Abstract
Multiple unique protein markers sorted to the inner nuclear membrane (INM) from the Autographa californica nucleopolyhedrovirus occlusion-derived virus (ODV) envelope were used to decipher common elements of the sorting pathway of integral membrane proteins from their site of insertion into the membrane of the endoplasmic reticulum (ER) through their transit to the INM. The data show that during viral infection, the viral protein FP25K is a partner for all known ODV envelope proteins and that BV/ODV-E26 (designated E26) is a partner for some, but not all, such proteins. The association with the ER membrane of FP25K, E26, and the cellular INM-sorting protein importin-α-16 is not static; rather, these sorting proteins are actively recruited to the ER membrane based upon requirements of the proteins in transit to the INM. Colocalization analysis using an ODV envelope protein and importin-α-16 shows that during viral infection, importin-α-16 translocates across the pore membrane to the INM and then is incorporated into the virus-induced intranuclear membranes. Thus, the association of importin-α-16 and INM-directed proteins appears to remain at least through protein translocation across the pore membrane to the INM. Overall, the data suggest that multiple levels of regulation facilitate INM-directed protein trafficking, and that proteins participating in this sorting pathway have a dynamic relationship with each other and the membrane of the ER.
Baculovirus infection results in a synchronous, amplified pulse of the synthesis of integral membrane proteins that integrate into the membrane of the endoplasmic reticulum (ER) and utilize the membranes of the nuclear envelope during transit to intranuclear membrane vesicles. These virus-induced intranuclear membrane vesicles ultimately become the envelope of the occlusion-derived virus (ODV) (6). The N-terminal 33 amino acids of the viral envelope protein ODV-E66 (designated E66) are sufficient to target this protein, and fusion proteins thereof, to the ODV envelope (14) and, in the absence of infection, target proteins to the inner nuclear membrane (INM) (15). Thus, this sequence has been termed an INM-sorting motif (INM-SM) (8). Knowledge of the INM-SM and the ability to generate fusion and mutant constructs provide unique tools to study the molecular mechanisms that regulate the trafficking of integral membrane proteins from their site of membrane insertion in the ER to the membranes of the nuclear envelope (8, 15).
The E66-derived INM-SM sequence functions as an N-terminal signal anchor and has two major targeting features. The first feature is a hydrophobic domain of approximately 18 amino acids that, during translation, is precisely positioned within the central channel of the ER translocon in close proximity to both Sec61α and TRAM (21). The second feature is a positively charged amino acid within four to eight amino acids from the end of the hydrophobic sequence that is positioned on the cytoplasmic face of the ER membrane (8). Chemical cross-linking studies show that when the nascent chain of the INM-directed protein resides within the central channel of the translocon, the positively charged amino acid of the INM-SM sequence is in close proximity to the cellular protein importin-α-16, and importin-α-16 remains associated with this sequence after release from the translocon and ER membrane integration (22). In infected cells, after the INM-directed protein has been released from the translocon and integrated into the ER membrane, the positively charged amino acid of the INM-SM sequence is proximal to the viral proteins BV/ODV-E26 (E26) or FP25K (8, 21). The sequence of molecular events that occur after the association of the INM-SM with cellular importin-α-16 and the subsequent association of the same region of the INM-SM with the viral protein E26 or FP25K is unknown, but clearly these data suggest that the sorting of INM-directed proteins begins during protein translation and continues after protein integration into the ER membrane.
The discovery of Spodoptera frugiperda importin-α-16 (and human KPNA4-16) was hailed as the discovery of a missing link to understanding the molecular pathway of integral membrane proteins directed to the INM (19). However, the discovery of importin-α-16 and its association with INM-directed proteins had greater implications. The identification of importin-α-16 led to the discovery of other isoforms of importin-α, KPNA4-26, KPNA1-12, and KPNA2-12, although nothing is known of their function (7). It is widely held that importin-α is a soluble protein; however, several reports now show that a subset of importin-α associates with cellular membranes (1, 12, 13). While little is known of the function of full-length, membrane-associated importin-α, we know that the truncated isoform importin-α-16 not only associates with the ER membrane but is positioned at the translocon such that it can optimally survey translating nascent chains, recognize INM-SM sequences, and associate with them. However, importin-α-16 and KPNA4-16 can recognize INM-SM sequences and associate with them posttranslationally. As such, importin-α-16 may serve to direct INM-SM-containing proteins into a common and specific INM-directed trafficking pathway regardless of their method of membrane integration. Until the identification of S. frugiperda importin-α-16 and its human homologue KPNA4-16, it was widely held that INM-directed proteins were not specifically targeted to the INM but rather diffused freely through the contiguous membranes of the ER and nuclear envelope and were enriched only at the INM after binding with nucleoplasmic components. Clearly, not only is our knowledge of the well-described adaptor protein importin-α incomplete, but the previous doctrine of the trafficking pathway of integral proteins to the INM is challenged by current data.
Data suggest that during baculovirus infection, cellular importin-α-16 and the viral proteins FP25K and E26 all function to sort and traffic ODV envelope proteins to the INM and viral envelope. However, how these proteins interact with each other or associate with the ER is unknown. Full-length importin-α is predominantly soluble, and it is likely that the ER membrane/translocon association of importin-α-16 is transient, although its membrane association is resistant to salt and alkali extraction (22). A transient membrane association is predicted for the viral proteins FP25K and E26; FP25K is predominantly a soluble protein, and the factors regulating its membrane association are unknown, while E26 membrane association apparently is regulated by a reversible palmitoylation event (9). The idea of proteins transiently associating with the molecular machinery of the translocon and providing a unique molecular activity is not new; signal peptidase, oligosaccharyltransferase, the signal recognition particle, and BiP all are well-described examples of such proteins (16). Based upon other known pathways involving the ER, it is reasonable to postulate that the cell has evolved mechanisms to keep such accessory proteins at optimal stoichiometric levels relative to the demand of molecular events occurring at the ER membrane. Thus, such accessory proteins may be recruited or displaced from the ER membrane as needed. Such dynamic interactions are difficult to detect and study in intact cells or by using routine in vitro assays; however, the study of amplified pathways of protein transport, like that produced by baculovirus infection, substantially increases the potential to detect such transient and dynamic interactions of known proteins.
