteristic pattern for specific interaction confirms MAVS oligomerization. The BRET signal at which the saturation curve plateaus (BRETmax) is a function of the distance and orientation between the donor and acceptor within oligomeric conformation, whereas the concentration of acceptor giving 50% of BRETmax (BRET50) is a reflection of the relative affinity between fusion proteins (24). As a negative control, we again used eYFP-TOM22 (Fig. 4A). Increasing concentrations of eYFP-TOM22 led to a linear and weak increase of BRET intensity, a characteristic pattern for random collisions between two molecules.
FIG. 4.
Oligomerization of MAVS identified by BRET and semiendogenous coimmunoprecipitation experiments. (A) BRET experiments with live 293T cells. Rluc-MAVS (BRET donor) was cotransfected with eYFP-MAVS (BRET acceptor) or eYFP-TOM22 (nonspecific control). Forty-eight hours later, energy transfer was initiated by the addition of the cell-permeable Rluc substrate coelenterazine h. Donor saturation curves were obtained by measuring the BRET in the presence of a fixed quantity of donor and increasing amounts of acceptor. The x axis shows the ratio between the fluorescence of the acceptor (YFP-YFP0, where YFP0 is the fluorescence value in cells expressing the BRET donor alone) and the luminescence of the donor. BRETmax is the maximal BRET signal reached by the saturation curve, whereas BRET50 is the concentration of acceptors giving 50% of BRETmax. Curves shown represent the means ± standard deviations of results from one representative experiment carried out in duplicate. The curves were fitted using a nonlinear regression equation, in which a single binding site was assumed (GraphPad Prism). (B) eYFP-MAVS or control eYFP-TOM22 constructs were transfected in 293T cells. After 48 h, cells were infected with Sendai virus for 4 h before cell lysates were immunoprecipitated (IP) with anti-GFP antibodies, followed by immunoblot analysis (WB) with anti-GFP (lower panels) and anti-MAVS (upper panel) antibodies.
To further corroborate MAVS oligomerization, the interaction of eYFP-MAVS with endogenous MAVS was also investigated under expression conditions used for BRET studies. We performed a semiendogenous coimmunoprecipitation experiment (Fig. 4B). 293T cells were cotransfected with eYFP-MAVS or eYFP-TOM22 (control) for 48 h, and cell lysates were immunoprecipitated with a GFP antibody (cross-reactive with eYFP). The eYFP immunoprecipitate was immunoblotted with a MAVS antibody (upper panel) and a GFP antibody (lower panels). As shown, endogenous MAVS coprecipitated with eYFP-MAVS but not with eYFP-TOM22, demonstrating that eYFP-MAVS proteins are specifically forming oligomers with endogenous MAVS under expression conditions used for BRET studies.
The MAVS TM domain determines its oligomerization.
To identify regions of MAVS mediating its oligomerization, BRET saturation curves were obtained for MAVS mutants lacking their CARD, TM domain, or mitochondrial targeting sequence (MAVSΔCARD, MAVSΔTM, or MAVSΔ535-540, respectively) and compared to curves from full-length MAVS. As shown in Fig. 5A, the BRET intensities for the four fusion proteins have distinctive characteristics (linear and saturation curves), and significant differences between the four mutants in BRETmax and BRET50 were observed. As reflected by the BRET50 values, deletion of the TM domain completely abolished the capacity of MAVSΔTM to oligomerize, while deletion of the CARD or the mitochondrial targeting sequence of MAVS only slightly reduced oligomer affinity. These results were confirmed with coimmunoprecipitation experiments, where FLAG- and myc-tagged MAVS, MAVSΔCARD, MAVSΔ535-540, or MAVSΔTM were cotransfected in 293T cells (Fig. 5F).
Functional studies were also performed with these MAVS deletion mutants; average (n-fold) activations of the IFN-β reporter and the NF-κB-dependent reporter are reported in Fig. 5B. MAVSΔCARD has been identified as a partially inactive form of MAVS (32). However, fusion with eYFP or Rluc at the N-terminal extremity of MAVSΔCARD was sufficient to restore its signaling activity to levels close to those of wild-type MAVS (Fig. 5B). This observation indicates that the MAVS CARD, which mediates the interaction with RIG-I, is dispensable for downstream signaling events. As shown earlier, MAVSΔTM is cytoplasmic and completely inactive, while MAVSΔ535-540, which is also delocalized from the mitochondria, enables activation of the IFN-β reporter and the NF-κB-dependent reporter to levels close to those of wild-type MAVS (Fig. 5B).
