Abstract
Chlamydiae are unusual obligate intracellular bacteria that cause serious infections in humans. Chlamydiae contain genes that appear to encode products with peptidoglycan biosynthetic activity. The organisms are also susceptible to antibiotics that inhibit peptidoglycan synthesis. However, chlamydiae do not synthesize detectable peptidoglycan. The paradox created by these observations is known as the chlamydial anomaly. The MurC enzyme of chlamydiae, which is synthesized as a bifunctional MurC-Ddl product, is expected to possess UDP-N-acetylmuramate (UDP-MurNAc):l-alanine ligase activity. In this paper we demonstrate that the MurC domain of the Chlamydia trachomatis bifunctional protein is functionally expressed in Escherichia coli, since it complements a conditional lethal E. coli mutant possessing a temperature-sensitive lesion in MurC. The recombinant MurC domain was overexpressed in and purified from E. coli. It displayed in vitro ATP-dependent UDP-MurNAc:l-alanine ligase activity, with a pH optimum of 8.0 and dependence upon magnesium ions (optimum concentration, 20 mM). Its substrate specificity was studied with three amino acids (l-alanine, l-serine, and glycine); comparable Vmax/Km values were obtained. Our results are consistent with the synthesis of a muramic acid-containing polymer in chlamydiae with UDP-MurNAc-pentapeptide as a precursor molecule. However, due to the lack of specificity of MurC activity in vitro, it is not obvious which amino acid is present in the first position of the pentapeptide.
Chlamydiae are an important group of obligate intracellular pathogens that cause serious infections in humans (25). They are distinguished from other eubacteria by a unique development cycle involving two morphological forms, one adapted to extracellular survival (the infectious elementary body [EB]) and the other adapted to intracellular multiplication (the reticulate body [RB]) (22, 25). In the development cycle, entry of EBs into host cells is followed by transformation to RBs, RB division, and expansion of the chlamydial microcolony, followed by differentiation back to EBs and release from the infected cell (22, 25). The fragility and pleomorphism of the RB contrasts with the rigidity and stability of the EB (7, 22). These differences might be explained by the presence of peptidoglycan in EBs, which would confer mechanical stability. However, numerous attempts to detect peptidoglycan in EBs have consistently failed (8, 9, 12) and the rigidity of EBs is probably conferred by two cross-linked cysteine-rich envelope proteins whose synthesis is initiated during differentiation of RBs to EBs (7, 22).
Despite the failure to detect peptidoglycan in chlamydiae, bioinformatic analysis of the Chlamydia trachomatis and Chlamydia pneumoniae genomes reveals a complete set of genes whose products are apparently capable of synthesizing peptidoglycan in these organisms (3, 14, 28). A number of these genes are known to be expressed in chlamydiae and encode functional products. Thus, chlamydiae express penicillin-binding proteins (1, 29), and transcripts of murA (encoding UDP-N-acetylglucosamine-enolpyruvate transferase) and murB (encoding UDP-N-acetylglucosamine-enolpyruvate reductase) have been detected during the chlamydial growth cycle (21; T. Hatch, presented at the Ninth International Symposium on Human Chlamydial Infection, 1998). Furthermore, genes for chlamydial penicillin-binding proteins have been cloned and expressed in Escherichia coli and Schizosaccharomyces pombe (3), and in vitro and in vivo functional activity of MurA from C. trachomatis has recently been demonstrated (21). The susceptibility of chlamydiae to penicillin, d-cycloserine, and bacitracin is also consistent with expression of peptidoglycan biosynthetic enzymes in these organisms and provides further evidence that peptidoglycan has some fundamental role in chlamydiae (3). Nevertheless, the exact role of peptidoglycan in the metabolism and growth of chlamydiae remains obscure.
Since peptidoglycan has never been recovered from chlamydiae, it has not been possible to confirm its presence or to examine its structure. A characteristic feature of peptidoglycan in other bacteria is the presence of a tetrapeptide side chain covalently attached to N-acetylmuramic acid (MurNAc) in the glycan strands (26). Therefore if peptidoglycan is synthesized in chlamydiae, it is expected that chlamydial MurC will possess ATP-dependent amino acid ligase activity to synthesize the UDP-N-acetylmuramoyl-amino acid precursor containing the first amino acid of the tetrapeptide side chain. In bacterial peptidoglycans l-alanine is most often encountered at position 1 of the tetrapeptide, but in some species glycine or l-serine is an alternative (26). This can result from variation in the substrate specificity of MurC, unusually high concentrations of the alternative amino acids in the intracellular pool, or a combination of both factors (20, 32).
