Abstract
The phage shock protein (psp) operon of Escherichia coli is induced by membrane-damaging cues. Earlier studies linked defects in secretion across the inner membrane to induction of the psp response. Here we show that defects in yidC and sec secretion induce psp but that defects in tat and srp have no effect. We have also determined the cellular location of PspB and PspD proteins.
The pspABCDE operon of Escherichia coli is specifically associated with a wide variety of membrane stresses. Expression of the pspABCDE operon is strongly induced by the infection of E. coli with filamentous phage f1 and, more specifically, by expression of f1 gene IV, which encodes protein IV (pIV). The pIV protein is a member of the secretin family that forms an outer membrane (OM) pore through which phage is extruded. Other stresses, including heat shock, osmotic shock, and exposure to organic solvents, induce psp (for a review, see reference 24). Overexpression of PulD, an OM secretin, also mediated up-regulation of PspA; however, this effect was reduced when the periplasmic chaperone PulS was coexpressed (14). Translocation of mutant forms of the OM protein maltoporin, encoded by lamB (7), and PhoE, a pore-forming OM protein (16), resulted in up-regulation of PspA. Furthermore, chromosomal mutations in secA, secD, and secF up-regulate expression of PspA, whereas mutations in secE and secY do not. For the secA51 mutation, it was speculated that proteins might be trapped inside the translocase due to interruption of SecA ATP hydrolysis functioning, but this was not confirmed experimentally (16). SecDF depletion leads to cells being unable to maintain the proton motive force (PMF) across the inner membrane (2). PspA is not part of the secretion apparatus but may act to maintain PMF, since in an E. coli mutant lacking pspA, PMF decreased specifically compared to the level in the wild type when a mutant form of PhoE was overproduced (17). Recent work has identified new participants in protein secretion in E. coli, which led us to reexamine the determinants for induction of the psp response. We wished to clarify whether psp specifically responds to Sec-related stress or to more general secretion stresses at the inner membrane.
Secretion of proteins destined for the periplasm or OM is mediated by several different systems in E. coli.
The SecYEG translocase translocates proteins across the inner membrane. Secretion requires energy (from ATP hydrolysis) and the proton gradient and electrochemical potential components of the PMF. Since earlier studies linking secretion problems to psp induction were carried out, several other components of the secretion apparatus, including the bacterial signal recognition particle (SRP), the Tat system, and YidC, have been identified. The bacterial SRP, which presents proteins to the Sec translocase, functions to target and integrate cytoplasmic membrane proteins. In E. coli, the SRP comprises ffh (48-kDa protein), ffs (4.5S RNA), and ftsY (the SRP receptor in the inner membrane) (3, 25, 27, 32). The Tat system (twin-arginine translocation pathway) transports a specific subset of fully folded E. coli proteins across the bacterial inner membrane (reviewed in reference 26). YidC copurifies with SecY and is part of the secYEG/secDF yajC complex. YidC interacts with the translocase through a bridging interaction with SecDF YajC. YidC can work with the Sec translocase but also functions independently to facilitate the insertion of Sec-dependent and Sec-independent proteins into the membrane bilayer (9). Some evidence suggests that the SecDF complex is involved in the release of proteins into the periplasmic space (21) and can prevent the backsliding of partly translocated proteins from the translocase to the cytoplasm (11). YidC is hypothesized to contribute to the clearing of the translocon channel. YidC depletion does not affect protein export unless a Sec-dependent protein is overexpressed. Its primary role is in membrane protein topogenesis (reviewed in reference 9). Yi et al. (35) have recently shown a role for YidC in the function of the F1Fo ATP synthase.