Because MAVS activity was previously proposed to be linked to its mitochondrial localization (32) and these results identify the ability of MAVS to form oligomers, mediated by the TM domain, as the main determinant for downstream signaling, we replaced the MAVS TM domain with a TOM22 TM domain (MAVS TM TOM22) to see the impact on oligomerization and signaling. While this TM domain swapping did not affect mitochondrial localization (data not shown), it did reduce oligomerization affinity about fourfold compared to the levels for wild-type MAVS (Fig. 5C), confirming the role of the MAVS TM domain in oligomerization. Interestingly, MAVS TM TOM22 signaling was affected; the activations of both the IFN-β reporter and the NF-κB-dependent reporter were significantly reduced (three- and fourfold, respectively) (Fig. 5D). The residual oligomerization and signaling observed with MAVS TM TOM22 indicate that MAVS ectodomains are playing a role in its oligomerization and/or that the TM domain of TOM22 has the ability to oligomerize. A compilation of affinity and signaling activities for the different mutants is presented in Fig. 5E, which shows a direct correlation between oligomer affinity and downstream signaling activity.
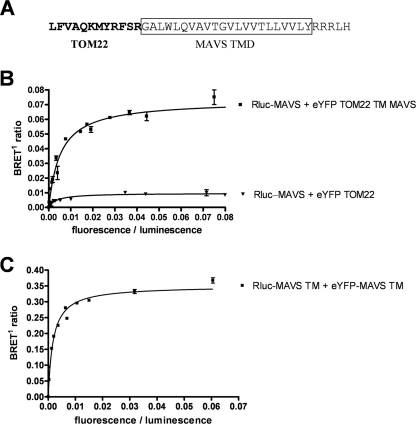
To determine if the MAVS TM domain alone is sufficient to promote oligomerization, we replaced the TOM22 TM domain with the MAVS TM domain, generating TOM22 TM MAVS. As shown in Fig. 4A and reproduced here in Fig. 6B, BRET saturation assays show no interaction between MAVS and TOM22, although they are both located at the external membranes of mitochondria. However, MAVS is able to interact with TOM22 TM MAVS, as seen by an increase in BRETmax and the formation of a saturation curve. Furthermore, we also observed a strong BRET saturation curve by using only the MAVS TM domain (aa 510 to 540) N-terminally tagged with either Rluc or eYFP (Fig. 6C) and confirmed that MAVS TM alone was sufficient to target the eYFP to the mitochondria (data not shown). Taken together, these results identify the TM domain of MAVS as the main determinant in MAVS oligomerization that is required for its downstream signaling function.
FIG. 6.
The MAVS TM domain is sufficient to promote oligomerization. (A) C-terminal sequence of TOM22 TM MAVS. The TOM22 sequence is in bold, and the MAVS TM domain is boxed. (B) BRET saturation assays between Rluc-MAVS and eYFP-TOM22 or eYFP-TOM22 TM MAVS were performed as described for Fig. 4A. (C) BRET saturation assays between Rluc-MAVS TM and eYFP-MAVS TM (aa 510 to 540) were performed as described for Fig. 4A.
Mutagenesis and computational modeling define MAVS TM domain dimerization interface.
To address the contribution of the core residues of the MAVS TM domain in a putative oligomer interface, we generated single-point mutations where each residue between aa 519 and 528 of MAVS was replaced with Trp (Fig. 7A). While the nonpolar side chain of Trp is compatible with an α-helical secondary structure, its size should introduce steric hindrance and disrupt helical interactions. A BRET saturation assay was performed for each mutant, and the relative affinities, calculated from the BRET50, are compiled in Fig. 7B. As expected, most of the single-point mutations had relatively small effects on MAVS oligomer affinity. However, mutations A521W and G524W decreased the affinity between MAVS partners by approximately 40%, and mutation V528W decreased this affinity by 70%. We mutated two aromatic residues to Ala (W517 and Y535) to investigate their potential role in MAVS oligomerization through stacking of aromatic side chains. While the Y535A mutant did not affect the oligomerization affinity of MAVS, mutation W517A decreased this affinity by about 50%, identifying Trp 517 as part of the MAVS oligomerization interface.