The goals of the present work were threefold: (i) use multiple viral ODV envelope proteins with INM-SM-like sequences to determine if the molecular events identified for the E66-derived INM-SM sequence also are general features of the molecular sorting pathways for other INM-directed proteins; (ii) use various recombinant viruses that amplify the quantity of INM-directed proteins or control the levels of expression of known sorting proteins (FP25K, E26, and importin-α-16) to determine if altered expression affects the quantity of sorting proteins present at the ER membrane; and (iii) use a recombinant baculovirus to determine if importin-α-16 transits to the INM.
MATERIALS AND METHODS
Insect cell lines, virus, and preparation of microsomal membranes.
Spodoptera frugiperda IPLB-Sf21-AE clonal isolate 9 (Sf9) cells were cultured in suspension at 27°C in TNMFH medium (23) supplemented with 10% fetal bovine serum. Cells were infected at a multiplicity of infection of 10, with time zero set as the time of virus addition. Viruses used were the following: Autographa californica multiple nucleopolyhedrosis virus (AcMNPV) (strain E2), ΔFP25K (2, 20), and the recombinant viruses importin-α-16-T7(polh), E66-His(polh), and E66-SM(polh). Sf9 microsomes were prepared from infected cells 33 h postinfection using techniques described previously (18, 24).
Generation of cross-linking cassette sequences for the multiple ORFs.
The sequence of the lysine-free portion of the cross-linking cassette containing the T7 epitope and His7 sequence was reported previously (8). The sequence encoding the linking amino acid sequence GANA includes a unique Nar1 site, and this site was used to insert annealed, complementary synthetic oligonucleotides encoding the N-terminal INM-SM sequence of each of the open reading frames (ORFs) into the cassette located in the in vitro translation vector pGEM4Z. All gene constructs were sequence confirmed.
SDS-PAGE and Western blot analyses.
Protein concentrations of microsomal membranes were determined by the method of Bradford (4), and all lanes were loaded at equal protein concentrations. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed according to Laemmli (17) using a 4% stack gel and 15% separating gel. Samples were incubated in 1.5% SDS, 0.5% β-mercaptoethanol, 25 mM Tris-HCl (pH 6.8), and 7% glycerol for 15 min at 65°C. Following electrophoresis, the gels were transferred onto Immobilon-P membranes (Millipore, Bedford, MA). The membranes were blocked with TTBS-BLOTTO (150 mM NaCl, 10 mM Tris, 0.1% Tween 20, pH 8.0, supplemented with 1% nonfat dry milk). Antibody was bound overnight (4°C), blots were washed twice with Tris-buffered saline (TBS), and horseradish peroxidase-linked immunoglobulin G (IgG) (1:10,000) was bound for 1 h at room temperature (RT). Blots were washed three times with TTBS, reacted for 1 min with ECL reagent (Amersham, Arlington Heights, IL), and exposed to X-ray film. The following antibodies and dilutions were used: anti-FP25K, no. 2804 (1:5,000); anti-T7, no. 69522 (1:5,000; Novagen); and anti-E26, no. 7554 (1:10,000) (9).
Immunofluorescence confocal microscopy.
Cells were prepared for microscopy as described previously (10). Cells were collected and resuspended in Grace's medium, and 2.8 × 105 cells were transferred to a 1-well cytofuge container (Statspin Technologies, Norwood, MA). After allowing attachment at room temperature for 5 min, the cells were fixed with 3.7% paraformaldehyde (made in phosphate-buffered saline [PBS] at RT) for 10 min. The cells were washed three times with PBS, incubated with methanol for 10 min, washed, incubated with Triton X-100 (0.5%; made in PBS) for 10 min at RT, and washed. The cells were incubated in blocking solution (1% porcine serum, 3% bovine serum albumin in PBS) for 1 h at RT. Primary antibodies were diluted in blocking solution and then incubated at 4°C. The cells were washed with PBS and incubated with secondary antibody (Alexa Fluor conjugates; Molecular Probes, Inc., Eugene, OR) diluted at 1:2,000 in blocking solution for 2 h at RT. The cells were washed again three times. When DNA staining was required, the cells were incubated with DAPI (4′,6′ diamidino-2-phenylindole) diluted at 0.1 μg/ml in PBS for 5 s and then washed three times in PBS. Ten microliters of Dako fluorescent mounting medium (Dako Corporation, Carpinteria, CA) was added and covered with a coverslip (no. 1.5). Slides were viewed using a Zeiss Axiovert 135 (Carl Zeiss MicroImaging, Inc., Thornwood, NY) with a CARV confocal module (Atto Bioscience, Rockville, MD). After at least 20 fields were viewed, representative cells/fields were collected using either the CARVer software (Atto Bioscience) or Zeiss Axiovision v. 3.1 (Carl Zeiss MicroImaging, Inc., Thornwood, NY). Confocal sections were collected at 0.75-μm intervals. The following antibodies and dilutions were used: E26, no. 7554 (1:5,000); and anti-T7, no. 69522 (1:5,000; Novagen).