One hypothesis for psp induction is that slowed translocation, or slowed folding of an OM protein into its correct conformation, may be a trigger for PspA synthesis. Therefore, we hypothesized that defects in the SRP, Tat, or YidC may trigger the induction of psp. We decided to further probe the nature of the inducing signal for psp through a combination of depletion experiments and defined mutations. Induction signals for psp are poorly defined, placing an emphasis on the need to establish which translocation systems might be sensed by the psp system. Crucially, several systems previously used to study the induction of psp utilized an overexpression of membrane proteins which might themselves interact with Psp proteins. All the mutants used in this study have been well characterized and are chromosomally located, and the assays that were applied rely on conventional growth conditions without overexpression of membrane proteins, except key positive control assays. Pairs of strains used were isogenic or very closely related.
Mutations in all the components of the bacterial SRP do not up-regulate pspA expression.
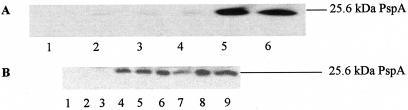
A reporter gene fusion construct (pSJ1) was made by PCR amplification of the pspA promoter with primers SE5 and SE6 (13) and then cloning into vector pMR25 to fuse the pspA promoter to lacZ. Verification was done by sequence analysis. The clone, pSJ1, allowed a specific assay for levels of pspA transcription, since stop codons are present in all three reading frames and a translational start site is provided for the promoterless lacZ gene (Table 1). The vector was present at one to five copies per cell. β-Galactosidase assays of the appropriate plasmid-containing strain, with tetracycline present at 10 μg/ml, were carried out with a 100-fold dilution of overnight culture into Luria-Bertani broth (LB) with the appropriate antibiotics (28). Cultures were grown at 37°C to an optical density at 600 nm (OD600) of 0.6 to 0.8, and the activity was assayed (23) and expressed as the number of units at an OD600 (Miller units). By classical induction with pIV harbored on pPMR129 (10), pspA up-regulation was confirmed (data not shown). pMR25 (control vector) or pSJ1 (pspA-lacZ) was transformed into HPT strains (Table 1), each of which harbors defined point mutations in various components of the bacterial SRP (33). β-Galactosidase activity was determined after growth overnight (data not shown) or after growth to an OD600 of between 0.6 and 0.8 (Table 1). In all cases, the levels of β-galactosidase observed were comparable to those of the appropriate parental strain. Hence, mutations that cause defects in the presentation of inner membrane proteins to the translocon (and hence, accumulation of proteins in the cytoplasm) do not confer any significant up-regulation of psp. pMR25 and pSJ1 were also introduced into MC4100 (wild type) and HDB45 (ffh::kan-1; resulting in pHDB4, where pHDB4 harbors ffh under trc promoter control) (4). HDB45 can be used to deplete levels of the SRP. Strains were all grown in the presence of 10 mM IPTG (isopropyl-β-d-thiogalactopyranoside) to maintain Ffh levels. Where necessary, cultures were washed with LB to remove IPTG and grown in LB without IPTG to determine the effect of depleting Ffh. Table 1 shows that depletion of Ffh in HDB45 did not up-regulate pspA expression, confirming that defects in the SRP fail to induce pspA. In contrast, both HPT73 and HPT93 up-regulated psp at least sixfold (compare the results for the HPT57 wild type harboring pSJ1 to those for HPT73 and HPT93 harboring pSJ1) (Table 1). Both strains harbor mutations in secM, which is not part of the SRP. Western blotting was used to determine whether PspA was detectably up-regulated at the protein level (Fig. 1). For generation of antibodies, the pspA gene from pET28b+ harboring a six-His-tagged pspA gene (13) was subcloned into pET29a+ with BamHI and NdeI to allow overexpression of native nontagged PspA. PspA was overexpressed by using E. coli B834(DE3) and partially purified by extraction with 1.1% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, essentially as described by Elderkin et al. (13). PspA was excised from a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel (15% polyacrylamide), eluted with 0.1 M sodium acetate and 0.1% (wt/vol) SDS, and then dried under a vacuum to lyophilize the protein. Naïve BALB/c mice were injected with 10 μg of PspA protein mixed 1:1 (vol/vol) with Freund's adjuvant (Sigma). Mice received a 10-μg booster injection of protein, as before, on days 20 and 40 after the primary injection. Serum was taken from mice on day 60 and was judged by Western blotting exactly as described previously (13) with a 1:10,000 dilution of PspA and a 1:8,000 dilution of horseradish peroxidase-conjugated rabbit anti-mouse antibody (Sigma) to detect bound PspA antibody from samples of whole-cell extract of E. coli B834(DE3) harboring pET29a+ (pspA). The wild type (HPT57), HPT73, and HPT93 were grown to saturation and then diluted to an OD600 of 0.05 in LB and grown at 37°C. Samples were taken at 2, 4, and 6 h postdilution and were analyzed by SDS-PAGE. Equal amounts of cells were loaded in each lane, and no significant differences in growth rate were noticed. Western blotting of samples using the PspA antibody clearly revealed significant up-regulation of PspA protein (Fig. 1B) in the secM mutants.
