Abstract
The group B streptococcus (GBS) is an important human pathogen that infects newborns as well as adults. GBS also provides a model system for studying adaptation to different host environments due to its ability to survive in a variety of sites within the host. In this study, we have characterized a transcription factor, MtaR, that is essential for the ability of GBS to survive in vivo. An isogenic strain bearing a kanamycin insertion in mtaR was attenuated for survival in a neonatal-rat model of sepsis. The mtaR mutant grew poorly in human plasma, suggesting that its utilization of plasma-derived nutrients was inefficient. When an excess of exogenous methionine (200 μg/ml) was provided to the mtaR mutant, its growth rate in plasma was restored to that of the wild-type strain. The mtaR mutant grew poorly in chemically defined medium (CDM) prepared with methionine at a concentration similar to that of plasma (4 μg/ml) but was able to grow normally in CDM prepared with a high concentration of methionine (400 μg/ml). Both the wild-type strain and the mtaR mutant were incapable of growth in CDM lacking methionine, indicating that GBS cannot synthesize methionine de novo. When the abilities of the strains to incorporate radiolabeled methionine were compared, the mtaR mutant incorporated fivefold less methionine than the wild-type strain during a 10-min period. Collectively, the results from this study suggest that the ability to regulate expression of a methionine transport system is critical for GBS survival in vivo.
The potential to identify new antibiotic targets has been a major driving force for the study of bacterial pathogens at the molecular level. The majority of research on bacterial pathogens has focused on uncovering the virulence mechanisms of bacteria, which are most commonly defined as bacterial activities that result in damage to the host or that allow the evasion of immune mechanisms. However, from the perspective of developing new antibiotics, many virulence factors do not provide effective targets because they are not essential for bacterial survival and growth. Historically, broad-spectrum antibiotics target essential bacterial metabolic processes, such as transcription and translation. It has been estimated that the antibiotics in current use target only 15 of ∼300 essential genes in bacterial genomes (8). A broader understanding of the basic metabolisms of bacterial pathogens will facilitate the development of new antibiotics.
The group B streptococcus (Streptococcus agalactiae) is a leading cause of invasive infections in human neonates. GBS is a normal member of the human vaginal flora, and ∼30% of adult females are colonized with GBS. GBS can be transmitted to the newborn during the childbirth process when the neonate aspirates GBS-containing amniotic fluid or vaginal contents. GBS infections of the neonate can manifest as pneumonia, sepsis, and meningitis. GBS infections in adults also occur and are especially common among the immunocompromised.
The GBS infection pathway dictates that the bacterium must grow in a wide variety of host environments. GBS is considered to be primarily an extracellular pathogen, though it appears to have the ability to persist to some degree within host cells, including macrophages (3). During the infection process, growth of GBS occurs in a variety of body sites, including the maternal vagina, amniotic fluid, and the neonatal lung, as well as the bloodstream. Growth of GBS to high densities has been reported in amniotic fluid, as well as in the lungs of neonates (16). However, it is not well understood how GBS adapts to the different host environments it encounters.
Much remains to be learned about how basic bacterial metabolic pathways function when the bacterium is within the host. Nutrient concentrations in vivo vary dramatically from those used to culture bacteria in vitro, and pathogenic bacteria will inevitably face nutrient limitations in various host compartments. The metabolic characteristics of GBS in vivo, however, have not been thoroughly explored. While some pathogens have the ability to synthesize all amino acids and other nutrients required for growth, GBS is auxotrophic for multiple amino acids and must rely on transport of many nutrients from the host environment during in vivo growth. This is apparent from inspection of the GBS genome (6), which reveals the absence of most biosynthetic pathways for amino acids, as well as the presence of a large number of genes encoding components of ABC transporters. Collectively, these observations indicate that GBS obtains most of its nutrients from the host; this suggests that the GBS metabolism is most likely finely tuned to the availability of nutrients within its host, which would be essential for its ability to cause disease.
In this study, we provide evidence that methionine availability can limit the growth of GBS in plasma. We demonstrate that a regulatory protein that controls methionine transport is essential for the ability of GBS to survive within a neonatal-rat host. Mutation of a gene encoding a LysR-type transcriptional regulatory protein, mtaR, resulted in a decrease in GBS growth rates in human plasma, and this phenotype was linked to a diminished ability to grow in defined media containing low levels of methionine. A methionine transport deficiency is implicated as one mechanism to explain the poor survival of the mutant in vivo. The findings of our study implicate methionine availability as an important determinant for the survival of GBS in vivo. Since MtaR homologues are found in the genomes of a number of other pathogenic bacteria, MtaR may also play an important role in the survival of other pathogens in vivo.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli strain XL1-Blue (Stratagene, La Jolla, Calif.) was used as a host for pACYC-derived and pTRKL2-derived (18) plasmids (Table 1). E. coli strain MC1061 was used as a host for pHY304-derived plasmids. COH1 is a virulent, highly encapsulated GBS isolate from a newborn with septicemia. Plasmid pHY304 (2), which was used for the construction of GBS mutants, is a derivative of pVE6007 (12) and replicates in a temperature-sensitive manner in E. coli and GBS. Plasmid pTRKL2 (18), which was used for complementation analysis, replicates with a p15A origin of replication in E. coli and replicates with a pAMβ1 origin of replication in GBS.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| S. agalactiae | ||
| COH1 | Type III GBS isolate from neonate with septicemia; highly virulent; wild-type reference strain | 14 |
| DS101 | mtaR::kan derivative of COH1; Kanr | This work |
| HY105 | cpsB::kan derivative of COH1; acapsular; Kanr | 30 |
| DS105 | Derivative of DS101 (mtaR::kan) bearing pDS8 (mtaR+); Kanr Ermr | This work |
| DS106 | Derivative of DS101 bearing pTRKL2; Ermr | This work |
| E. coli | ||
| MC1061 | Host strain for propagation of Ermr plasmids | 28 |
| XL1-Blue | Host strain for general cloning | Stratagene |
| Plasmids | ||
| pVE6007 | E. coli-GBS shuttle vector; temperature-sensitive replication; pWV origin of replication; Cmr | 12 |
| pACYC177 | Medium-copy-number plasmid utilized for cloning | New England Biolabs (Beverly, Mass.) |
| pCIV2 | Source of Ω Km-2 fragment used for mutagenesis | 17 |
| pHY304 | Derivative of pVE6007; temperature sensitive; Ermr | 2 |
| pTRKL2 | E. coli-GBS shuttle vector; low-copy-number plasmid; pAMβ1 origin of replication; Ermr | 18 |
| pDS6 | pHY304 bearing mtaR::kan allele; used to construct DS101; Kanr Ermr | This study |
| pDS8 | pTRKL2 bearing wild-type mtaR gene; used for complementation of DS101; Ermr | This study |
Bacterial growth and antibiotics.