Furthermore, we assessed the ability of each of these 12-point mutations in the TM domain of MAVS to activate IFN-β and NF-κB (Fig. 7B). Interestingly, the most-pronounced decreases in activation of the IFN-β reporter and the NF-κB-dependent reporter (about 40%) were observed with G524W and V528W mutants, confirming their involvement in the interaction interface.
The clear prediction of a TM α-helix by all tested methods for the MAVS segment comprising aa 516 to 535, together with the 25% sequence identity and 85% sequence similarity to the TM α-helix of protein J of the photosystem II reaction center (Fig. 7C), supports the three-dimensional molecular homologous model of the MAVS TM domain shown in Fig. 7D. In this model, residues W517, A521, G524, and V528 are located on the same side of the TM α-helix, supporting formation of MAVS dimers (and not of higher-order oligomers). Moreover, the importance of small residues A521 and G524 in the dimer formation strongly suggests their involvement at the dimer interface. Because the combination of computational and mutagenesis methods has been shown to provide accurate models of TM helix interactions (23), the monomer model was used to calculate a MAVS TM domain dimer by using the protein-protein docking program HADDOCK (6), in which it was assumed that residues W517, A521, G524, and V528 of each monomer are involved in the dimer interaction and that the dimer exhibits a C2 symmetry. Figure 7E shows a representative dimer model putatively positioned in a 1-palmitoyl-2-oleoyl-3-sn-glycero-3-phospholcholine (POPC) phospholipid bilayer. This positioning assumes that the basic Arg residues, as well as the aromatic residues Tyr and Trp, are preferentially located at the membrane interface formed by the polar head of phospholipids (11). In this TM domain dimer model, the side chains of V520 and L531 extend the interface cluster formed by residues A521, G524, and V528. In addition, the W517 residues of both monomers participate in the dimer interface via the stacking of their side chains. The dimer interface thus involves the side chains and backbones of six residues, all making favorable van der Waals interactions.
Mitochondrial localization of MAVS is not required for activation of MAVS by RIG-I upon viral infection.
Our mutagenesis analysis of the MAVS TM domain identified a mutation (Q519W) located on the opposite side of the TM α-helix dimer interface which is sufficient to delocalize MAVS from the mitochondria (data not shown) without affecting MAVS oligomerization and downstream signaling (Fig. 7B). The importance of glutamine 519 for targeting MAVS to the mitochondria was confirmed by another mutant (Q519L), which delocalized MAVS from the mitochondria (Fig. 8A).
To assess a potential role for mitochondrial localization of MAVS in activation of MAVS by RIG-I upon viral infection, we used a 293T cell line stably expressing shRNA against MAVS mRNA. Endogenous MAVS expression was decreased by more than 95% (Fig. 8B, compare lanes 1 and 2). Knockdown of MAVS was sufficient to almost completely abrogate activation of the IFN-β promoter and the NF-κB-dependent promoter, activation which is normally observed upon Sendai virus infection (Fig. 8C and D). We then transfected the knockdown cell line with plasmids encoding eYFP-MAVS or eYFP-MAVS Q519L with a mutated mRNA sequence preventing recognition by the shRNA. The expression levels of MAVS fusion proteins were adjusted according to the endogenous MAVS normally present in 293T cells to prevent constitutive activation and to allow RIG-I-dependent activation following Sendai virus infection (Fig. 8B). IFN-β and NF-κB activations following Sendai virus infection were similar to those in wild-type 293T cells when eYFP-MAVS or eYFP-MAVS Q519L was reintroduced in shRNA knockdown cells. Strikingly, reintroduction of eYFP-MAVS Q519L led to a constitutive activation of MAVS, a phenomenon that was not observed with the Q519W mutant (data not shown), suggesting that the addition of a leucine hydrophobic residue could potentially stabilize MAVS oligomerization and translate into its constitutive activity.
Altogether, these results demonstrate that mitochondrial localization of MAVS is not essential for activation of MAVS by RIG-I following Sendai virus infection.
The HCV NS3/4A protease disrupts MAVS oligomers and downstream signaling.