TABLE 1.
Survey of induction of pspA by chromosomal mutations in secretion loci of E. coli
| Straina | β-Galactosidase assay result (Miller units)b | Reference |
|---|---|---|
| JS7131(pMR25) ara | 6 ± 0 | 29 |
| JS7131(pSJ1) ara | 59 ± 0.4 | 29 |
| JS7131(pMR25) glu | 8 ± 0.29 | 29 |
| JS7131(pSJ1) glu | 277 ± 1.9 | 29 |
| MC4100(pMR25) | 15.4 ± 0.7 | 8 |
| MC4100(pSJ1) | 183 ± 3.5 | 8 |
| BILKO (MC4100 ΔtatC)(pMR25) | 17 ± 0.3 | 6 |
| BILKO (MC4100 ΔtatC)(pSJ1) | 180 ± 0.17 | 6 |
| ELV15 (MC4100 ΔtatA)(pMR25) | 33 ± 0.9 | 30 |
| ELV15 (MC4100 ΔtatA)(pSJ1) | 176 ± 2.6 | 30 |
| J1M1 (MC4100 ΔtatE)(pMR25) | 31 ± 0.8 | 30 |
| J1M1 (MC4100 ΔtatE)(pSJ1) | 160 ± 0.9 | 30 |
| JARV15 (MC4100 ΔtatA ΔtatE)(pMR25) | 34 ± 0.4 | 30 |
| JARV15 (MC4100 ΔtatA ΔtatE)(pSJ1) | 172 ± 1.7 | 30 |
| HPT57 (MC1000 phoA+ phoR leu+ 102::malF-lacZ102)(pMR25) | 3.6 ± 0.7 | 33 |
| HPT57 (MC1000 phoA+ phoR leu+ 102::malF-lacZ102)(pSJ1) | 39 ± 0.8 | 33 |
| HPT-UV46 (ffs-46) (pMR25) | 4.6 ± 0.8 | 33 |
| HPT-UV46 (ffs-46) (pSJ1) | 45.6 ± 2.9 | 33 |
| HPT-UV70 (ftsY70) (pMR25) | 5 ± 2.3 | 33 |
| HPT-UV70 (ftsY70) (pSJ1) | 26 ± 0.8 | 33 |
| HPT-UV73 (secM73) (pMR25) | 5.2 ± 0.2 | 33 |
| HPT-UV73 (secM73) (pSJ1) | 418 ± 0.8 | 33 |
| HPT-UV77 (ffh-77) (pMR25) | 2.2 ± 1.3 | 33 |
| HPT-UV77 (ffh-77) (pSJ1) | 34 ± 0.65 | 33 |
| HPT-UV87 (ffh-87) (pMR25) | 4.8 ± 0.3 | 33 |
| HPT-UV87 (ffh-87) (pSJ1) | 23.1 ± 0.84 | 33 |
| HPT-UV89 (ffh-89) (pMR25) | 2.9 ± 0.9 | 33 |
| HPT-UV89 (ffh-89) (pSJ1) | 49 ± 2.2 | 33 |
| HPT-UV93 (secM93) (pMR25) | 7.4 ± 2.3 | 33 |
| HPT-UV93 (secM93) (pSJ1) | 420 ± 9.9 | 33 |
| HPT-UV103 (ffh-103) (pMR25) | 4.9 ± 0.49 | 33 |
| HPT-UV103 (ffh-103) (pSJ1) | 37.7 ± 0.27 | 33 |
| MC4100(pMR25)c | 1.36 ± 0.79 | 4 |
| MC4100(pMR25)d | 2.57 ± 0.8 | 4 |
| MC4100(pSJ1)c | 37.3 ± 0.8 | 4 |
| MC4100(pSJ1)d | 38.4 ± 0.6 | 4 |
| HDB45(pMR25)c | 3.0 ± 0.58 | 4 |
| HDB45(pMR25)d | 3.3 ± 0.86 | 4 |
| HDB45(pSJ1)c | 37.1 ± 0.35 | 4 |
| HDB45(pSJ1)d | 36.4 ± 1.09 | 4 |
Where present, the pMR25 vector numbered one to five copies per cell (C. Mohr and R. Roberts, unpublished data) (a gift from Dickon Alley). ara or glu, cultures were grown with LB plus 0.2% arabinose or glucose, respectively. pSJ1 is pMR25 with a pspA-lacZ transcriptional fusion. JS7131 strains were supplied by T. Palmer. The BILKO, ELV15, J1M1, and JARV15 strains, as well as the MC4100(pMR25) and MC4100(pSJ1) strains described in reference 8, were supplied by R. Dalbey. The HPT strains were supplied by J. Beckwith. The HBD strains and the MC4100(pMR25) and MC4100(pSJ1) strains described in reference 4 were supplied by Harris Bernstein.