Chemical cross-linking and immunoprecipitation.
In vitro translations were performed in the presence of appropriate mRNA, nuclease-treated rabbit reticulocyte lysate (Promega), amino acid mixture minus methionine (Promega), RNasin (Promega), approximately eight equivalents of microsomes, and [35S]Met (8, 24). Following translation, samples were pooled and subsequently realiquoted so that all treatments were performed on identical samples. Membranes were sedimented through a 0.5 M sucrose cushion in a Beckman Coulter Optima TLA ultracentrifuge at 4°C for 3 min at 100,000 rpm in a TLA 100 rotor. The resulting membrane pellet was resuspended in 50 μl of cross-linking buffer (25 mM sodium phosphate, 150 mM NaCl, pH 7.0); one sample, representing the non-cross-linked control, was put aside; and the remaining samples were treated with the cross-linking reagent Bis(sulfosuccinimidyl)suberate, (BS3; 2.5 mM; Pierce). Samples were incubated at RT for 30 min. For Talon purification, cross-linked samples were solubilized (in 4 M urea-0.5% [wt/vol] SDS for 30 min at 37°C) and incubated with 20 μl of Dynabeads Talon per the manufacturer's instructions (Dynal Biotech). The bound material was eluted using SDS sample buffer and resolved using SDS-PAGE. For immunoprecipitation, the cross-linked samples were solubilized with radioimmunoprecipitation assay buffer (50 mM Tris, pH 8.0, 100 mM NaCl, 1% NP-40, 1% desoxycholate, 0.1% SDS) and incubated with primary antibody overnight (4°C), followed by incubation with protein A/G agarose (2 h at 4°C; Sigma). The protein A/G-bound complexes were recovered by sedimentation, washed, separated using SDS-15% PAGE, and visualized using X-ray film.
RESULTS
Experimental rationale and design.
An advantage of using viral models to study cellular events is that viruses often provide multiple unique protein markers sorted to the same destination. A comparison of the molecular events that control the sorting of these viral proteins destined for the same cellular location can collectively reveal general features of a common trafficking pathway. In addition, the use of baculovirus and recombinant viruses thereof provides the opportunity to selectively increase the quantity of proteins targeted to the INM and intranuclear membrane vesicles derived from the INM. In this manner, the stoichiometric ratios of INM-directed proteins can be increased relative to the quantity of sorting factors present in the cell (or vice versa). Considering that microsomal membranes are easily prepared from infected or uninfected Sf9 cells, the effects that such alterations in protein expression have on other proteins associated with the ER membrane can be detected. Finally, the use of recombinant baculoviruses can reveal the cellular destination(s) of uncharacterized proteins. Considering that much is known about the destination of ODV envelope proteins and intermediate states of ODV envelopment (6), we can compare the cellular destination of known ODV envelope proteins to that of the less well-characterized protein importin-α-16. In this way, we can predict whether importin-α-16 disassociates from the ODV protein on the cytoplasmic face of the nuclear envelope and is released into the cytoplasm or continues through the pore complex to the INM.
Proximity with the INM-SM-like sequences and FP25K or E26 is not consistent.
The perusal of the AcMNPV genome reveals eight genes that encode proteins containing N-terminal domains similar to the targeting sequence identified from E66 (Fig. 1). With the exception of orf91 (which is still uncharacterized), all of these ORFs encode ODV envelope proteins (6). To determine whether these sequences utilize molecular events similar to those described for E66, constructs were made that allowed chemical cross-linking analyses similar to that previously performed for the E66-derived INM-SM sequence. Each of the viral ORF sequences had the appropriate positively charged amino acid in the N-terminal region replaced with a lysine, and this sequence was fused to a cassette that contained a T7 epitope, a lysine-free sequence of amino acids, and a C-terminal His7 tag (the cassette sequence was fully detailed previously [8]) (Fig. 1). Thus, each fusion protein has an approximate mass of 15 to 18 kDa, and the lysine(s) flanking the hydrophobic sequence is the only amino acid(s) available as a substrate for the chemical cross-linking reagent BS3, a lysine-lysine cross-linker (11.4-Å linking arm).
FIG. 1.
INM-SM cassettes used for cross-linking experiments. The N-terminal amino acids of each ORF encoding an ODV envelope protein and containing the features of the INM-SM sequence are shown. The characterized INM-SM sequence derived from E66 is shown for reference (highlighted). Each N-terminal sequence was fused to the same cassette as that shown for E66 containing the amino acids GANA as the linker sequence, followed by a T7 epitope, a lysine-free sequence, and a C-terminal His7 tag. The hydrophobic sequences are highlighted in yellow, and the associated positively charged amino acids are shown in light red. The positively charged amino acid(s) mutated to lysine to serve as bait for the BS3 cross-linking reagent is noted above the sequence in dark red. All of the resultant fusion proteins have molecular masses of 15 to 18 kDa.