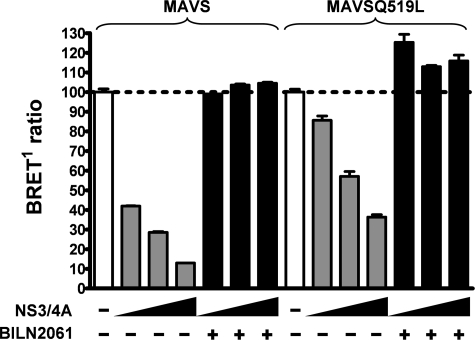
The HCV NS3/4A protease cleaves MAVS at a membrane-proximal site (Cys 508) and blocks the activation of IRF3 and NF-κB to evade antiviral immunity in infected cells (22, 25). We confirmed these observations with live cells and showed that coexpression of the NS3/4A protease alongside our eYFP-MAVS fusion protein resulted in cytoplasmic detection of the eYFP signal (data not shown). The status of MAVS oligomerization was then evaluated by BRET, in the presence of increasing expression of the NS3/4A protease construct (Fig. 9). We observed a dose-dependent reduction of the BRET signal, achieving a plateau in inhibition at 90% with the highest level of NS3/4A expression. This reduction in BRET signal is blocked by a highly specific HCV NS3/4A protease inhibitor (BILN 2061) (18), confirming the restoration of MAVS oligomerization. A less drastic inhibition of MAVS oligomerization by the NS3/4A protease was observed when a MAVS mutant delocalized from the mitochondria (MAVS Q519L) was used (Fig. 8A). This decreased inhibition of MAVS oligomerization likely arises from a reduced NS3/4A cleavage efficiency, as NS3/4A is a membranous protein primarily localized to the mitochondrial membrane (data not shown). Our data underline a putative mechanism for the inactivation of MAVS-dependent IFN-β production by the HCV NS3/4A protease, which could eventually lead to viral persistence.
FIG. 9.
HCV NS3/4A protease activity impairs MAVS oligomerization. 293T cells were cotransfected with Rluc-MAVS or Rluc-MAVS Q519L, eYFP-MAVS, and increasing amounts of expression plasmid encoding HCV NS3/4A protease (50, 100, and 250 ng) in the presence or absence of an HCV protease inhibitor (BILN2061). The BRET1 ratio is expressed in percentage, where 100% is the BRET1 ratio measured in the absence of NS3/4A.
DISCUSSION
In this study, fluorescence microscopy, FRET and BRET analyses, and coimmunoprecipitation experiments all demonstrated MAVS oligomerization in live cells. On the basis of our results and previously published works, we propose a model for the early events of the RIG-I/MAVS pathway, as follows. Upon infection and recognition of viral RNA, RIG-I undergoes a conformational shift promoting its oligomerization (31). RIG-I oligomers may then interact with MAVS through their respective CARDs to dislodge NLRX1 (a recently identified negative regulator which interacts with the MAVS CARD (27) and to induce oligomerization of MAVS dimers. This MAVS complex then triggers recruitment of various signaling components, leading to activation of IRF3 and NF-κB transcriptional factors (reviewed in reference 26). In line with this signaling model, FRET experiments demonstrated the direct interaction of a RIG-I cellular subset with MAVS as well as MAVS oligomerization at the mitochondrial membrane in live Huh7 cells.
Strikingly, this proposed mechanism for the activation of MAVS is reminiscent of the activation of another mitochondrial, CARD-containing protein, caspase-9. In this pathway, the recognition of cytoplasmic cytochrome c by APAF-1 triggers APAF-1 oligomerization and allows its interaction with pro-caspase-9 at the mitochondrial membrane (1, 37). The interaction of APAF-1 oligomers and pro-caspase-9 is mediated by the CARD of each interactor and enables the formation of a pro-caspase-9 dimer, which is then autocleaved, activating the caspase-9 dimer, which then leads to the formation of the apoptosome (29).
Using mutagenesis studies combined with a powerful BRET approach for detection of membrane protein-protein interaction in live cells, we identified the MAVS TM domain as the main determinant of dimerization. Among 12 single-amino-acid mutations of the MAVS TM domain, 4 mutations (W517A, A521W, G524W, and V528W) reduced the affinity between the MAVS partner by 40% to 70%. Importantly, these four residues are on consecutive α-helical turns that define a putative dimerization interface on the same side of the TM α-helix of MAVS (Fig. 7D). Identification of a single helix-helix interaction interface strongly supports formation of MAVS dimers (and not of higher-order oligomers). As illustrated in the proposed MAVS TM domain dimer model (Fig. 7E), short side chains from A521, G524, and V528, together with additional residues (V520 and L531), form a closely packed helix interface between two MAVS proteins, while the aromatic side chain of W517 could play a role in the regulation of this dimerization through conformational changes allowing stacking of its aromatic side chain. The corresponding interaction surface is 380 Å2, and the crossing angle between the two monomer helix axes is 31°. All of these values are comparable to those reported for the glycophorin A TM domain dimers (seven interface residues, 400 Å2 and 40o) (23), indicating that our theoretical dimer model can be used as a useful template for further investigation.