E. coli was cultured in Luria-Bertani medium. For routine culture, GBS strains were grown in Todd-Hewitt Broth (THB). For growth of GBS, antibiotics were added when necessary at the following concentrations: kanamycin, 500 μg/ml, and erythromycin, 5 μg/ml. The C-48 chemically defined medium (CDM) of Willett and Morse (29) was utilized for cultivation of GBS with defined nutrients. The concentrations of nutrients utilized in the present study were identical to those reported previously, except that the methionine concentrations were varied as specified in Results. To evaluate growth in CDM, GBS was initially grown to log phase (optical density at 600 nm[OD600], 0.3) in THB. The cells were harvested by centrifugation, washed twice in an equivalent volume of phosphate-buffered saline (PBS) (130 mM NaCl, 3 mM KCl, 1.5 mM KH2PO4, 4.3 mM Na2HPO4, pH 7.4), and diluted 1 to 50 in CDM. Growth was monitored spectrophotometrically at a wavelength of 600 nm. For growth experiments with human plasma, plasma was obtained from healthy human volunteers after consent. Prior to growth in plasma, GBS was grown in THB to log phase (OD600, 0.3), collected by centrifugation, and washed twice in an equal volume of PBS. The resulting suspension was diluted to ∼2 × 104 CFU/ml in PBS, and 75 μl was used to inoculate 750 μl of human plasma (∼1,000-CFU/ml initial concentration of GBS) in a 2-ml microcentrifuge tube. The tubes were then incubated in hybridization tubes with slow rolling at 37°C. At designated intervals, growth was quantified by plating to Todd-Hewitt agar. For experiments in which plasma was supplemented with CDM components, the CDM components were used at the concentrations specified previously (29).
DNA and RNA techniques.
Standard methodologies were utilized for cloning, PCR amplification, and Northern and Southern analyses (23). RNA was isolated by the method of Yim and Rubens (31) with minor modifications. Northern hybridization was performed with antisense RNA probes, utilizing the digoxigenin nonradioactive labeling and detection system (Roche, Indianapolis, Ind.).
Construction of a GBS mtaR mutant.
The mtaR and flanking regions (including an IS861 element and a cpsA region) were amplified from COH1 genomic DNA using Expand High Fidelity polymerase (Roche). PCR primers that incorporate XhoI and BamHI sites for cloning were designed to amplify the mtaR region. The resulting PCR product was digested with XhoI and BamHI and was ligated to similarly digested pACYC177, generating the plasmid pDS4. The region flanking mtaR was sequenced to insure that no mutations were introduced during PCR amplification. The kanamycin resistance cassette from pCIV2 was amplified using primers that incorporate PvuII sites for cloning and was digested with PvuII. Plasmid pDS4 was then digested with BsaBI and ligated to the PvuII-cut kanamycin resistance fragment, inserting the cassette into the mtaR gene. The plasmid insert, bearing the mtaR::kan allele, was liberated with PstI and XhoI and ligated to similarly cut pHY304, generating plasmid pDS6, which was transformed directly into GBS strain COH1 (4).
The following procedure was then utilized to derive a strain bearing a replacement of the wild-type mtaR allele with the mtaR::kan allele described above. The GBS strain carrying pDS6 was cultured at 30°C overnight in THB with selection for kanamycin. The culture was then washed in THB without antibiotics, diluted 1/20 in THB without antibiotics, and incubated at 37°C until the culture was saturated. This dilution-and-outgrowth procedure was repeated for a total of five cycles. After the last outgrowth, dilutions of the culture were plated onto kanamycin-containing THB agar. GBS colonies from the kanamycin plates were then screened for sensitivity to erythromycin. A Kmr Ems clone was designated DS101 and was selected for further study. The presence of the mtaR::kan allele on the chromosome of DS101 was verified by Southern blotting.
Construction of GBS strains used for complementation studies on mtaR.