From genome examination, it appears that in chlamydiae MurC is synthesized as part of a fused MurC-Ddl product (30). In this paper we report the results of experiments which establish that the MurC domain can be functionally expressed in E. coli and that it catalyzes in vitro the addition of l-alanine, l-serine, or glycine to UDP-MurNAc with comparable efficiencies.
MATERIALS AND METHODS
Cloning of C. trachomatis murC.
The murC domain from the murC-ddl open reading frame of C. trachomatis L2 was amplified with introduced restriction sites by using the PCR primers MurCF1 (5′-CTCTATGAGCTCATGATG-3′ [SacI site underlined]) and MurCR1 (5′-CGGAAATCTAGAGTTCGC-3′ [XbaI site underlined]). MurCF1 binds at position 894943 and MurCR1 binds at position 896347, based on the GenBank nucleotide sequence NC_000117. The resulting 1,404-bp PCR product was ligated into the cloning vector pCR4-TOPO (Invitrogen, Paisley, United Kingdom). The insert was excised by using the restriction enzymes SacI and XbaI (New England Biolabs, Herefordshire, United Kingdom) and cloned into the compatible sites of the vector pTrc99A (Pharmacia). The resulting construct was designated pCTMurC. The insert was sequenced by using the primers PTRC99AF (5′-CCGACATCATAACGGTTCTGGC-3′) and PTRC99AR (5′-ATCAGACCGCTTCTGCGTTCTG-3′).
For overexpression studies, primers MurCF2 (5′-ATGAAAAGCTTGTTTTACC-3′) and MurCR2 (5′-TTTCCCTCCACAAATAATTCC-3′) were used to amplify the murC domain from the same template. MurCF2 binds at position 894958 and MurCR2 binds at 896325 of the GenBank nucleotide sequence NC_000117. The resulting 1,367-bp PCR product was ligated into the vector pGEM-T Easy (Promega, Southampton, United Kingdom). The insert was excised by using the restriction enzyme NotI (New England Biolabs) and ligated into NotI-digested pET30a (Novagen, Nottingham, United Kingdom). The resulting construct pCTHisSMurC was sequenced.
Complementation analysis with C. trachomatis murC.
The E. coli MurC temperature-sensitive mutant H1119 (18) was transformed with constructs pTrc99A and pCTMurC. Transformants were grown at the permissive temperature (30°C) before being shifted to the restrictive temperature (42°C). Cultures were incubated with and without the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) 5 min before the temperature shift. The growth rate was measured by recording the A600 at suitable time intervals.
Overexpression and purification of C. trachomatis MurC.
For recombinant overexpression and purification of the C. trachomatis MurC domain, plasmid pCTHisSMurC was transformed into E. coli BL21(DE3) (Promega) and grown in 10 liters of Terrific broth (24) at 37°C until an A600 of 0.6 was reached. Cultures were induced with 1 mM IPTG and grown for a further 3 h before bacteria were harvested by centrifugation. The cell pellet was resuspended in 100 ml of lysis buffer (50 mM sodium phosphate [pH 8], 300 mM NaCl, 5 mM 2-mercaptoethanol) containing 10 mM imidazole. Cells were then lysed by sonication (20 cycles of 20 s on and 60 s off; 4°C). Cell debris was pelleted by centrifugation at 100,000 × g in an ultracentrifuge for 1 h at 4°C. The clear supernatant was applied to an Ni-nitrilotriacetic acid (Ni-NTA) Superflow agarose-filled column (10 by 100 mm; Amersham) equilibrated with 10 mM imidazole in lysis buffer, according to the manufacturer's instructions. The column was washed with at least 10 column volumes of the same buffer and then with 60 and 155 mM imidazole in lysis buffer. The His-tagged protein eluted at 155 mM imidazole. Peak fractions containing MurC activity were pooled and concentrated by ultrafiltration (molecular weight cutoff, 10,000). The concentrated enzyme was loaded on a Superdex 200 pg column (16 by 600 mm; Amersham), equilibrated with storage buffer (50 mM Tris-HCl [pH 7.8], 3 mM dithiothreitol, 20% glycerol), at 0.75 ml min−1. Peak fractions were again concentrated in storage buffer and stored at −20°C.
Analysis of purified C. trachomatis MurC.
Purified MurC enzyme was analyzed on a Q-TOF mass spectrometer (Micromass UK Ltd., Altrincham, Cheshire, United Kingdom) in order to verify its identity by measurement of its molecular mass. The purified protein was also analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie brilliant blue staining.