Assays were in triplicate using bacteria grown in LB supplemented with the appropriate antibiotic (tetracycline at 10 μg/ml). Results are expressed as means ± standard errors of the means.
Grown with IPTG.
Grown without IPTG.
FIG. 1.
(A) JS3171(paraBAD-yidC) samples were grown to saturation in LB with 0.2% arabinose and then washed twice in 5 ml of LB to remove arabinose. Cells were diluted to an OD600 of 0.05 and grown in LB with 0.2% arabinose (lanes 1 to 3) or LB with 0.2% glucose (lanes 4 to 6). Samples were taken at 2 h (lanes 1 and 3), 4 h, (lanes 2 and 4), or 6 h (lanes 3 and 6) postdilution. Western blot analysis used a mouse PspA antibody and horseradish peroxidase detection. PspA is clearly induced and is indicated. (B) Plasmid pMR25 or pSJ1 was introduced into HPT57 (wild type), HPT73 (secM point mutant), and HPT93 (secM point mutant). Following growth to saturation, cells were diluted to an OD600 of 0.05 and grown for 2 h (lanes 1, 4, and 7), 4 h (lanes 2, 5, and 8), or 6 h (lanes 3, 6, and 9). Samples were prepared for Western blotting using mouse PspA antibody and horseradish peroxidase detection. Lanes 1 to 3 contain HPT57 (wild type), lanes 4 to 6 contain HPT73 (secM73), and lanes 7 to 9 contain HPT93 (secM93).
Depletion of YidC dramatically up-regulates pspA expression.
Depletion of YidC was used in these experiments, since yidC is essential. JS7131 contains a single copy of yidC integrated into the attB locus of MC1061 under the control of the araBAD promoter (29). JS7131 harboring pMR25 (vector control) or pSJI was grown to saturation in LB (plus 0.2% arabinose) and then washed twice with 5 ml of LB to remove arabinose. Cells were diluted to an OD600 of 0.05 in either LB plus 0.2% arabinose or LB plus 0.2% glucose and grown for 4 h. Triplicate samples were assayed for β-galactosidase activity as described above for Table 1. An approximately 4.7-fold up-regulation of pspA transcription was observed following YidC depletion. Following depletion of YidC, exactly as before, cell samples were analyzed by SDS-PAGE, and Western blotting clearly showed up-regulation of PspA protein at 2 and 4 h postdilution to remove arabinose (Fig. 1A).