The mtaR gene and surrounding region, including the 5′ end of the adjacent cpsA gene and the 3′ end of the IS861 element, were amplified from GBS strain COH1 using Expand High Fidelity DNA polymerase. The resulting amplification product was purified, digested with XmnI and BglII, and ligated to pTRKL2 (18) digested with EcoRV and BamHI, generating the plasmid pDS8. Plasmid pDS8 was introduced into strain DS101 by electroporation, generating GBS strain DS105.
Competition and LD50 experiments in a neonatal-rat model.
The ability of GBS to survive within a neonatal rat was measured by a competition assay and 50% lethal-dose (LD50) analysis. For in vivo competition assays, wild-type and mtaR mutant GBS were cultured in THB to an OD600 of 0.3, harvested by centrifugation, washed with an equal volume of PBS, and resuspended in PBS. It was determined that for both the wild-type and mutant strains, equivalent numbers of wild-type and mutant CFU were present at an OD600 of 0.3. The wild-type and mtaR mutant strains were mixed in equivalent numbers and diluted in PBS to a density of ∼2.1 × 105 CFU/ml, and 100 μl was injected into the peritoneal cavities of newborn (24- to 48-h-old) rat pups. The following day, the spleens were harvested and homogenized in PBS, and dilutions of the homogenates were plated for CFU. The mutant was distinguished from the wild-type strain by its Kanr phenotype. The survival of the mutant in relation to the survival of the wild type was expressed as a competitive index, defined as follows: [(CFU of mutant recovered from spleen)/(CFU of wild type recovered from spleen)]/[(CFU of mutant input)/(CFU of wild-type input)]. For LD50 analysis, wild-type (COH1), mutant (DS101), and mtaR-complemented strains were grown to an OD600 of 0.3 in THB. Tenfold dilutions of the test strains were prepared and used to inoculate five or six rat pups per dilution for each strain tested.
Measurement of amino acid transport.
The transport of amino acids was evaluated using a procedure based on that of Kadner (10). GBS was grown in CDM supplemented with methionine at 400 μg/ml to ensure equivalent growth of wild-type and mtaR mutant GBS. Cultures of GBS were grown at 37°C and harvested at an OD600 of 0.30. Ten milliliters of each culture was harvested by centrifugation, and the cells were washed three times in CDM without methionine and leucine and supplemented with chloramphenicol at 50 μg/ml. The resulting suspension was adjusted to an OD600 of 0.36 in the same medium. The culture was preincubated at either 4 or 37°C. When the transport of methionine was measured, an equivalent volume of CDM containing 10 μCi of l-[methyl-3H]methionine (Amersham-Pharmacia, Piscataway, N.J.)/ml, 2 μM total l-methionine, and 6.6 μM unlabeled leucine was added to the cell suspension. For measuring the transport of leucine, an equivalent volume of CDM was added to the cells to achieve a final concentration of 2 μCi of l-[U-14C]leucine (Amersham-Pharmacia)/ml, 6.6 μM total l-leucine, and 2 μM total l-methionine. After incubation at either 4 or 37°C for 10 min, 600 μl of the cell suspension was filtered in triplicate onto AcetatePlus membranes (Osmonics, Minnetonka, Minn.). The membrane filters were washed with 10 ml of PBS and dried. Radioactivity was then determined by scintillation counting after the addition of 10 ml of BCS-NA scintillation fluid (Amersham-Pharmacia).
RESULTS
MtaR encodes a LysR-type transcriptional regulator that is conserved in pathogenic streptococci.
The sequence analysis of the GBS mtaR gene was originally reported by Koskiniemi and coworkers (11), who named the gene cpsY (see below) based upon its physical linkage to the capsular polysaccharide biosynthesis cluster. The sequence of mtaR predicts that it encodes a transcriptional regulator from the LysR family (24). MtaR shows the typical characteristics of LysR-type transcriptional regulators, including a putative DNA-binding domain at its N terminus and an effector-binding region at its C terminus (24). Koskiniemi et al. suggested that MtaR may regulate the adjacent polysaccharide synthesis locus (cps) (11). However, no experimental evidence was provided to support this hypothesis.
Inactivation of mtaR.
To test whether MtaR influenced expression of the GBS capsular polysaccharide, we disrupted the wild-type copy of mtaR with a kanamycin resistance cassette. The mtaR::kan mutation was constructed as described in Materials and Methods and subcloned to a temperature-sensitive E. coli-GBS shuttle plasmid (pHY304 [2]), generating pDS6. Subsequently, pDS6 was electroporated into GBS strain COH1, which is a heavily encapsulated, highly virulent type III clinical isolate. The plasmid was cured, and a strain bearing an allelic replacement of mtaR was isolated, as detailed in Materials and Methods. Southern blots of genomic DNA (not shown) confirmed the presence of the desired mutation on the chromosome of a representative isolate, designated DS101.
Cell surface-associated polysaccharides from COH1 and DS101 were quantified by competitive enzyme-linked immunosorbent assay. No significant difference was observed between the two strains (data not shown). As an independent test to determine if transcription of the cps region was altered in DS101, we evaluated cps transcript accumulation with Northern blots. No difference in cps transcript accumulation was observed when total RNAs extracted from log-phase COH1 and DS101 were compared (data not shown). Thus, under the in vitro conditions tested, we found no evidence that MtaR controls transcription of the adjacent cps operon.