When assessing the ability of these 12 single-amino-acid mutations in the MAVS TM domain to activate IRF3 and NF-κB, we observed decreases in signaling activity for only G524W and V528W, two residues implicated in the dimer interface. However, mutation of the other two residues of this interaction interface (W517A and A521W) did not affect signaling. These discrepancies might reflect the central role of V528 in the formation of MAVS dimers. Thus, mutation of this V528 residue or surrounding residues, like G524, could potentially have a dual effect: altering MAVS oligomerization and inducing a significant conformational change that blocks recruitment of a yet undefined partner required for optimal downstream activation of IRF3 and NF-κB.
Like all tail-anchored proteins in the outer membrane of the mitochondria, MAVS has to be synthesized by cytoplasmic ribosomes before being imported to the mitochondria by a noncleavable targeting sequence (30). This targeting sequence is not a discrete signal but depends mainly on the degree of positive charge flanking the TM domain, a phenomenon extensively studied for Bcl-xL (16). Here, we show that MAVSΔ535-540, where the last six residues of MAVS (YRRRLH) predicted to serve as the mitochondrial targeting sequence are deleted, is delocalized from mitochondria. Strikingly, this MAVSΔ535-540 mutant is still a potent inducer of IFN-β, demonstrating that mitochondrial localization of MAVS is not essential for downstream activation of IRF3 and NF-κB.
Furthermore, we identified glutamine 519, which is located on the opposite side of the TM α-helix dimer interface, as a key residue in targeting of MAVS to mitochondria. Surprisingly, replacement of this hydrophilic Q519 residue for hydrophobic leucine or tryptophan significantly destabilizes the predicted TM domain structure (free energy of −800 kcal/mol for Q519, compared to −500 kcal/mol for L519 or W519). In our predicted TM domain model, the polar side chain of Q519 moves out of the membrane to be exposed to the cytoplasm (Fig. 7D). We can thus propose that this peculiar positioning of Q519 in the MAVS TM domain is required for its targeting to the mitochondrial membrane. We also demonstrated that MAVS Q519 mutants can be activated upon Sendai virus infection (Fig. 8), demonstrating that mitochondrial localization of MAVS is not essential for activation of MAVS by RIG-I.
Altogether, our results thus demonstrate that the inability of MAVSΔTM to induce IFN-β cannot be explained by the delocalization of MAVSΔTM from the mitochondria, as was previously proposed (32). We show here that, rather, it is the inability of MAVSΔTM to oligomerize upon RIG-I interaction that explains its loss in signaling activity. However, it is not excluded that mitochondrial localization could be important for other functions of MAVS, like its proapoptotic activity following reovirus infection (14).
Finally, it was previously reported that HCV NS3/4A protease cleaves MAVS from mitochondria to block the activation of IFN-β and NF-κB and the induction of antiviral effector genes in infected cells (22, 25). In this report, we showed an impaired oligomerization of MAVS upon NS3/4A expression, an effect mediated by the serine protease activity of NS3/4A (Fig. 9). This inhibitory effect of the NS3/4A protease was significantly reduced for the MAVS Q519L mutant, which is delocalized from the mitochondria, and likely reflects altered efficiency of cleavage by NS3/4A, which is predominantly localized at the mitochondria (data not shown). Our results suggest that the MAVS dimer is the target of NS3/4A-mediated interference of antiviral innate immunity resulting in the inability of MAVS to oligomerize and to recruit signaling intermediates that activate IRF3 and NF-κB. It is noteworthy that RIG-I is overexpressed about ninefold in HCV-infected chimpanzees (5), a phenomenon that could lead to constitutive oligomerization of RIG-I and downstream signaling through the MAVS oligomer to induce a strong antiviral response (31). Accumulation of the NS3/4A protease, reaching a threshold upon HCV infection, could block the RIG-I/MAVS pathway to allow HCV viral persistence. This model for viral persistence seems to be validated by liver biopsy samples from patients with chronic HCV infection (22). These observations validate the TM domain of MAVS as a therapeutic target for the identification of chemical compounds affecting its mitochondrial localization. Such compounds would decrease the sensitivity of MAVS to NS3/4A-dependent cleavage and lead to the constitutive production of IFN-β.