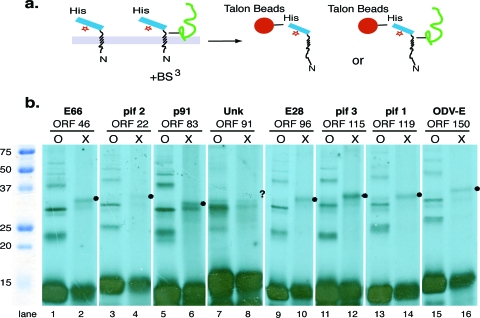
To determine if these N-terminal INM-SM-like sequences cross-linked with any protein, each construct was translated and radiolabeled in vitro in the presence of microsomal membranes prepared from AcMNPV-infected cells and exposed to the chemical covalent cross-linking reagent BS3. To enrich the INM-SM-containing bait protein and potential cross-linked protein complexes, the samples were bound to His-binding Talon beads (Fig. 2a), and the bound protein or covalently linked protein complex was separated using SDS-PAGE. When the samples were analyzed, cross-linked adducts with similar molecular masses were detected for each of the fusion proteins except for that of orf91 (Fig. 2b, lanes 2, 4, 6, 8, 10, 12, 14, and 16) (Orf22 is difficult to observe in this experiment, and the cross-linked adduct is detected more clearly in Fig. 3g). Repeated trials testing the ability of Orf91 to cross-link with another protein failed to detect a cross-linked adduct, and since Orf91 is uncharacterized, it was not pursued further.
FIG. 2.
Cross-linking of ODV envelope protein cassettes to proteins within microsomal membranes. (a) Schematic of the experimental protocol utilized for chemical cross-linking assays. After in vitro translation and the generation of a radiolabeled protein, the sample was treated with BS3, the microsomal membranes were denatured, and the bait cassette or covalently linked associated complex was enriched using His-binding Talon beads. (b) The enriched samples were separated using SDS-PAGE. O represents the un-cross-linked control, X represents enriched cross-linked samples, and • notes the cross-linked adducts. No detectable cross-linked adduct was detected for Orf91, but the expected site of an adduct comparable to that detected for the other ORFs is noted with a question mark.
FIG. 3.
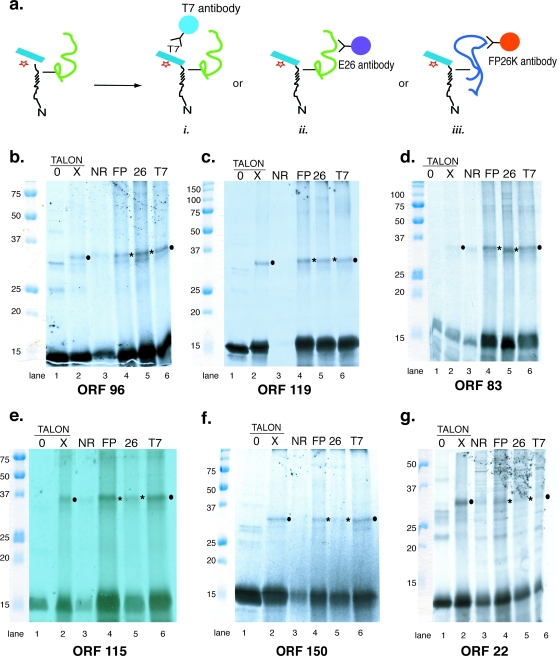
Cross-linking of ODV envelope protein cassettes to FP25K and E26. (a) Schematic of the experimental protocol utilized for chemical cross-linking assays. After in vitro translation and treatment with BS3, the microsomal membranes were denatured and bound to Talon beads (as depicted in Fig. 2) or were precipitated using antisera to T7 epitope (i), antibody to E26 (ii), or antibody to FP25K (iii). (b to g) Autoradiographs showing the results of cross-linking experiments for the various ORFs. Binding to Talon beads is shown in lanes 1 and 2 (0, negative control with no treatment with BS3; X, treatment with BS3). Lanes 3 to 6 show results from immunoprecipitation experiments (lane 3, negative control with normal rabbit serum; lane 4, antibody to FP25K; lane 5, antibody to E26; lane 6, antibody to T7 epitope). Cross-linked adducts enriched using the bait sequence to enrich the complex are noted with filled circles. Cross-linked adduct enriched using antibody to E26 or FP25K is noted by asterisks.
To determine if the identity of the cross-linked protein was E26 or FP25K, immunoprecipitation experiments using antiserum to these proteins were performed (Fig. 3a, images ii and iii). For E26, two well-characterized antibodies are available. One of these (no. 7554) only recognizes viral E26 and does not cross-react with the cellular protein importin-α-16 (9). This antibody was used throughout. As a control, the enrichment of the cross-linked adduct using Talon beads was repeated (Fig. 2a) along with the immunoprecipitation protocol using antibody to the T7 epitope (Fig. 3a, image i). A control sample (not exposed to BS3) was bound to Talon beads to determine the background binding of non-cross-linked proteins, and normal rabbit serum was used to determine background reactivity for the immunoprecipitation experiments. We note that when the fusion cassette is exposed to BS3, some portion of it nonspecifically precipitates with normal rabbit serum. So in these lanes, instead of seeing a blank lane, it is routine to see some amount of bait protein. If the bait has cross-linked with an adduct, a trace amount of the adduct also may be visible. We have no explanation for why the chemically treated cassette bait sequence has a tendency to precipitate with random IgGs, but this result has been reported previously for the use of multiple fusion proteins exposed to chemical cross-linking reagents (7, 22).
The results from Orf96, Orf119, Orf83, and Orf115 (Fig. 3b, c, d, and 3e, respectively) showed similar results. Each of the fusion cassette sequences cross-linked with a protein that was enriched on the His-binding Talon beads and precipitated using T7 antibody (Fig. 3b, c, d, and e, lanes 2 and 6). The cross-linked adduct for each of these proteins was easily detected when precipitated using E26 or FP25K antibody (Fig. 3b, c, d, and e, lanes 4 and 5). These data suggest that the INM-SM cassette sequences of Orf96, Orf119, Orf83, and Orf116 independently associate with both of the viral proteins FP25K and E26 when they are located in the ER membrane of the infected cell.