Mutations in tat loci do not affect pspA expression.
E. coli strain MC4100 harboring in-frame deletions in tatA, tatE, tatAE, and tatC were all assessed for pspA expression. Mutations in tatC and tatAE completely block translocation of tat substrates, while deletions in either tatA or tatE lead to partial blocking of tat substrate transport since their functions overlap (30; T. Palmer, personal communication). pMR25 (vector control) and pSJ1 were introduced into tat mutants and a wild-type reference strain (MC4100). All strains were grown to an OD600 of 0.6 to 0.8 or saturation. PspA was not up-regulated under any of the conditions tested (Table 1). Western blotting was employed to confirm these results; again no up-regulation of PspA protein was observed (data not shown). We did not test psp induction under anaerobic conditions, since tat genes are constitutively expressed, indicating a requirement for the Tat export apparatus under all growth conditions (15).
Which Psp protein(s) could monitor stress at the inner membrane?
Recent evidence indicated that regulation of the psp operon proceeds through protein-protein interactions (1). PspA is a cytoplasmic protein associated with the inner membrane (12). PspE is a periplasmic protein (22), while PspC is known to span the inner membrane once with an N-terminus-in, C-terminus-out topology (17). PspB is an inner membrane protein with one membrane-spanning domain, but the topological arrangement has not been proven. PspD was identified as a peripherally bound inner membrane protein by Western blotting and cell fractionation studies (1). To confirm the cellular location of PspB and PspD, we used a gene fusion approach, a well-characterized method of determining the topology of proteins (20). We fused lacZ or phoA to the C termini of PspD and PspB and used enzyme assays to determine the activities of the fusion proteins produced (Table 2). Since both PspB and PspD fused to lacZ gave reproducible β-galactosidase activity, whereas fusion of either to phoA gave no detectable alkaline phosphatase activity, the C termini of these proteins must be located in the cytoplasm. Western blotting with an antibody to LacZ (Cp Laboratories) demonstrated the production of stable fusion proteins of the expected size (data not shown). Hence, alterations in PMF (or improperly inserted membrane proteins) might be sensed by PspA, PspB, PspC, or PspD. A previous study showed that both pspB and pspC are required for induction of the psp response but that pspE is dispensable for induction (24). Eviction-exchange mutagenesis was used to delete pspD from the chromosome of E. coli MG1655. A PCR product containing an in-frame deletion of pspD was constructed as described by Link et al. (19) with primers listed on the website http://arep.med.harvard.edu/labgc/adnan/projects/EcoliKOprimers/EcoliKOprimers.html in which the SalI restriction site is replaced with a BamHI site. The PCR product was digested with BamHI and NotI and ligated into the same sites in plasmid pSR47s (modified sacB vector supplied by Paul Casaz). Eviction-exchange mutagenesis was carried out as described by Link et al. (19) by using E. coli MG1655 CGSC 7740 (5) as the parent strain for the gene deletion. pspD deletion mutants were detected by colony PCR with the same primers used to make the original PCR product. Amplified DNA from putative mutants was sequenced to verify that the deletion was in frame. Induction of pspA with pPMR129 (which harbors pIV under lac promoter control) was not affected in a strain with pspD deleted (Table 2). Hence, although it remains a possibility that PspD may influence the kinetics of induction by pIV or other inducers, it seems likely that either PspA, PspB, or PspC (or a combination) is the primary sensor of membrane stresses.
TABLE 2.