Determination of the concentration of MurC.
The protein concentration in the active fractions (Table 1) was determined by quantitative amino acid analysis of acid hydrolysates (6 M HCl containing 1/2,000 [vol/vol] 2-mercaptoethanol; 105°C, 24 h) with an LC2000 amino acid analyzer (Biotronik, Frankfurt/Main, Germany).
TABLE 1.
Purification of recombinant C. trachomatis MurC
| Sample | Concn (mg ml−1) | Total protein (mg) | Purification (fold) | Activity (nmol min−1 mg of protein−1) |
|---|---|---|---|---|
| Soluble fraction | 13.2 | 1,320 | 1 | 5.8 ± 1.0a |
| Nickel-NTA eluate | 0.8 | 17.3 | 76b | 24.8 ± 1.7a |
| Gel filtration eluate | 1.1 | 5.4 | 244b | 73.8 ± 5.1c |
Mean ± standard deviation from two enzymatic determinations
Owing to the presence of chromosomal E. coli MurC in the soluble fraction, this value may be underestimated
Vmax value.
Enzyme assays.
The l-alanine-adding activity was assayed by monitoring the formation of UDP-MurNAc-l-[14C]Ala as described by Liger et al. (17). The final reaction mixture (50 μl) contained 200 mM Tris-HCl (pH 8.6); 40 mM (NH4)2SO4; 20 mM MgCl2; l-[14C]alanine (3.4 kBq; 5.5 GBq mmol−1) (Amersham); various concentrations of ATP, UDP-MurNAc, and unlabeled l-alanine; and enzyme dilution [in 20 mM potassium phosphate (pH 7.2)-40 mM (NH4)2SO4-1 mM EDTA-2.5 mM 2-mercaptoethanol-15% glycerol]. The exact concentrations of the variable substrates are provided in Table 2. The mixture was incubated at 37°C for 30 min, and the reaction was stopped by addition of 10 μl of glacial acetic acid followed by lyophilization. The residue was dissolved in 100 μl of 50 mM ammonium formate (pH 4.3), and 80 μl was injected on a Nucleosil 5 C18 column (4.6 by 150 mm; Alltech France, Templemars, France) with the same buffer at 0.6 ml min−1 as the mobile phase. Detection was performed with a radioactive flow detector (model LB506-C1; Berthold France, La Garenne-Colombes, France) and the Quicksafe Flow 2 scintillator (Zinsser Analytic, Maidenhead, United Kingdom) at 0.6 ml min−1. Quantitation was carried out with the Winflow software (Berthold). Data were fitted to the equation v = VA/(K + A) by using the Prism program (Graphpad, San Diego, Calif.) (where v is the experimentally determined rate, V is the maximum velocity, A is the substrate concentration, and K is the Michaelis constant). For the determination of the kinetic constants for the alternative substrates, l-[14C]serine (5.5 GBq mmol−1) and [14C]glycine (3.96 GBq mmol−1) (both from CEA, Saclay, France) were used. pH dependence was determined in this assay at 100 mM sodium HEPES in the pH range of 7 to 9.
TABLE 2.
Kinetic parameters of recombinant C. trachomatis MurC
| Substratea | Km (mM) (mean ± SD) | Vmax (nmol/min−1 mg of protein−1) | Vmax/Km (nmol min−1 mM−1 mg of protein−1) |
|---|---|---|---|
| UDP-MurNAcb | 0.196 ± 0.065 | —c | 376 |
| ATPb | 0.162 ± 0.040 | —c | 456 |
| l-Alanine | 0.124 ± 0.037 | 73.8 ± 5.1 | 595 |
| l-Serine | 0.242 ± 0.067 | 76.6 ± 4.4 | 316 |
| Glycine | 1.17 ± 0.19 | 206 ± 10 | 176 |
Concentrations of the fixed substrates: ATP, 2.5 mM; UDP-MurNAc, 2 mM; l-alanine, 10 mM. The concentration range of the varied substrates was 0 to 5 mM, except that for glycine, which was 0 to 10 mM.
Determined with l-alanine as the amino acid substrate.
The Vmax is the same as that for l-alanine.
Substrate preparation.