The mtaR gene shows strong identity with two uncharacterized open reading frames from other important human pathogens. The organization of the mtaR region is presented in Fig. 1. When we searched the available databases for homologues of MtaR by using BLAST (1), we found homology to open reading frames from two other pathogenic streptococci. MtaR showed 78% identity and 86% similarity to the product of an open reading frame (Spy0898) from the genome of an M1 strain of Streptococcus pyogenes. MtaR also showed 75% identity and 84% similarity to the predicted product of an open reading frame (SP0676) from the genome of a virulent isolate of Streptococcus pneumoniae, TIGR4 (26). S. pneumoniae also produces a polysaccharide capsule, but the S. pneumoniae mtaR homologue is not physically linked to its capsule-biosynthetic locus (26). MtaR shows lower levels of similarity to a large number of other members of the LysR family (23).
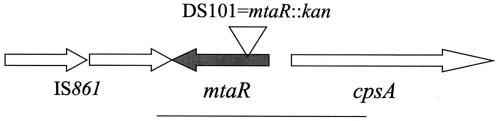
FIG. 1.
The mtaR region of GBS. The open arrows depict the coding regions of the genes described in this study and indicate the direction of transcription. The cps gene cluster, which directs synthesis of the extracellular capsular polysaccharide of GBS, is transcribed divergently from mtaR. The IS861 element (22) is adjacent to mtaR in strain COH1 (14). The open triangle depicts the location of the kanamycin omega cassette in GBS strain DS101 (mtaR::kan). The solid bar represents the DNA fragment bearing mtaR that was used for complementation of DS101 (mtaR mutant).
MtaR is required for GBS survival in vivo in a neonatal rat.
We considered the possibility that, since mtaR does not regulate expression of the GBS capsular polysaccharide, it may instead regulate a distinct cellular process that is important for the virulence of the organism. A number of transcription factors from the LysR family regulate virulence-related genes in diverse species of bacteria (24). To test if MtaR contributes to GBS virulence, we compared the ability of DS101 (mtaR mutant) to survive in an animal model in relation to its isogenic parent strain, COH1. Neonatal rats were infected by intraperitoneal injection with a mixture of the wild type and mutant in a ratio of 1:1. The next day, spleens were harvested from infected animals and the surviving bacteria were enumerated. In three separate experiments, the mtaR mutant survived much more poorly than the wild-type strain (competitive-index values, 0.0000136, 0.00042, and 0.00000019; see Materials and Methods for a definition of competitive index). We also conducted LD50 analysis using the neonatal-rat sepsis model. The wild-type (COH1) LD50 was 1.0 × 103, while the mtaR mutant (DS101) displayed an LD50 of 1.1 × 106, a decrease in virulence of ∼3 log units. These results suggest that MtaR is essential for survival within the host.
MtaR is necessary for wild-type growth in human plasma.
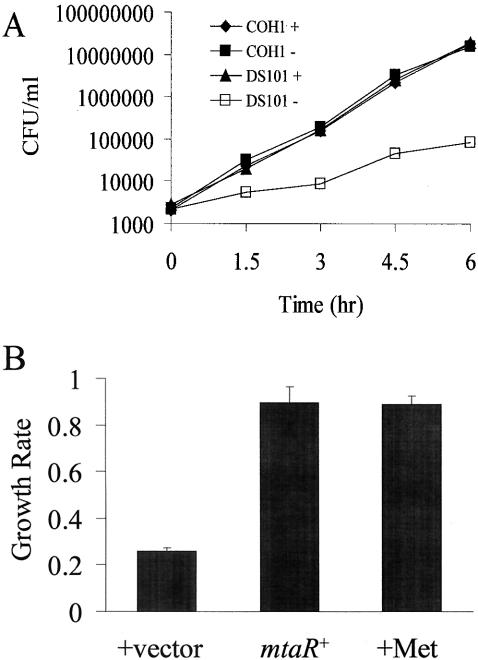
In the sepsis model that was utilized in our experiments, bacteria access the bloodstream shortly after intraperitoneal injection. Thus, we hypothesized that poor survival in the animal may reflect a defect in the ability of the mutant to grow in blood. To determine if the attenuated survival of DS101 was due to poor growth in blood, we tested its ability to grow in human plasma in vitro. The use of plasma, as opposed to whole blood, allowed the evaluation of bacterial growth in the absence of the bactericidal effect of blood-derived immune cells. When COH1 (wild type) and DS101 (mtaR mutant) were compared, the growth rate of DS101 was substantially less (∼2.5-fold) than that of the wild type (Fig. 2A). Control experiments in which the growth rates of the strains in THB (data not shown) and in the CDM of Willett and Morse (29) were compared revealed no difference in growth rates (see below). We also tested another mutant bearing the kanamycin resistance element, HY105 (cpsB::kan), to verify that the presence of the kanamycin cassette was not the cause of the slow growth of the strain; no growth defect was present for the strain in plasma (data not shown). Together, these results suggested that mutation of mtaR resulted in a methionine-related metabolic defect that is observable specifically in plasma.
FIG. 2.