Roles of PspB and PspD in the phage shock responsea
| Strain | β-Galactosidase assay result (Miller units) | Alkaline phosphatase assay result (U/h) |
|---|---|---|
| DH5α(vector) | 57.2 ± 0.05 | 6 ± 0.05 |
| DH5α(vector + pspD) | 1968 ± 26 | 7 ± 0.02 |
| DH5α(vector + pspB) | 686 ± 8 | 7 ± 0.02 |
| MG1655(pSJ1, pGZ119EH, NI) | 183.4 ± 6.4 | |
| MG1655(pSJ1, pGZ119EH, I) | 242.9 ± 6.4 | |
| MG1655(pSJ1, pPMR129, NI) | 196 ± 6.6 | |
| MG1655(pSJ1, pPMR129, I) | 657.2 ± 9.3 | |
| MG1655 ΔpspD(pSJ1, pGZ119EH, NI) | 191.0 ± 3.3 | |
| MG1655 ΔpspD(pSJ1, pGZ119EH, I) | 197.24 ± 2.2 | |
| MG1655 ΔpspD(pSJ1, pPMR129, NI) | 199.6 ± 4.0 | |
| MG1655 ΔpspD(pSJ1, pPMR129, I) | 700.6 ± 18.6 |
E. coli strain DH5α was the host for gene fusion constructs. This strain lacks endogenous β-galactosidase activity and has minimal alkaline phosphatase activity. The vector was pLKC480 for fusions made to lacZ (34), and pspB and pspD were amplified with primers and clones containing appropriate restriction sites (verified by sequencing; primers are available on request). The vector was pBAF for fusions made to phoA (13a), and cloning sites and primers were as described above for fusions to lacZ. Assays were in triplicate using bacteria grown in LB supplemented with the appropriate antibiotic (ampicillin at 100 μg/ml), and results are means ± standard errors. The pGZ119EH vector is described in the work of Lessel et al. (18). Strains were grown with no induction (NI) in LB supplemented with the appropriate antibiotics. For strains grown with induction (I), IPTG at a concentration of 1 mM was used throughout growth to induce the expression of pIV (encoding the secretin) from pPMR129. pPMR129 is vector pGZ119EH containing the pIV gene from phage f1 under lac promoter control (10).
Implications.
Transport of proteins across the bacterial inner membrane may impose stress on the cell, perhaps through modulation of the PMF. Using a simple reporter system for pspA expression and a specific antibody to detect PspA levels, we have shown that while stress(es) imposed by depletion of YidC and impaired functioning of SecM leads to an increased synthesis of PspA (and, in concert, pspBCDE, since these genes are arranged as an operon), defects in the bacterial SRP and Tat systems have no effect on psp. Since the tat system can open a large pore while maintaining the PMF intact (31) and mutations in the SRP affect only the presentation of substrates to the Sec translocon, this study confirms that the elimination of PMF is a good candidate for psp up-regulation. PMF is, however, required for translocation through the Tat pore; perhaps the mutations in the Tat apparatus analyzed in this study did not lead to the dissipation of PMF. Alternatively, perhaps even though export in a tatA background is severely affected (T. Palmer, personal communication), the number of exported proteins affected is significantly lower than when the sec machinery is corrupted. The Psp system may sense problems specifically at the Sec translocon. The effects of secA, secD, secF, and secM upon the induction of the psp operon are clear, yet secY and secE mutations do not up-regulate psp. The basis for this differential effect is not yet clear, but it might be related to some redundancy in the roles that secY and secE can play in the cell. It has been suggested that YidC acts with the Sec translocon, and we hypothesize that one or more of the Psp proteins may sense alterations in the PMF directly, or perhaps they interact with members of the Sec translocon or YidC itself. SecM has been suggested to participate directly in the insertion of membrane proteins into the inner membrane. The secM mutants used in this study affected membrane protein insertion into the inner membrane but had no effect on SecA levels (33). YidC can also independently insert membrane proteins into the inner membrane. So the possibility also exists that a Psp protein(s) can sense incorrectly inserted proteins directly, perhaps by close association with SecM, SecD, or SecF and YidC. The differences in psp expression between secM and srp mutants is striking. An involvement of SecM in the regulation of SecA may be one basis for the clear effects of SecM upon psp expression. Although secM and srp mutations have the property of having weak effects upon membrane protein insertion, the secM mutant has the additive effect of interfering with protein translocation. By utilizing chromosomal mutations in several components of the secretion apparatus, as opposed to determining the effects of overexpressed proteins on the psp system, we have established a firm link between secretion-imposed stress and the psp system of E. coli. Recent reports that regulation of the phage shock response proceeds through protein-protein interactions speculated that PspC was the most likely candidate for the prime sensor of membrane integrity (1). PspD is dispensable for induction of psp (this study), leaving PspA, PspB, or PspC as likely sensors of membrane stress. The challenge now is to identify how the Psp protein(s) senses stress and to work out how PspA can act to restore PMF and hence aid the secretion of proteins across the bacterial inner membrane.