UDP-MurNAc was prepared by using purified E. coli MurA and MurB in a complete reaction with the substrates UDP-N-acetylglucosamine (UDP-GlcNAc), phosphoenolpyruvate, and NADPH. The reaction was set up as described by Jin et al. (13) with some modifications. Typically, a reaction was in a 15-ml total volume at 37°C by adding 3 mg of MurA to a mixture containing 100 mM sodium HEPES (pH 8), 3 mM dithiothreitol, 20 mM UDP-GlcNAc, 30 mM NADPH, and 30 mM phosphoenolpyruvate. At 10 and 60 min, 3 mg of MurB was added, and the disappearance of NADPH was monitored at 340 nm. After completion of the reaction (at 2 h), the enzymes were removed by ultrafiltration with a membrane with a molecular mass cutoff of 5,000 Da. The filtrate was loaded onto a Q-Sepharose column (50 by 200 mm; Amersham) equilibrated with buffer A (20 mM ammonium acetate [pH 5]); after a 2-column-volume washing with 30% buffer B (1 M ammonium acetate [pH 5]) in buffer A, the product was eluted with 40% buffer B in buffer A. The peak containing UDP-MurNAc was freeze-dried three or four times to remove trace amounts of the buffer and then dissolved in water. The compound was characterized by Q-TOF mass spectrometry; the m/z ratio of 678.0 determined for the [M − H]− ion was in agreement with the calculated value. The yield was ≥90%.
RESULTS
Complementation analysis of C. trachomatis MurC in an E. coli temperature-sensitive MurC mutant.
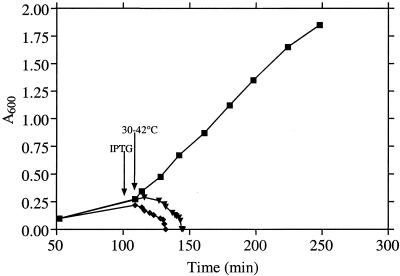
The murC domain of the C. trachomatis murC-ddl open reading frame was cloned into the IPTG-inducible expression vector pTrc99A to create pCTMurC. The vector and pCTMurC were transformed separately into the temperature-sensitive E. coli strain H1119, harboring a defective murC gene. The aim was to determine whether the chlamydial gene in pCTMurC was active and able to complement the temperature-sensitive defect in the E. coli MurC mutant. Growth of E. coli H1119(pTrc99A) to early logarithmic phase at 30°C, followed by a shift to 42°C, led to rapid lysis of the culture (Fig. 1). A similar profile was obtained with cultures of H1119(pCTMurC) to which no IPTG had been added (Fig. 1). In contrast, cultures of H1119(pCTMurC) to which IPTG was added at the time of the temperature shift continued growing at 42°C (Fig. 1). Thus, IPTG-induced expression of chlamydial MurC encoded by pCTMurC permitted complementation of the temperature-conditional UDP-MurNAc:l-alanine ligase (MurC) defect in E. coli. Furthermore, it can be concluded that the Ddl domain of the fused chlamydial MurC-Ddl protein was not essential for the UDP-MurNAc:l-alanine ligase activity of the MurC domain.
FIG. 1.
Growth of E. coli H1119(pTrc99A) with IPTG (♦), H1119(pCTmurC) without IPTG (▾), and H1119(pCTmurC) with IPTG (▪). Cultures were grown at 30°C, and IPTG (1 mM) was added at 110 min as indicated by the arrow. Cultures were then shifted to 42°C, and the culture absorbance (600 nm) was monitored for a further 140 min.
Overexpression and purification of the chlamydial MurC domain.
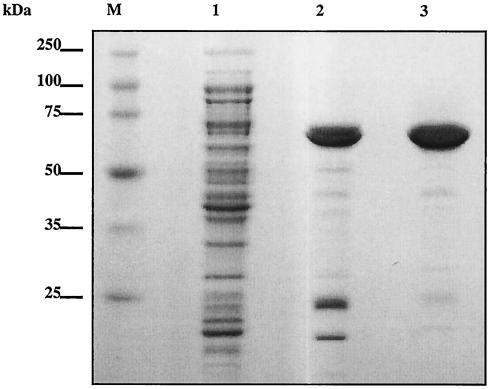
We subcloned the murC domain of the murC-ddl gene for recombinant expression in the pET system. The N-terminal murC domain encoding amino acid residues 2 to 457 of MurC-Ddl was cloned into the pET30a vector, which introduces a His tag and an S tag at the N terminus of the recombinant protein. T7 RNA polymerase-based expression was performed in E. coli BL21(DE3). Expression was induced with 1 mM IPTG, and the tagged protein was purified by Ni-NTA chromatography followed by gel filtration chromatography. Enzyme purification was monitored by SDS-PAGE (Fig. 2). The recombinant MurC exhibited a molecular mass of approximately 65 kDa. Contaminating low-molecular-mass proteins from the eluate of the Ni-NTA column were removed by gel filtration chromatography, resulting in a pure preparation of the enzyme. The protein concentration determined by quantitative amino acid analysis demonstrated that 0.54 mg of purified enzyme was isolated per liter of bacterial culture (Table 1.). This relatively low expression level may be due to differences in codon usage between Chlamydia and E. coli (15). Overall, a 244-fold purification was achieved with this protocol. The purified protein exhibited a specific activity of approximately 74 nmol of UDP-MurNAc-l-Ala formed/min/mg of protein.