Altogether, our data demonstrate that MAVS oligomers, rather than its mitochondrial localization, play a central role in the formation of a multiprotein membrane-associated signaling complex and enable downstream activation of IRF3 and NF-κB in antiviral innate immunity.
Acknowledgments
We thank Nathalie Grandvaux for providing the IFN-β-LUC promoter, Rafick Pierre Sékaly for the NF-κB-LUC-dependent promoter, Marc J. Servant for the IRF3-LUC-dependent promoter, and Masato Yano for the TOM22 cDNA. We are grateful to Michel Bouvier, Billy Breton, and Martin Audet for their help with setting up BRET assays and Antoine Loquet for his help with HADDOCK calculations. We thank Hawley Rigsby and Nathalie Grandvaux for critical reviews of the manuscript.
This work was supported by a Université de Montréal Novartis/Canadian Liver Foundation Hepatology Research chairmanship (D.L.), the French Centre National de la Recherche Scientifique (CNRS), and the Agence Nationale pour la Recherche sur le SIDA et les Hépatites Virales (ANRS).
Footnotes
Published ahead of print on 26 November 2008.
REFERENCES
- 1.Acehan, D., X. Jiang, D. G. Morgan, J. E. Heuser, X. Wang, and C. W. Akey. 2002. Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol. Cell 9423-432. [DOI] [PubMed] [Google Scholar]
- 2.Angers, S., A. Salahpour, E. Joly, S. Hilairet, D. Chelsky, M. Dennis, and M. Bouvier. 2000. Detection of beta 2-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET). Proc. Natl. Acad. Sci. USA 973684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastiaens, P. I., I. V. Majoul, P. J. Verveer, H. D. Soling, and T. M. Jovin. 1996. Imaging the intracellular trafficking and state of the AB5 quaternary structure of cholera toxin. EMBO J. 154246-4253. [PMC free article] [PubMed] [Google Scholar]
- 4.Bérubé, P., B. Barbeau, R. Cantin, R. P. Sékaly, and M. Tremblay. 1996. Repression of human immunodeficiency virus type 1 long terminal repeat-driven gene expression by binding of the virus to its primary cellular receptor, the CD4 molecule. J. Virol. 704009-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bigger, C. B., B. Guerra, K. M. Brasky, G. Hubbard, M. R. Beard, B. A. Luxon, S. M. Lemon, and R. E. Lanford. 2004. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. J. Virol. 7813779-13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dominguez, C., R. Boelens, and A. M. Bonvin. 2003. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 1251731-1737. [DOI] [PubMed] [Google Scholar]
- 7.Dull, T., R. Zufferey, M. Kelly, R. J. Mandel, M. Nguyen, D. Trono, and L. Naldini. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 728463-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira, K. N., T. M. Iverson, K. Maghlaoui, J. Barber, and S. Iwata. 2004. Architecture of the photosynthetic oxygen-evolving center. Science 3031831-1838. [DOI] [PubMed] [Google Scholar]
- 9.Foy, E., K. Li, R. Sumpter, Jr., Y. M. Loo, C. L. Johnson, C. Wang, P. M. Fish, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc. Natl. Acad. Sci. USA 1022986-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grandvaux, N., M. J. Servant, B. tenOever, G. C. Sen, S. Balachandran, G. N. Barber, R. Lin, and J. Hiscott. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 765532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Granseth, E., G. von Heijne, and A. Elofsson. 2005. A study of the membrane-water interface region of membrane proteins. J. Mol. Biol. 346377-385. [DOI] [PubMed] [Google Scholar]
- 12.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 182714-2723. [DOI] [PubMed] [Google Scholar]
- 13.Hiscott, J., J. Lacoste, and R. Lin. 2006. Recruitment of an interferon molecular signaling complex to the mitochondrial membrane: disruption by hepatitis C virus NS3-4A protease. Biochem. Pharmacol. 721477-1484. [DOI] [PubMed] [Google Scholar]
- 14.Holm, G. H., J. Zurney, V. Tumilasci, S. Leveille, P. Danthi, J. Hiscott, B. Sherry, and T. S. Dermody. 2007. Retinoic acid-inducible gene-I and interferon-beta promoter stimulator-1 augment proapoptotic responses following mammalian reovirus infection via interferon regulatory factor-3. J. Biol. Chem. 28221953-21961. [DOI] [PubMed] [Google Scholar]