The data obtained from the fusion cassette sequences of Orf150 and Orf22 also showed that these proteins cross-linked with a protein adduct (Fig. 3f and g, lanes 2 and 6, respectively), and antibody to FP25K convincingly precipitated the cross-linked adduct (Fig. 3f and g, lanes 4). However, E26 antibody precipitated an amount of cross-linked adduct that was discernible but barely increased relative to that of the control lane using normal rabbit IgG (Fig. 3f and g, compare lanes 3 and 5). This result was consistent and reproducible. So while the cross-linking data show that Orf150 and Orf22 are in close proximity to FP25K, an interaction with E26 is less certain.
Quantity of FP25K, E26, and importin-α-16 in the ER membrane is dynamic.
It is possible that the abundance of either E26 or FP25K associated with the ER membrane is determined by the quantity of ODV envelope protein in transit (i.e., E26 and FP25K are recruited to the ER membrane when demand for the sorting of INM-directed proteins is increased). Considering that cellular importin-α-16 clearly is interacting in the pathway of INM-directed protein trafficking, its concentration in the ER membrane also may affect, or be affected by, the events of virus infection. To test this, microsomal membranes were prepared from recombinant virus-infected cells that either had increased amounts of INM-directed proteins expressed or that had an altered expression of the sorting factor importin-α-16 or FP25K. For comparison, control microsomal membranes were prepared from uninfected or AcMNPV-infected Sf9 cells. Two recombinant viruses were used that abundantly expressed INM-directed proteins; one of these had increased quantities of the viral ODV envelope protein E66 (tagged with a C-terminal His7 sequence [E66HIS]), while the second increased the quantity of the E66-derived INM-SM cassette (E66SM) (Fig. 4a). Several recombinant viruses were used that altered the expression levels of the sorting factors FP25K and importin-α-16. Recombinant virus was generated that abundantly expressed a T7 epitope-tagged version of Sf9 importin-α-16 (Fig. 4a), while the affects of deleting the sorting factor FP25K were studied using a ΔFP25K deletion recombinant virus (2, 20). While it would be desirable to include a virus with E26 deleted, such is not possible because E26 is essential (9). Microsomal membranes were prepared from cells infected at equivalent multiplicities of infection and harvested at 33 h postinfection. Approximately equal quantities of microsomal membranes were analyzed using SDS-PAGE (Fig. 4b). The microsomal membranes were analyzed by Western blotting and reacted against various antisera.
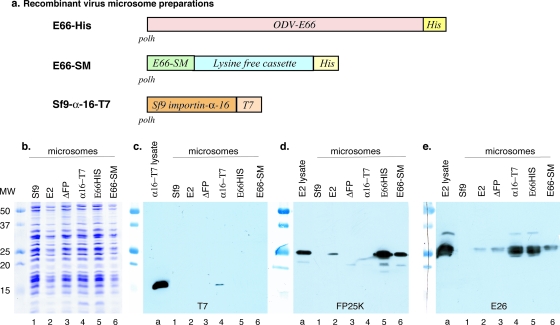
FIG. 4.
Association of E26, FP25K, and importin-α-16 with enriched ER membranes. (a) Schematic of the recombinant viruses used for infection and the preparation of in vitro translation-competent microsomal membranes. In every case, the recombinant gene was inserted into the polyhedrin gene locus under the control of the polyhedrin promoter. (b) Coomassie blue-stained SDS-PAGE gel showing that the microsomal preparations were loaded at approximately equal concentrations. (c to e) SDS-PAGE-separated gels were blotted and analyzed using antibody to T7 epitope (c), FP25K antibody (d), and E26 antibody (e). In panels c to e, lane a shows the control from a total cell lysate obtained from cells infected with the relevant virus. Lanes 1 to 6 represent microsomal membranes prepared from control Sf9 cells (lane 1) and from cells infected with wild-type AcMNPV (lane 2), ΔFP25K virus (lane 3), importin-α-16-T7 virus (lane 4), E66HIS virus (lane 5), and E66-SM virus (lane 6).
When the membranes were tested using T7 antibody, tagged importin-α-16 was detected in total cell lysates and microsomal membranes prepared from virus expressing importin-α-16-T7 (Fig. 4c, lanes a and 4, respectively). Using the T7 epitope-specific antibody, no nonspecific bands were detected in any of the microsomal membrane preparations (Fig. 4c, lanes 1 to 3 and 5 and 6). When the microsomes were tested for the presence of FP25K, FP25K was clearly detected in total cell lysate and in microsomal membranes prepared from AcMNPV-infected cells (Fig. 4d, lanes a and 2, respectively). As expected, FP25K was not detected in microsomes prepared from ΔFP recombinant virus-infected cells (Fig. 4d, lane 3). The relative amount of FP25K was increased in microsomes prepared from cells infected with viruses in which the quantity of the INM-directed protein E66 or the E66-derived INM-SM fusion cassette was increased (Fig. 4d, compare lane 2 to lanes 5 and 6). When the membranes prepared from cells infected with the recombinant virus expressing an increased quantity of Sf9 importin-α-16 were analyzed, the amount of FP25K had decreased to undetectable levels (Fig. 4d, lane 4), suggesting that importin-α-16 could be: (i) displacing FP25K or nullifying the need for FP25K within the microsomal membranes by providing an equivalent function, or (ii) interfering with another protein interaction responsible for FP25K membrane association.