FIG. 2.
Purification of recombinant C. trachomatis MurC. Lane 1, soluble fraction of the lysed E. coli BL21(DE3) strain transformed with pCTHisSMurC and induced with 1 mM IPTG (10 μg); lane 2, active fraction from the Ni-NTA agarose column (3 μg); lane 3, active fraction from the Superdex 200 pg column (3 μg); lane M, molecular mass markers. Samples were loaded on an SDS-10% polyacrylamide gel; the gel was stained with Coomassie brilliant blue.
Characterization of purified MurC.
The calculated molecular mass of the His- and S-tagged MurC enzyme is 57,760.9 Da. The apparent molecular mass determined by SDS-PAGE was 65 kDa. This was somewhat higher than expected. Nevertheless, the molecular mass measured by mass spectrometry was 57,763.1 Da, which is very close to the calculated one. Such discrepancies between apparent molecular masses determined by SDS-PAGE analysis and true molecular masses are often observed. The value found by mass spectrometry indicated that the N-terminal methionine residue was not removed after translation. This was also the case for the overproduced MurC from E. coli (17).
Requirements for the incorporation of l-alanine into UDP-MurNAc-l-alanine.
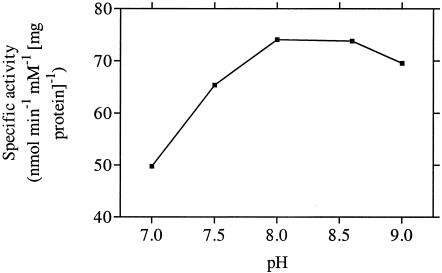
Since the complementation studies indicated that chlamydial MurC possessed l-alanine-adding activity, the overall properties of the enzyme with respect to this activity were determined. A pH optimum of between 8.0 and 9.0 was observed (Fig. 3). The reaction of MurC was dependent on magnesium chloride, with maximal enzymatic activity at 20 mM MgCl2. It was stimulated slightly (by 20%) by ammonium sulfate at concentrations above 25 mM.
FIG. 3.
Dependence of C. trachomatis UDP-MurNAc:l-alanine ligase activity on pH.
Kinetic parameters of chlamydial MurC and comparison of relative catalytic efficiencies of l-alanine, l-serine, and glycine.
E. coli MurC possesses the ability to add l-alanine, l-serine, or glycine to UDP-MurNAc (5, 11, 17). The purified chlamydial MurC also showed ligase activity with radioactive l-alanine, l-serine, or glycine as a substrate, consistent with the formations of UDP-MurNAc-l-Ala, UDP-MurNAc-l-Ser, and UDP-MurNAc-Gly, respectively. The kinetic parameters for chlamydial MurC are presented in Table 2. The enzyme showed comparable Km and Vmax values for l-Ala and l-Ser and hence showed similar Vmax/Km ratios (595 and 316 nmol min−1 mM−1 mg of protein−1, respectively). The Vmax/Km ratio for glycine (176 nmol min−1 mM−1 mg of protein-1) was lower than those for alanine and serine.
DISCUSSION
Bacterial peptidoglycan is a giant macromolecule with a periodic structure whose basic unit is a disaccharide of alternating β-1,4-linked N-acetylglucosamine and N-acetylmuramic acid residues (23, 26). The linear GlcNAc-MurNAc backbone is cross-linked laterally through the formation of peptide bonds between neighboring pentapeptide side chains attached to MurNAc (23, 26). The synthesis of the pentapeptide side chain proceeds via a well-defined sequence of cytoplasmic reactions (23). In E. coli, a group of ATP-dependent ligases sequentially add l-alanine (MurC), d-glutamic acid (MurD), meso-diaminopimelic acid (MurE), and a preformed d-alanyl-d-alanine dipeptide (MurF) to UDP-MurNAc, thus forming UDP-MurNAc-pentapeptide (31) (Fig. 4).