Growth of GBS in plasma. (A) Growth of wild-type (COH1) and MtaR− (DS101) GBS in plasma with and without methionine. GBS was grown in THB to an OD600 of 0.3, washed twice in PBS, and resuspended to a final OD600 of 0.3. After a dilution of 1/1,000 with PBS, 75 μl of the suspension was used to seed 675 μl of plasma obtained from a healthy adult human volunteer, achieving a bacterial density of ∼1,000 CFU/ml. The plasma was either supplemented with methionine at 200 mg/ml (+) or not supplemented (−). The plasma containing the bacteria was then incubated at 37°C with end-over-end rotation, and dilutions were plated for CFU at the indicated time points. The experiment was conducted twice with similar results; one replicate is shown. (B) Complementation of growth defect of MtaR− GBS. The growth rates in plasma of GBS strain DS101 (MtaR−) and DS105 (DS101 bearing a plasmid that contains a wild-type mtaR gene) were compared. As a positive control, DS101 was incubated in the presence of methionine (400 μg/ml). Growth experiments were conducted similarly to the experiment described in the legend to panel A, except the average growth rate over a 3-h period was calculated. Shown are the growth rates of DS101 with the vector portion of the complementing plasmid alone (+vector) (strain DS106), DS101 bearing a vector with the wild-type mtaR gene (mtaR+) (strain DS105), and DS101 grown in plasma supplemented with methionine (+Met). The error bars represent the standard deviations of three growth experiments.
Inactivation of mtaR results in a requirement for increased exogenous methionine.
One hypothesis to explain the poor growth of DS101 in plasma is that the mutant strain experiences a nutritional deficiency in plasma. As noted above, the mutant strain grew comparably to the wild-type strain in the CDM of Willett and Morse (29). CDM contains 20 amino acids at levels that are higher than those required for growth of GBS. The normal growth of the mtaR mutant in CDM suggested that some component(s) of the CDM was deficient in plasma. We thus screened the individual components of the CDM for a compound that, when added to plasma, could stimulate growth of the mutant. We found that methionine, when added to plasma at the concentration present in CDM (400 μg/ml), increased the growth rate of the mutant to a rate equivalent to that of the wild-type strain (Fig. 2A). As a control, we tested if methionine specifically stimulated the growth of the mutant strain or if instead it stimulated GBS growth in general under the conditions present in plasma. We thus cultured the wild-type strain with and without the addition of methionine at 400 μg/ml. The addition of methionine did not change the growth rate of the wild-type strain.
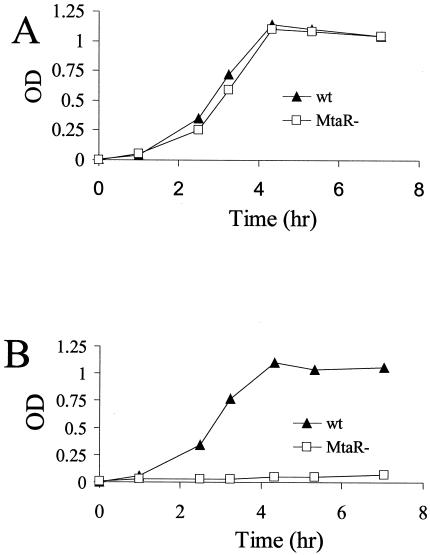
The stimulation of the growth of the mtaR mutant by methionine supplementation strongly suggested that the mtaR mutant had a defect in some aspect of methionine metabolism. However, the metabolism of methionine by GBS has not been well characterized, so we performed growth studies in the CDM of Willett and Morse (29). As mentioned above, both strains grow normally in CDM with 400 μg of methionine/ml (Fig. 3A). When methionine was completely eliminated from CDM, neither the wild-type nor the mutant strain grew over the time frame of the experiment (data not shown). Thus, COH1 was incapable of de novo synthesis of methionine, at least when cultured in CDM.
FIG. 3.
Growth of GBS in chemically defined medium (CDM). GBS was grown in THB to log phase (OD600, 0.3), washed twice in PBS, and resuspended to a final OD600 of 0.3. The resulting suspension was diluted 1/100 and was used to inoculate CDM (29). The cultures were incubated statically at 37°C. Growth over time was monitored spectrophotometrically by measuring the OD600. Shown are the growth rates of GBS strains in CDM with methionine at 400 (A) and 4 (B) μg/ml. The experiment was conducted three times with similar results, and one replicate is shown. wt, wild type.
The concentration of free methionine present in adult human plasma is ∼4 μg/ml, which is ∼50-fold lower than the concentration of methionine in CDM (29). The mtaR mutant was next evaluated for its ability to grow at low methionine concentrations. In CDM prepared with methionine at 4 μg/ml, the wild-type strain grew well, but the mutant strain did not grow over the time frame of the experiment (Fig. 3B). Collectively, the results of the CDM growth experiments indicate that GBS must scavenge methionine from an exogenous source and that the mtaR mutation renders the mutant unable to utilize exogenous methionine at physiological concentrations.
We next verified by complementation analysis that the virulence and growth defects that were observed were attributable to the mtaR mutation. A low-copy-number plasmid (pDS8 [see Materials and Methods]) bearing the wild-type mtaR allele was introduced into DS101. The introduction of pDS8 (mtaR+) to DS101 decreased the LD50 of the resulting strain to 1.5 × 104, a decrease of ∼72-fold (compared to the mtaR mutant). We are not sure why the introduction of pDS8 did not restore the virulence of DS101 to the level of COH1. We have observed that pTRKL2-based plasmids are stable in GBS during animal passage and do not alter the virulence of plasmid-bearing strains. However, it is possible that non-wild-type levels of MtaR are expressed from pDS8 due to its episomal nature and that this decreases the virulence of DS105 relative to COH1.