- 15.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 1433-38. [DOI] [PubMed] [Google Scholar]
- 16.Kaufmann, T., S. Schlipf, J. Sanz, K. Neubert, R. Stein, and C. Borner. 2003. Characterization of the signal that directs Bcl-x(L), but not Bcl-2, to the mitochondrial outer membrane. J. Cell Biol. 16053-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6981-988. [DOI] [PubMed] [Google Scholar]
- 18.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St. George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426186-189. [DOI] [PubMed] [Google Scholar]
- 19.Li, X. D., L. Sun, R. B. Seth, G. Pineda, and Z. J. Chen. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. USA 10217717-17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, R., J. Lacoste, P. Nakhaei, Q. Sun, L. Yang, S. Paz, P. Wilkinson, I. Julkunen, D. Vitour, E. Meurs, and J. Hiscott. 2006. Dissociation of a MAVS/IPS-1/VISA/Cardif-IKKɛ molecular complex from the mitochondrial outer membrane by hepatitis C virus NS3-4A proteolytic cleavage. J. Virol. 806072-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, R., Y. Mamane, and J. Hiscott. 2000. Multiple regulatory domains control IRF-7 activity in response to virus infection. J. Biol. Chem. 27534320-34327. [DOI] [PubMed] [Google Scholar]
- 22.Loo, Y. M., D. M. Owen, K. Li, A. K. Erickson, C. L. Johnson, P. M. Fish, D. S. Carney, T. Wang, H. Ishida, M. Yoneyama, T. Fujita, T. Saito, W. M. Lee, C. H. Hagedorn, D. T. Lau, S. A. Weinman, S. M. Lemon, and M. Gale, Jr. 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 1036001-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacKenzie, K. R., J. H. Prestegard, and D. M. Engelman. 1997. A transmembrane helix dimer: structure and implications. Science 276131-133. [DOI] [PubMed] [Google Scholar]
- 24.Mercier, J. F., A. Salahpour, S. Angers, A. Breit, and M. Bouvier. 2002. Quantitative assessment of beta 1- and beta 2-adrenergic receptor homo- and heterodimerization by bioluminescence resonance energy transfer. J. Biol. Chem. 27744925-44931. [DOI] [PubMed] [Google Scholar]
- 25.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 4371167-1172. [DOI] [PubMed] [Google Scholar]
- 26.Meylan, E., J. Tschopp, and M. Karin. 2006. Intracellular pattern recognition receptors in the host response. Nature 44239-44. [DOI] [PubMed] [Google Scholar]
- 27.Moore, C. B., D. T. Bergstralh, J. A. Duncan, Y. Lei, T. E. Morrison, A. G. Zimmermann, M. A. Accavitti-Loper, V. J. Madden, L. Sun, Z. Ye, J. D. Lich, M. T. Heise, Z. Chen, and J. P. Ting. 2008. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature 451573-577. [DOI] [PubMed] [Google Scholar]
- 28.Patterson, G. H., D. W. Piston, and B. G. Barisas. 2000. Forster distances between green fluorescent protein pairs. Anal. Biochem. 284438-440. [DOI] [PubMed] [Google Scholar]
- 29.Pop, C., J. Timmer, S. Sperandio, and G. S. Salvesen. 2006. The apoptosome activates caspase-9 by dimerization. Mol. Cell 22269-275. [DOI] [PubMed] [Google Scholar]
- 30.Rapaport, D. 2003. Finding the right organelle. Targeting signals in mitochondrial outer-membrane proteins. EMBO Rep. 4948-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saito, T., R. Hirai, Y. M. Loo, D. Owen, C. L. Johnson, S. C. Sinha, S. Akira, T. Fujita, and M. Gale, Jr. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. USA 104582-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122669-682. [DOI] [PubMed] [Google Scholar]
- 33.Sumpter, R., Jr., Y. M. Loo, E. Foy, K. Li, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 792689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19727-740. [DOI] [PubMed] [Google Scholar]
- 35.Yano, M., N. Hoogenraad, K. Terada, and M. Mori. 2000. Identification and functional analysis of human Tom22 for protein import into mitochondria. Mol. Cell. Biol. 207205-7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5730-737. [DOI] [PubMed] [Google Scholar]
- 37.Zou, H., Y. Li, X. Liu, and X. Wang. 1999. An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 27411549-11556. [DOI] [PubMed] [Google Scholar]