Acknowledgments
We thank Jon Beckwith, Harris Bernstein, Tracy Palmer, Ross Dalbey, and Marjorie Russel, who kindly provided strains and plasmids used in this work. We are grateful to Paul Casaz for the gift of plasmid pSR47 and tips for the gene knockout methodology.
This work was supported by funding from the Wellcome Trust to M.B. L.J.L. is the recipient of a Wellcome Trust studentship.
REFERENCES
- 1.Adams, H., W. Teertstra, J. Demmers, R. Boesten, and J. Tommassen. 2003. Interactions between phage-shock proteins in Escherichia coli. J. Bacteriol. 185:1174-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arkowitz, R. A., and W. Wickner. 1994. SecD and SecF are required for the proton electrochemical gradient stimulation of preprotein translocation. EMBO J. 13:954-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein, H. D., M. A. Poritz, K. Strub, P. J. Hoben, S. Brenner, and P. Walter. 1989. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature 340:482-486. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, H. D., and J. B. Hyndman. 2001. Physiological basis for conservation of the signal recognition particle targeting pathway in Escherichia coli. J. Bacteriol. 183:2187-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Bogsch, E. G., F. Sargent, N. R. Stanley, B. C. Berks, C. Robinson, and T. Palmer. 1998. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem. 273:18003-18006. [DOI] [PubMed] [Google Scholar]
- 7.Carlson, J. H., and T. J. Silhavy. 1993. Signal sequence processing is required for the assembly of LamB trimers in the outer membrane of Escherichia coli. J. Bacteriol. 175:3327-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casadaban, M. J., and S. N. Cohen. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. USA 76:4530-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, M., K. Xie, F. Jiang, L. Yi, and R. E. Dalbey. 2002. YidC, a newly defined evolutionarily conserved protein, mediates membrane protein assembly in bacteria. Biol. Chem. 383:1565-1572. [DOI] [PubMed] [Google Scholar]
- 10.Daefler, S., M. Russel, and P. Model. 1997. Module swaps between related translocator proteins pIV(f1), pIV(IKe) and PulD: identification of a specificity domain. J. Mol. Biol. 266:978-992. [DOI] [PubMed] [Google Scholar]
- 11.Duong, F., and W. Wickner. 1997. The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J. 16:4871-4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dworkin, J., G. Jovanovic, and P. Model. 2000. The PspA protein of Escherichia coli is a negative regulator of σ54-dependent transcription. J. Bacteriol. 182:311-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elderkin, S., S. Jones, J. Schumacher, D. Studholme, and M. Buck. 2002. Mechanism of action of the Escherichia coli phage shock protein PspA in repression of the AAA family transcription factor PspF. J. Mol. Biol. 320:23-37. [DOI] [PubMed] [Google Scholar]
- 13a.Fulkerson J. F., and Mobley H. C. 2000. Membrane topology of the NixA nickel transporter of Helicobacter pylori: two nickel transport-specific motifs within transmembrane helices II and III J. Bacteriol. 182:1722-1730. [DOI] [PMC free article] [PubMed]
- 14.Hardie, K. R., S. Lory, and A. P. Pugsley. 1996. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 15:978-988. [PMC free article] [PubMed] [Google Scholar]