FIG. 4.
Biosynthesis of UDP-MurNAc-pentapeptide in E. coli.
The murC gene from C. trachomatis is predicted to encode UDP-MurNAc:l-alanine ligase activity (3, 10). However, the C. trachomatis and E. coli murC products are not highly related at the amino acid sequence level (4). Therefore, it is uncertain whether chlamydial MurC would function in E. coli. The complementation analysis reported here clearly demonstrates that the MurC domain of the fused chlamydial MurC-Ddl protein displays UDP-MurNAc:l-alanine ligase activity when expressed in E. coli. Furthermore, since a recombinant, enzymatically active chlamydial murC product could be purified from E. coli, the enzyme can clearly tolerate the presence of a nonhomologous environment that presumably does not adversely affect the folding properties of the protein.
The Vmax/Km ratios reported here for chlamydial MurC (176 to 595 nmol min−1 mM−1 mg of protein−1) are very low compared to those found for the E. coli enzyme (565,000, ∼13,000, and ∼4,500 nmol min−1 mM−1 mg of protein−1 for l-Ala, l-Ser, and Gly, respectively [17]). However, they are higher than those determined for MurC from Mycobacterium tuberculosis and Mycobacterium leprae (3 to 86 nmol min−1 mM−1 mg of protein−1 [20]). Whether such low values represent the true in vivo functioning of chlamydial and mycobacterial MurC enzymes or are underestimated (e.g., due to lack of an in vivo activator or, in the case of chlamydiae, to better catalytic efficiency of the MurC-Ddl protein) is unknown. Nevertheless, it is possible that the low catalytic activity of MurC from chlamydiae and mycobacteria is sufficient for the requirements of these slow-growing, primarily intracellular bacteria.
MurC enzymes from conventional bacteria have the capacity to add l-alanine, l-serine, or glycine to UDP-MurNAc in vitro (5, 11, 17, 20, 32). In general, the in vitro and in vivo results correlate well; i.e., the best amino acid substrate in vitro is the one which is added in vivo. However, in M. leprae, the correlation is not found: although l-Ala is favored in vitro (Vmax/Km = 46 and 3 nmol min−1 mM−1 mg of protein−1 for l-Ala and Gly, respectively), Gly is preferentially incorporated in vivo (20). In the case of chlamydial MurC, where the lack of specificity for l-Ala, l-Ser, and Gly is demonstrated (Table 2), it is not immediately obvious which UDP-MurNAc:l-amino acid ligase activity is displayed by the enzyme within the normal chlamydial host. This activity could be influenced by the concentration of alternative amino acids in the chlamydial cell pool, about which nothing is known.
With respect to pH and magnesium dependence, chlamydial MurC displayed properties similar to those of the well characterized E. coli enzyme (11, 17). On the basis of the pH dependency of chlamydial MurA, which is extensively inactivated above pH 7.0, McCoy et al. (21) have proposed that changes in the pH from around 6.0 early in the chlamydial infection cycle to more alkaline conditions at later stages may serve to control the enzymatic activity of MurA within the inclusion body. This proposed alkaline-induced control system for chlamydial MurA activity is unlikely to apply for MurC, since this enzyme displayed optimal activity at pH values of above 7.0 (Fig. 3).
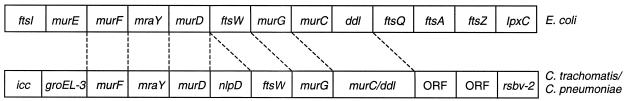
Although muramic acid has never been detected in chlamydiae, our observations add to the growing body of evidence that this molecule occurs in these organisms, probably as a component of trace quantities of peptidoglycan (3, 21). Another similarity to organisms known to synthesize peptidoglycan is exhibited by the clustering and order of chlamydial murF, mraY, murD, ftsW, murG, and murC-ddl, which is analogous to the presence and relative positions of these genes in the 2-min region of the E. coli chromosome (Fig. 5). However, correlation with the E. coli 2-min region is not absolute, since murE maps to this region in E. coli but does not in chlamydiae and since murD and ftsW in chlamydiae are separated by nlp (Fig. 5).
FIG. 5.
Comparison of the genomic regions containing the mur genes in E. coli and in C. trachomatis and C. pneumoniae. ORF, open reading frame encoding a product of unknown function. The diagram is not drawn to scale.