E26 was easily detected in the total cell lysate and microsomal membranes prepared from AcMNPV-infected cells (Fig. 4e, lanes a and 2). The relative amount of E26 within the membranes does not appear to be greatly influenced by the presence or absence of FP25K; in ΔFP25K microsomes, E26 was present in quantities approximately equal to those in AcMNPV-infected cells (Fig. 4b and e, compare lanes 2 and 3), and when FP25K itself was expressed at high levels, the level of FP25K or E26 did not substantially increase in the microsomal membranes (this experiment was performed as an independent data set and is not shown here). An increase in the quantity of E26 was detected in microsomes prepared from the recombinant virus abundantly expressing the E66SM cassette, but a much larger increase was detected in membranes prepared from cells generating abundant copies of full-length E66 (Fig. 4e, lanes 6 and 5, respectively). The quantity of E26 also was increased in membranes prepared from cells infected with virus expressing abundant quantities of Sf9 importin-α-16 (Fig. 4e, lane 4).
E26 and importin-α-16 are targeted to the same cellular destination during infection.
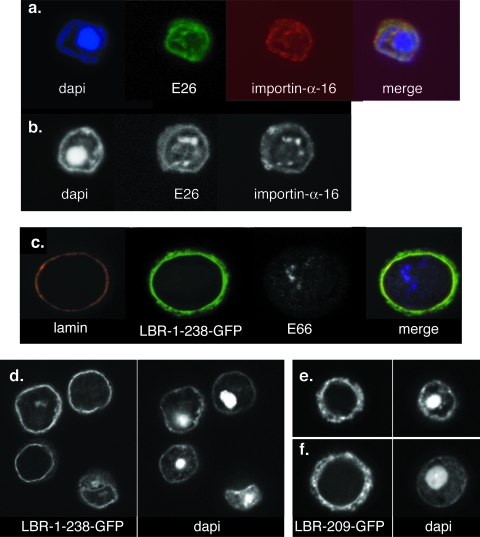
While we know that cellular importin-α-16 associates with the INM-directed sequence at the time of translation and remains with the protein after membrane integration, we have no insights into the time of the dissociation of importin-α-16 with the INM-directed protein. If it disassociates from its cargo prior to translocation across the pore membrane, then one would expect to detect it within the cytoplasm. A very strong case can be made for the conclusion that the INM serves as a precursor for the virus-induced intranuclear membranes (6). Therefore, if importin-α-16 is detected within the foci of intranuclear microvesicles, it must have utilized the INM during its transit to these membranes. ODV envelope proteins are known to be enriched within the virus-induced intranuclear membranes, so the use of a recombinant virus expressing an epitope-tagged version of importin-α-16 and colocalization studies using ODV envelope marker proteins provide a straightforward assay to determine if importin-α-16 passes the pore membrane to the INM. E26 was chosen as the ODV envelope protein marker for the intranuclear membranes (3), and recombinant virus expressing importin-α-16-T7 was used for infection and confocal analyses. The perusal of many cells showed that E26 and importin-α-16 were targeted to the same regions within the infected cells (Fig. 5a, b). These sites include membranes at the periphery of the nucleus, and foci of microvesicles that accumulate within the infected cell are predicted to serve as the precursors of the ODV envelope (6). The trafficking of importin-α-16 to the intranuclear membranes is not random. When the mammalian INM protein lamin B receptor (LBR; an N-terminal region of the transmembrane sequence 1 that was fused to green fluorescent protein [GFP], as described previously [11]) was abundantly expressed in Sf9 cells infected with recombinant virus, the LBR remained at the periphery of the nucleus, and only a small amount of protein could be detected in the virus-induced intranuclear membranes (Fig. 5c, d). This pattern of localization clearly is different from the ER/ONM localization that is detected for another LBR construct that has the INM-SM sequence deleted (Fig. 5e, f) (the construct was described previously [7]). These results suggest that during infection, importin-α-16 remains with the ODV envelope protein from its time of membrane integration in the ER membrane throughout its transit to the INM and the virus-induced intranuclear microvesicles.
FIG. 5.
Confocal microscopy showing the colocalization of E26 and importin-α-16 in infected Sf9 cells. (a and b) Cells were infected with the recombinant virus expressing importin-α-16-T7 (as described in the legend to Fig. 4a). At 33 h postinfection, cells were harvested and prepared for confocal microscopy. E26 was visualized using primary antibody to E26, importin-α-16-T7 was detected using antibody specific for the T7 epitope, and DNA (the overall location of the nucleus) was visualized using DAPI. Single Z sections representing 0.75-μm slices are shown. (c and d) Cells were infected with recombinant virus expressing an LBR construct consisting of the N-terminal region of the LBR through transmembrane sequence 1 fused to GFP (11). Cells were harvested at 33 h postinfection and prepared for confocal microscopy. The nuclear region was detected using DAPI, and LBR was detected using GFP autofluorescence. LBR remains at the periphery of the nucleus. (e and f) Cells were infected with recombinant virus expressing an LBR construct consisting of the LBR transmembrane sequence 1 with the INM-SM-like sequence deleted and then fused to GFP (7). Cells were harvested at 33 h postinfection and prepared for confocal microscopy. The nuclear region was detected using DAPI, and LBR was detected using GFP autofluorescence. When the INM-SM sequence of LBR is deleted, the protein orients in the membrane incorrectly (7) and then locates throughout the ER.