The complemented strain (DS105) was also tested for the ability to grow in plasma. The introduction of mtaR into DS101 restored the ability of the mutant to grow normally in plasma, to a rate that was comparable to that of DS101 grown in plasma supplemented with methionine (Fig. 2B). Thus, the mtaR::kan mutation was responsible for the attenuated virulence and the growth defect in plasma displayed by DS101.
MtaR is necessary for efficient methionine transport.
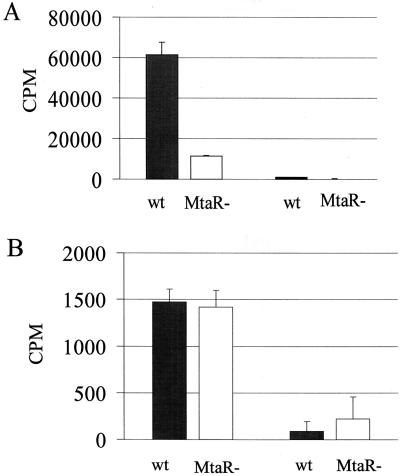
The growth defect of the mtaR mutant in medium with low levels of methionine could be readily explained if DS101 were defective in its ability to transport methionine. To test this hypothesis, we compared the abilities of DS101 and COH1 to incorporate [3H]methionine. DS101 and COH1 were grown in CDM supplemented with high levels of methionine (400 μg/ml) to ensure that both strains grew equivalently. After growth to mid-log phase (OD600, 0.3), the cells were washed in CDM lacking methionine and leucine and supplemented with chloramphenicol (50 μg/ml) to stop protein synthesis, as described in Materials and Methods. After being washed, the cells were incubated in CDM at 37°C with [3H]methionine or [14C]leucine. After 10 min, the cells were filtered and washed with PBS, and the radioactivity in the filters was determined by scintillation counting. Figure 4 depicts the amount of labeled amino acid incorporated by the cells. When incubated at a temperature of 37°C, the wild-type strain incorporated approximately fivefold more [3H]methionine than the mtaR mutant over a 10-min period. When an identical experiment was performed at 4°C, there was an ∼47-fold decrease in [3H]methionine incorporation by the wild-type strain. This suggested that the incorporation of methionine by GBS was an enzymatic process, which typically shows temperature dependence. As a control, [14C]leucine incorporation was measured under conditions identical to those used to measure [3H]methionine transport. Wild-type and mtaR mutant GBS incorporated similar amounts of [14C]leucine (Fig. 4B); more leucine was incorporated at 37 than at 4°C. The results of the amino acid incorporation experiments together indicated that the mtaR mutation confers upon GBS a defect in its ability to efficiently transport methionine.
FIG. 4.
Methionine incorporation by GBS. Wild-type (wt) and mtaR mutant (MtaR−) GBS were grown to log phase in CDM supplemented with methionine at 400 μg/ml. After being washed in PBS, the GBS cells were suspended in CDM at equivalent cell densities. After the cell suspension was prewarmed, [3H]methionine (1 μM total methionine) or [14C]leucine (3.3 μM total leucine) was added to initiate the assay. After incubation for 10 min at 4 or 37°C, 600 μl of the cell suspension was filtered, in triplicate, to AcetatePlus membranes (Osmonics), and the filters were washed with 10 ml of PBS by using a vacuum manifold (Millipore, Bedford, Mass.). The filter membranes were dried, and the radioactivity on the filters was determined by scintillation counting. The background radioactivity was determined by performing the experiment with CDM in place of the cell suspension and was subtracted from the results. The radioactivity (CPM) incorporated for COH1 (solid bars) and DS101 (open bars) is depicted. The error bars represent the standard deviations of triplicate samples. The two left-hand bars of each panel represent the results obtained when the incubation was performed at 37°C; the two right-hand bars represent the results obtained at 4°C. (A) [3H]methionine incorporation; (B) [14C]leucine incorporation.
DISCUSSION
In this study, we provide evidence that a putative regulatory gene, mtaR, is required for the growth of GBS in human plasma and CDM in the presence of physiological methionine levels (4 μg/ml). Since GBS is auxotrophic for methionine, and since mutation of mtaR confers an inability to efficiently transport methionine, we conclude that MtaR is essential for methionine scavenging.
It was surprising that the mtaR mutant was not able to scavenge methionine through the degradation of plasma proteins. In order to utilize extracellular proteins as sources of amino acids, gram-positive bacteria, such as Lactococcus lactis, hydrolyze proteins via an extracellular protease, transport the resulting peptides via peptide permeases, and degrade the peptides to amino acids with multiple peptidases. Inspection of the genome of another GBS serotype III strain, NEM316, reveals the presence of two putative extracellular proteases, four exported peptidases, three oligopeptide transporters, and 21 genes encoding intracellular peptidases. This suggests that GBS has the capacity to utilize extracellular proteins as nutritional sources. Several possibilities could explain the inability of the GBS mtaR mutant to grow at wild-type rates when utilizing plasma proteins as sources of methionine. One possibility is that utilization of plasma proteins is not as efficient as utilization of free amino acids and thus may not support rapid growth. Alternatively, GBS may lack an oligopeptidase with the proper specificity to release methionine from plasma-derived oligopeptides. Finally, in addition to regulating the transport of free methionine, MtaR may be required for the expression of proteases, transport systems for oligopeptides, oligopeptidases, or some other component required for utilization of proteins as a source of methionine.