Although the function of peptidoglycan in chlamydiae is unknown, we have previously suggested that it could have a role in the division of chlamydial RBs (3). In other bacteria, FtsZ plays an essential role as a cytoskeletal element during cell division (6, 19). The fact that an ftsZ homologue has not been found in the chlamydial genome (14, 28) reinforces the concept that chlamydiae divide by a mechanism that is different from that of other bacteria. Since chlamydiae are obligate intracellular bacteria, they may be able to use the host cell's machinery for their own division (6). However, the hypothesis that peptidoglycan is involved in RB division is consistent with the recent identification of a unique, possibly nonproteinaceous, division-specific antigen in chlamydiae (2, 6). Earlier observations also suggest that peptidoglycan may have a role in RB division, because the addition of penicillin to chlamydial cultures leads to the formation of large RB-like forms that resume development when penicillin is removed (3).
Apart from the intriguing debate on the role of peptidoglycan in chlamydiae, there is another interesting aspect concerning the activities of MurC and Ddl in the proposed chlamydial biosynthetic pathway for the polymer. In other bacteria, MurC and Ddl are synthesized as separate proteins that catalyze different steps in the synthesis of the peptidoglycan precursor molecule UDP-MurNAc-pentapeptide (Fig. 4). In contrast, chlamydial MurC and Ddl activities reside in a single fused protein (30). Such a feature is usually associated with bifunctional enzymes that process sequential reactions in a pathway and for which substrate exchange between the two enzymatic domains through channeling within the protein may be advantageous (16, 27). Therefore, it is surprising to find MurC and Ddl as a fusion protein, since they do not catalyze sequential reactions. The significance of this phenomenon in chlamydiae is unknown. Perhaps the synthesis of this unusual enzyme is related to the novel requirement for peptidoglycan in the division of chlamydial RBs. Another possibility might be the formation of a multienzyme complex by all of the chlamydial Mur enzymes.
Acknowledgments
We thank Alison Ashcroft (University of Leeds, Leeds, United Kingdom) for assistance with mass spectrometry and Nicola Wallis (GlaxoSmithKline, Collegeville, Pa.) for purified E. coli MurA and MurB.
This work was supported by a grant to I.C. from the Wellcome Trust.
REFERENCES
- 1.Barbour, A. G., K. Amano, T. Hackstadt, L. Perry, and H. D. Caldwell. 1982. Chlamydia trachomatis has penicillin-binding proteins but not detectable muramic acid. J. Bacteriol. 151:420-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, W. J., and D. D. Rockey. 2000. Identification of an antigen localized to an apparent septum within dividing chlamydiae. Infect. Immun. 68:708-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chopra, I., C. Storey, T. J. Falla, and J. H. Pearce. 1998. Antibiotics, peptidoglycan synthesis and genomics: the chlamydial anomaly revisited. Microbiology 144:2673-2678. [DOI] [PubMed] [Google Scholar]
- 4.El Zoeiby, A., F. Sanschagrin, J. Lamoureux, A. Darveau, and R. C. Levesque. 2000. Cloning, over-expression and purification of Pseudomonas aeruginosa murC encoding uridine diphosphate N-acetylmuramate:l-alanine ligase. FEMS Microbiol. Lett. 183:281-288. [DOI] [PubMed] [Google Scholar]
- 5.Emanuele, J. J. J., H. Jin, B. L. Jacobson, C. Y. Chang, H. M. Einspahr, and J. J. Villafranca. 1996. Kinetic and crystallographic studies of Escherichia coli UDP-N-acetylmuramate:l-alanine ligase. Protein Sci. 5:2566-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Erickson, H. P. 2000. Dynamin and FtsZ. Missing links in mitochondrial and bacterial division. J. Cell Biol. 148:1103-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everett, K. D., and T. P. Hatch. 1995. Architecture of the cell envelope of Chlamydia psittaci 6BC. J. Bacteriol. 177:877-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox, A., J. C. Rogers, J. Gilbart, S. Morgan, C. H. Davis, S. Knight, and P. B. Wyrick. 1990. Muramic acid is not detectable in Chlamydia psittaci or Chlamydia trachomatis by gas chromatography-mass spectrometry. Infect. Immun. 58:835-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrett, A. J., M. J. Harrison, and G. P. Manire. 1974. A search for the bacterial muropeptide component, muramic acid, in Chlamydia. J. Gen. Microbiol. 80:315-318. [DOI] [PubMed] [Google Scholar]