DISCUSSION
Previous studies deciphering the molecular mechanisms of the sorting and trafficking of INM-directed proteins predict a common pathway. For both viral and cellular INM-directed proteins, this pathway is mediated in part by a cellular protein belonging to the importin-α family (S. frugiperda importin-α-16 or human KPNA4-16). In AcMNPV-infected cells, two viral proteins, E26 and FP25K, participate in this pathway, and both of these proteins function in association with the INM-SM sequence after the INM-directed protein has been integrated into the ER membrane and released from the translocon (21). The data set used to decipher this molecular pathway still is relatively small, and it includes several viral ODV envelope proteins (E66, E25, and INM-SM sequence fusion proteins) and the cellular proteins LBR and nurim (7). An important goal of this study was to the determine critical features of this common pathway by expanding the data set to include additional INM-directed proteins. Since addressing this question using mammalian cells or in vitro approaches is difficult, we turned once again to the baculovirus to provide the necessary insights. As a first approach, multiple ODV envelope proteins, each containing a characteristic INM-SM-like sequence, were tested for their ability to cross-link with the viral sorting factors FP25K and E26. All of the known ODV envelope proteins containing an N-terminal, INM-SM-like sequence cross-linked with FP25K, suggesting that FP25K provides a very important activity in this sorting pathway.
What is known of the function of FP25K in the INM-directed protein sorting process? Like cellular importin-α, most of the FP25K protein in the cell is soluble, and only a very small proportion of the total protein associates with cellular membranes. The function of the soluble FP25K is unknown; however, membrane-associated FP25K has been implicated as a protein that facilitates the transit of ODV envelope proteins from the outer nuclear membrane to the INM. When FP25K is absent, instead of the rapid transit of E66 to intranuclear membrane vesicles, E66 accumulates in punctate regions in the outer nuclear membrane and is not detected at the INM. Only after prolonged infection can E66 be detected within the intranuclear membranes (5, 20). When another ODV envelope protein, E25, is studied in ΔFP25K infected cells, its trafficking to intranuclear membranes also is delayed, and E25 slowly accumulates in the INM, where it aggregates in punctate clusters (20). Thus, the efficiency and rate of trafficking of the envelope proteins E66 and E25 are substantially affected in the absence of FP25K, and the transit across the pore membrane to the INM appears to be the step most impaired. However, the observation that a small amount of these proteins eventually transits to the intranuclear membrane vesicles in the absence of FP25K suggests that other proteins mediate this process and that the activity of these other proteins is sufficient to retain the production of viable virus. As such, other proteins (cellular or viral) may provide redundant activity that normally is provided at optimal levels by FP25K. An analysis of the dynamic interactions of sorting factors within microsomal membranes show that when importin-α-16 is abundantly present in ER membranes, FP25K is no longer recruited to these membranes. This result suggests that when additional copies of importin-α-16 are present in the ER membrane, FP25K is no longer required. Importin-α-16, then, may be a reasonable candidate to provide the redundant function for FP25K. If that is true and FP25K facilitates the transit of INM-directed proteins across the pore membrane to the INM, then one would predict that importin-α-16 also performs such a function. Colocalization analysis using the known ODV envelope protein E26 and importin-α-16 show that importin-α-16 is trafficked to the nuclear envelope and the virus-induced intranuclear membrane during viral infection. As such, importin-α-16 is appropriately positioned to have functional activity during protein translocation across the pore membrane to the INM.
The data suggest that multiple levels of regulation facilitate INM-directed protein trafficking, and that proteins participating in this sorting pathway have a dynamic relationship with each other. The association of FP25K, E26, and cellular importin-α-16 with the ER membrane is not static; rather, their presence at the ER membrane appears to be actively recruited and based upon the requirements of the INM-directed protein in transit. While the presence of increased amounts of importin-α-16 in the ER membrane results in the displacement of FP25K, the opposite relationship is observed between importin-α-16 and E26. When levels of importin-α-16 are increased in the ER membrane, there is a concomitant increase in the levels of E26. While these results suggest a synergistic relationship between E26 and importin-α-16, they do not provide substantial new insights into the functional activity of E26 in the infected cell. The complexity of these observations suggests that while there are common mechanisms within the pathway of directing proteins to the INM, individual INM-directed proteins have specific requirements, and multiple proteins facilitate and direct each stage of integral membrane protein movement through their passage from the ER membrane to the INM.
There is always the impulse to conclude a study of a molecular sequence of events by drawing a simple diagram to explain them. However, such models repeatedly expand until each pathway becomes a complex series of precisely positioned and interacting proteins that are not only responsive to the elements in the main pathway but also sensitive to the environment and global requirements of the cell. It appears that the molecular pathway of INM-directed proteins ultimately will prove to be such a complicated and multifaceted pathway. Current data suggest that the sorting of INM-directed proteins begins during the translation and interaction of the nascent chain with the translocon, and translocon- or ER-associated proteins are sensitive to the cellular requirements for such sorting. Sorting continues once the protein has been integrated into the ER and released from the translocon, and studies using baculovirus suggest that multiple sorting factors facilitate this process. As the INM-directed protein translocates across the pore membrane to the INM, additional sorting events occur. In baculovirus-infected cells, FP25K has a functional role during this event. Considering that viruses are regional experts in the molecular pathways of their host cell and that insights derived from the study of baculovirus-infected cells thus far can be correlated with similar events in human cells, one can only expect that continuing insights derived by the study of the baculovirus will provide substantial steps forward to our understanding of the molecular events that occur during the sorting and trafficking of INM-directed proteins to the INM in mammalian cells.
Acknowledgments
We thank Art Johnson (Texas A&M University System Health Science Center, College Station) and Suraj Saksena for the development and teaching of the technique to generate in vitro translation-competent microsomal membrane preparations from Sf9 and virus-infected cells, as well as German Rosas-Acosta for the development of the E66HIS recombinant virus.
This work was supported in part by Texas AgriLife Research Project TEXO8078 (to M.D.S.).
Footnotes
Published ahead of print on 19 November 2008.