- 15.Jack, R. L., F. Sargent, B. C. Berks, G. Sawers, and T. Palmer. 2001. Constitutive expression of Escherichia coli tat genes indicates an important role for the twin-arginine translocase during aerobic and anaerobic growth. J. Bacteriol. 183:1801-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kleerebezem, M., and J. Tommassen. 1993. Expression of the pspA gene stimulates efficient protein export in Escherichia coli. Mol. Microbiol. 7:947-956. [DOI] [PubMed] [Google Scholar]
- 17.Kleerebezem, M., W. Crielaard, and J. Tommassen. 1996. Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the protonmotive force under stress conditions. EMBO J. 15:162-171. [PMC free article] [PubMed] [Google Scholar]
- 18.Lessl, M., D. Balzer, R. Lurz, V. L. Waters, D. G. Guiney, and E. Lanka. 1992. Dissection of IncP conjugative plasmid transfer: definition of the transfer region Tra2 by mobilization of the Tra1 region in trans. J. Bacteriol. 174:2493-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manoil, C. 1991. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 34:61-75. [DOI] [PubMed] [Google Scholar]
- 21.Matsuyama, S., Y. Fujita, and S. Mizushima. 1993. SecD is involved in the release of translocated secretory proteins from the cytoplasmic membrane of Escherichia coli. EMBO J. 12:265-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mielke, D. L., and M. Russel. 1992. A modified TnphoA useful for single-strand sequencing. Gene 118:93-95. [DOI] [PubMed] [Google Scholar]
- 23.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24.Model, P., G. Jovanovic, and J. Dworkin. 1997. The Escherichia coli phage-shock-protein (psp) operon. Mol. Microbiol. 24:255-261. [DOI] [PubMed] [Google Scholar]
- 25.Poritz, M. A., H. D. Bernstein, K. Strub, D. Zopf, H. Wilhelm, and P. Walter. 1990. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science 250:1111-1117. [DOI] [PubMed] [Google Scholar]
- 26.Robinson, C., and A. Bolhuis. 2001. Protein targeting by the twin-arginine translocation pathway. Nat. Rev. Mol. Cell Biol. 2:350-356. [DOI] [PubMed] [Google Scholar]
- 27.Romisch, K., J. Webb, J. Herz, S. Prehn, R. Frank, M. Vingron, and B. Dobberstein. 1989. Homology of 54K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature 340:478-482. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Samuelson, J. C., M. Chen, F. Jiang, I. Moller, M. Wiedmann, A. Kuhn, G. J. Phillips, and R. E. Dalbey. 2000. YidC mediates membrane protein insertion in bacteria. Nature 406:637-641. [DOI] [PubMed] [Google Scholar]
- 30.Sargent, F., E. G. Bogsch, N. R. Stanley, M. Wexler, C. Robinson, B. C. Berks, and T. Palmer. 1998. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 17:3640-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sargent, F., B. C. Berks, and T. Palmer. 2002. Assembly of membrane-bound respiratory complexes by the Tat protein-transport system. Arch. Microbiol. 178:77-84. [DOI] [PubMed] [Google Scholar]
- 32.Struck, J. C., H. Y. Toschka, and V. A. Erdmann. 1988. Nucleotide sequence of the 4.5S RNA gene from Thermus thermophilus HB8. Nucleic Acids Res. 16:9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian, H., and J. Beckwith. 2002. Genetic screen yields mutations in genes encoding all known components of the Escherichia coli signal recognition particle pathway. J. Bacteriol. 184:111-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiedeman, A. A., and J. M. Smith. 1988. Lac24 gene fusion cassettes with kanR resistance. Nucleic Acids Res. 16:3587. [DOI] [PMC free article] [PubMed]
- 35.L. Yi, F. Jiang, M. Chen, B. Cain, A. Bolhuis, and R. E. Dalbey. 2003. YidC is strictly required for membrane insertion of subunits a and c of the F-1f-OATP synthase and SecE of the SecUEG translocase. Biochemistry 42:10537-10544. [DOI] [PubMed]