- 10.Ghuysen, J. M., and C. Goffin. 1999. Lack of cell wall peptidoglycan versus penicillin sensitivity: new insights into the chlamydial anomaly. Antimicrob. Agents Chemother. 43:2339-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gubler, M., Y. Appoldt, and W. Keck. 1996. Overexpression, purification, and characterization of UDP-N-acetylmuramyl:l-alanine ligase from Escherichia coli. J. Bacteriol. 178:906-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.How, S. J., D. Hobson, and C. A. Hart. 1984. Studies in vitro of the nature and synthesis of the cell wall of Chlamydia trachomatis. Curr. Microbiol. 10:269-274. [Google Scholar]
- 13.Jin, H., J. J. J. Emanuele, R. Fairman, J. G. Robertson, M. E. Hail, H. T. Ho, P. J. Falk, and J. J. Villafranca. 1996. Structural studies of Escherichia coli UDP-N-acetylmuramate:l-alanine ligase. Biochemistry 35:1423-1431. [DOI] [PubMed] [Google Scholar]
- 14.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 15.Kanaya, S., Y. Yamada, Y. Kudo, and T. Ikemura. 1999. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 238:143-155. [DOI] [PubMed] [Google Scholar]
- 16.Liang, P. H., and K. S. Anderson. 1998. Substrate channeling and domain-domain interactions in bifunctional thymidylate synthase-dihydrofolate reductase. Biochemistry 37:12195-12205. [DOI] [PubMed] [Google Scholar]
- 17.Liger, D., A. Masson, D. Blanot, J. van Heijenoort, and C. Parquet. 1995. Over-production, purification and properties of the uridine-diphosphate-N-acetylmuramate:l-alanine ligase from Escherichia coli. Eur. J. Biochem. 230:80-87. [DOI] [PubMed] [Google Scholar]
- 18.Lugtenberg, E. J., and A. von Schijndel-van Dam. 1972. Temperature-sensitive mutants of Escherichia coli K-12 with low activities of the l-alanine adding enzyme and the d-alanyl-d-alanine adding enzyme. J. Bacteriol. 110:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutkenhaus, J., and S. G. Addinall. 1997. Bacterial cell division and the Z ring. Annu. Rev. Biochem. 66:93-116. [DOI] [PubMed] [Google Scholar]
- 20.Mahapatra, S., D. C. Crick, and P. J. Brennan. 2000. Comparison of the UDP-N-acetylmuramate:l-alanine ligase enzymes from Mycobacterium tuberculosis and Mycobacterium leprae. J. Bacteriol. 182:6827-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCoy, A. J., R. C. Sandlin, and A. T. Maurelli. 2003. In vitro and in vivo functional activity of Chlamydia MurA, a UDP-N-acetylglucosamine enolpyruvyl transferase involved in peptidoglycan synthesis and fosfomycin resistance. J. Bacteriol. 185:1218-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moulder, J. W. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55:143-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogers, H. J., H. R. Perkins, and J. B. Ward. 1980. Microbial cell walls and membranes. Chapman and Hall, London, United Kingdom.
- 24.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Schachter, J. 1992. Chlamydia, p. 1633-1641. In S. L. Gorbach, J. G. Bartlett, and N. R. Blacklow (ed.), Infectious diseases. W. B. Saunders, Philadelphia, Pa.
- 26.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spivey, H. O., and J. Ovádi. 1999. Substrate channeling. Methods 19:306-321. [DOI] [PubMed] [Google Scholar]
- 28.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 29.Storey, C., and I. Chopra. 2001. Affinities of beta-lactams for penicillin binding proteins of Chlamydia trachomatis and their antichlamydial activities. Antimicrob. Agents Chemother. 45:303-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vandahl, B. B., S. Birkelund, H. Demol, B. Hoorelbeke, G. Christiansen, J. Vandekerckhove, and K. Gevaert. 2001. Proteome analysis of the Chlamydia pneumoniae elementary body. Electrophoresis 22:1204-1223. [DOI] [PubMed] [Google Scholar]
- 31.van Heijenoort, J. 2001. Recent advances in the formation of the bacterial peptidoglycan monomer unit. Nat. Prod. Rep. 18:503-519. [DOI] [PubMed] [Google Scholar]
- 32.Wyke, A. W., and H. R. Perkins. 1975. The specificity of enzymes adding amino acids in the synthesis of the peptidoglycan precursors of Corynebacterium poinsettiae and Corynebacterium insidiosum. J. Gen. Microbiol. 88:159-168. [DOI] [PubMed] [Google Scholar]