REFERENCES
- 1.Andrade, R., R. Alonso, R. Peña, J. Arlucea, and J. Aréchaga. 2003. Localization of importin α (Rch1) at the plasma membrane and subcellular redistribution during lymphocyte activation. Chromosoma 11287-95. [DOI] [PubMed] [Google Scholar]
- 2.Beames, B., and M. D. Summers. 1988. Comparison of host cell NA insertions and altered transcription at the site of insertions in few polyhedra baculovirus mutants. Virology 162206-220. [DOI] [PubMed] [Google Scholar]
- 3.Beniya, H., S. C. Braunagel, and M. D. Summers. 1998. Autographa californica nuclear polyhedrosis virus: subcellular localization and protein trafficking of BV/ODV-E26 to plasma membrane, intranuclear membranes and viral envelopes. Virology 24064-75. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 5.Braunagel, S. C., J. K. Burks, G. Rosas-Acosta, R. L. Harrison, H. Ma, and M. D. Summers. 1999. Mutations within the Autographa californica nucleopolyhedrovirus FP25K gene decreases the accumulation of ODV-E66 and alters its intranuclear transport. J. Virol. 738559-8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braunagel, S. C., and M. D. Summers. 2007. Molecular biology of the baculovirus occlusion-derived virus envelope. Curr. Drug Targets 81084-1095. [DOI] [PubMed] [Google Scholar]
- 7.Braunagel, S. C., S. T. Williamson, Q. Ding, X. Wu, and M. D. Summers. 2007. Early sorting of inner nuclear membrane proteins is conserved. Proc. Natl. Acad. Sci. USA 1049307-9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braunagel, S. C., S. T. Williamson, S. Saksena, Z. Zhong, W. K. Russell, D. H. Russell, and M. D. Summers. 2004. Trafficking of ODV-E66 is mediated via a sorting motif and other viral proteins: facilitated trafficking to the inner nuclear membrane. Proc. Natl. Acad. Sci. USA 1018372-8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burks, J. K., M. D. Summers, and S. C. Braunagel. 2007. BV/ODV-E26: a palmitoylated, multifunctional protein of Autographa californica nucleopolyhedrovirus. Virology 361194-203. [DOI] [PubMed] [Google Scholar]
- 10.Charlton, C. A., and L. E. Volkman. 1991. Sequential rearrangement and nuclear polymerization of actin in baculovirus-infected Spodoptera frugiperda cells. J. Virol. 651219-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellenberg, J., E. D. Siggia, J. E. Moreira, C. L. Smith, J. F. Presley, H. J. Worman, and J. Lippincott-Schwartz. 1997. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 1381193-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guillemain, G., J. J. Münoz-Alonso, A. Cassany, M. Loizeau, A.-M. Faussat, A.-F. Burnol, and A. Leturque. 2002. Karyopherin α2: a control step of glucose-sensitive gene expression in hepatic cells. Biochem. J. 364201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hachet, V., T. Köcher, M. Wilm, and I. W. Mattaj. 2004. Importin α associates with membrane and participates in nuclear envelope assembly in vitro. EMBO J. 231526-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong, T., S. C. Braunagel, and M. D. Summers. 1994. Transcription, translation, and cellular localization of PDV-E66: a structural proteins of the PDV envelope of Autographa californica nuclear polyhedrosis virus. Virology 204210-222. [DOI] [PubMed] [Google Scholar]
- 15.Hong, T., M. D. Summers, and S. C. Braunagel. 1997. N-terminal sequences from Autographa californica nuclear polyhedrosis virus envelope proteins ODV-E66 and ODV-E25 are sufficient to direct reporter proteins to the nuclear envelope, intranuclear microvesicles and the envelope of the occlusion-derived virus. Proc. Natl. Acad. Sci. USA 944050-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, A. E., and M. A. van Waes. 1999. The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 15799-842. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227680-695. [DOI] [PubMed] [Google Scholar]
- 18.Liao, S., J. Lin, H. Do, and A. E. Johnson. 1997. Both lumenal and cytosolic gating of the aqueous ER translocon pore is regulated from inside the ribosome during membrane protein integration. Cell 9031-41. [DOI] [PubMed] [Google Scholar]
- 19.Rexach, M. F. 2006. A sorting importin on Sec61. Nat. Struct. Mol. Biol. 13476-478. [DOI] [PubMed] [Google Scholar]
- 20.Rosas-Acosta, G., S. C. Braunagel, and M. D. Summers. 2001. Effects of deletion and overexpression of the Autographa californica nuclear polyhedrosis virus FP25K gene on synthesis of two occlusion-derived virus envelope proteins and their transport into virus-induced intranuclear membranes. J. Virol. 7510829-10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saksena, S., Y. Shao, S. C. Braunagel, M. D. Summers, and A. E. Johnson. 2004. Cotranslational integration and initial sorting at the endoplasmic reticulum translocon or proteins destined for the inner nuclear membrane. Proc. Natl. Acad. Sci. USA 10112537-12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saksena, S., M. D. Summers, J. K. Burks, A. E. Johnson, and S. C. Braunagel. 2006. Importin-α-16 is a translocon-associated protein involved in sorting membrane proteins to the nuclear envelope. Nat. Struct. Mol. Biol. 13500-508. [DOI] [PubMed] [Google Scholar]
- 23.Summers, M. D., and G. E. Smith. 1987. A manual of methods for baculovirus vectors and insect cell culture procedures. Texas Agric. Exp. Stn. Bull. 19871555. [Google Scholar]
- 24.Walter, P., and G. Blobel. 1983. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 9684-93. [DOI] [PubMed] [Google Scholar]