The ability of bacterial pathogens to adapt to in vivo environments, to acquire nutrients, and to grow in vivo is of fundamental importance to the disease process. In order to survive in vivo, pathogens must be able to adapt and utilize nutrients that are present. Despite the essential role of nutrition in the persistence of pathogens, surprisingly little research has been devoted to understanding what nutritional limitations pathogenic bacteria face in vivo and how the bacteria adapt to these limitations. Notable exceptions are the mechanisms of iron acquisition and its regulation, which have been well studied in a number of pathogenic bacteria. Animals have evolved sophisticated mechanisms to sequester iron in order to restrict the growth of pathogenic bacteria, and bacteria have countered by evolving strategies to acquire iron from their hosts. As for other nutrients, tryptophan availability may restrict the growth of pathogenic bacteria in eukaryotic cells. It has been suggested that macrophages can limit intracellular growth of bacteria such as Toxoplasma gondii (20) and Legionella pneumophila (5) by catabolizing tryptophan. Auxotrophic mutants of Listeria monocytogenes have been examined for virulence; a threonine auxotroph displayed an LD50 that was 1 log10 unit higher in a mouse model of infection. A number of in vivo expression technology and signature-tagged mutagenesis studies have implicated nutrition-related genes as important in virulence. Taken together, these observations suggest that nutrients can be expected to limit bacterial growth in the host environment, and pathogenic bacteria must have evolved mechanisms to counter these limitations. However, the nature of these nutritional limitations needs to be studied in more detail.
It is not clear if the decreased virulence of DS101 is due only to a defect in methionine transport. The nearly 3-log-unit decrease in virulence is striking, and while it appears that MtaR regulates methionine transport, it may directly regulate other virulence traits of GBS, and this may account for such a prominent attenuation in virulence. Another possibility is that slow growth of the mtaR mutant due to a methionine transport defect predisposes the mutant to clearance by the immune system.
The survival of a GBS in the sepsis model is primarily influenced by two factors: first, the growth rate of the GBS, and second, the ability of the GBS to resist clearance by the immune system. Interestingly, recent studies have shown that the two traits are linked; the ability of GBS to resist immune clearance is directly related to its growth rate. In vitro, growth rates of GBS affect the expression of several known GBS virulence factors. Paoletti and coworkers have shown that in vitro, fast-growing GBS resists opsonophagocytic killing more readily than does slow-growing GBS (19). Rapid growth rates stimulate high-level expression of the antiphagocytic polysaccharide capsule (19); this is one potential mechanism to explain the resistance of fast-growing GBS to immune clearance. A number of other potential GBS virulence factors appear to be regulated in response to the growth rate (21), and the invasiveness of GBS strains for epithelial cells is also regulated by the growth rate (13). Taken together, these studies suggest that after entry into the bloodstream, GBS must grow rapidly or face clearance by the immune system. Thus, it is also possible that the slow growth of the mtaR mutant leads to decreased expression of virulence factors, predisposing it to clearance by the immune system. Further studies are needed to determine the impact of MtaR on the expression of other virulence factors.
It is noteworthy that the genomes of two other important human pathogens, S. pyogenes and S. pneumoniae, contain highly similar MtaR homologues. As obligate human pathogens, S. pyogenes and S. pneumoniae also rely on the transport of nutrients from the host, and like GBS, the other two streptococcal species are auxotrophic for multiple amino acids. We hypothesize that MtaR may also play a role in the regulation of nutrient uptake in these species in vivo.
What is the methionine transport system that is regulated by MtaR?
The data presented in this study strongly indicate that MtaR is required for high-level expression of a methionine uptake system. Since mtaR encodes a protein that is similar to transcriptional regulatory proteins, the data presented in this study can be explained if MtaR normally effects activation of a gene encoding a methionine transport system. An alternative explanation is that loss of mtaR results in expression of a protein that blocks methionine transport. Members of the LysR family of transcriptional regulators, such as MtaR, activate or repress gene expression by binding directly to DNA sites and either activating or repressing transcription. The gene encoding the postulated methionine transport system controlled by MtaR, however, is unknown. One could hypothesize that MtaR regulates a gene located 5′ to IS861. However, when the genomes of the type III (6) and type V (27) GBS isolates are inspected, there are no open reading frames that encode products similar to amino acid transporters in the immediate vicinity of the IS element. Additionally, there has been no functional characterization of methionine transporters in gram-positive bacteria, so our ability to speculate on the nature of the gene encoding the transporter is limited. The methionine transport systems of E. coli (10) and Salmonella enterica serovar Typhimurium (7) are, to our knowledge, the only prokaryotic methionine transporters that have been described functionally. E. coli synthesizes two systems, a high-affinity methionine transport system from the ABC transporter family (MetD) (15) and a low-affinity system encoded by an unknown gene (9). To our knowledge, no homologues of methionine transporters for gram-positive bacteria have been characterized in the literature. We found no close homologues of the metD system of E. coli and S. enterica serovar Typhimurium in the genome of type III GBS or in the genomes of other gram-positive bacteria that have been released to date. This may indicate that gram-positive bacteria utilize different methionine transport systems than gram-negative bacteria.
What is the signal that is recognized by MtaR?
MtaR shows similarity to the LysR family of transcriptional regulatory proteins, which incorporate signal sensing, DNA binding, and transcriptional regulatory activities in a single protein. Proteins from this family are typically cytoplasmic and recognize intracellular signals, in contrast to members of the two-component family of transcriptional regulatory proteins (5), which frequently recognize extracellular signals. Thus, we expect MtaR to respond to an intracellular molecule. Inspection of the MtaR sequence reveals that the signal-sensing domain of the LysR family, which is located at the C terminus, is present in MtaR. The putative signal-sensing domain of MtaR, however, does not show strong similarity to the corresponding region of other LysR-type regulators; this may indicate that MtaR senses a signal distinct from those sensed by the other characterized LysR family members.
In summary, we have identified a regulatory protein from GBS that is distributed among pathogens, including S. pyogenes and S. pneumoniae. The finding that close homologues of MtaR exist in other important human pathogens suggests that MtaR may play a role in the pathogeneses of other gram-positive bacteria. In GBS, MtaR is essential for in vivo survival in a sepsis model and is also essential for normal growth in plasma and normal methionine transport. Taken together, our results suggest that methionine availability in blood may limit the growth of GBS and potentially other pathogens.
Acknowledgments
We thank Robert Kadner, Gary Roberts, Glen Tamura, and Donald Chaffin for helpful suggestions.
This work was supported by NIH National Research Service Award fellowship 5 F32 AI10426-03 to D.S.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Chaffin, D. O., S. B. Beres, H. H. Yim, and C. E. Rubens. 2000. The serotype of type Ia and III group B streptococci is determined by the polymerase gene within the polycistronic capsule operon. J. Bacteriol. 182:4466-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornacchione, P., L. Scaringi, K. Fettucciari, E. Rosati, R. Sabatini, G. Orefici, C. von Hunolstein, A. Modesti, A. Modica, F. Minelli, and P. Marconi. 1998. Group B streptococci persist inside macrophages. Immunology 93:86-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Framson, P. E., A. Nittayajarn, J. Merry, P. Youngman, and C. E. Rubens. 1997. New genetic techniques for group B streptococci: high-efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl. Environ. Microbiol. 63:3539-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gebran, S. J., Y. Yamamoto, C. Newton, T. W. Klein, and H. Friedman. 1994. Inhibition of Legionellapneumophila growth by gamma interferon in permissive A/J mouse macrophages: role of reactive oxygen species, nitric oxide, tryptophan, and iron(III). Infect. Immun. 62:3197-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 7.Grundy, C. E., and P. D. Ayling. 1992. Fine structure mapping and complementation studies of the metD methionine transport system in Salmonella typhimurium. Genet. Res. 60:1-6. [DOI] [PubMed] [Google Scholar]
- 8.Hutchinson, C. A., et. al. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165-2169. [DOI] [PubMed] [Google Scholar]
- 9.Kadner, R. J. 1974. Methionine transport in Escherichia coli: physiological and genetic evidence for two uptake systems. J. Bacteriol. 119:401-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadner, R. J. 1974. Transport systems for l-methionine in Escherichia coli. J. Bacteriol. 117:232-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koskiniemi, S., M. Sellin, and M. Norgren. 1998. Identification of two genes, cpsX and cpsY, with putative regulatory function on capsule expression. FEMS Immunol. Med. Microbiol. 21:159-168. [DOI] [PubMed] [Google Scholar]
- 12.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malin, G. 2001. Use of a dynamic in vitro attachment and invasion model system to determine influence of growth rate on invasion of respiratory epithelial cells by group B streptococcus. Proc. Natl. Acad. Sci. USA 98:13335-13340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin, T. R., C. E. Rubens, and C. B. Wilson. 1988. Lung antibacterial defense mechanisms in infant and adult rats: implications for the pathogenesis of group B streptococcal infections in the neonatal lung. J. Infect. Dis. 157:91-100. [DOI] [PubMed] [Google Scholar]
- 15.Merlin, C., G. Gardner, S. Durand, and M. Masters. 2002. The Escherichia coli metD locus encodes an ABC transporter which includes Abc (MetN), YaeE (MetI), and YaeC (MetQ). J. Bacteriol. 184:5512-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nizet, V., P. Ferrieri, and C. E. Rubens. 2000. Molecular pathogenesis of group B streptococcal disease in newborns, p. 180-221. In Streptococcal infections: clinical aspects, microbiology, and molecular pathogenesis. Oxford University Press, New York, N.Y.
- 17.Okada, N., R. T. Geist, and M. G. Caparon. 1993. Positive transcriptional control of mry regulates virulence in the group A streptococcus. Mol. Microbiol. 7:893-903. [DOI] [PubMed] [Google Scholar]
- 18.O'Sullivan, D. J., and T. R. Klaenhammer. 1993. High- and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening. Gene 137:227-231. [DOI] [PubMed] [Google Scholar]
- 19.Paoletti, L., R. A. Ross, and K. D. Johnson. 1994. Cell growth rate regulates expression of group B streptococcus type III polysaccharide. Infect. Immun. 64:1220-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfefferkorn, E. R. 1984. Interferon γ blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host cells to degrade trypotophan. Proc. Natl. Acad. Sci. USA 81:908-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross, R., L. C. Madoff, and L. C. Paoletti. 1999. Regulation of cell component production by growth rate in the group B Streptococcus. J. Bacteriol. 181:5389-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rubens, C. E., L. M. Heggen, and J. M. Kuypers. 1989. IS861, a group B streptococcal insertion sequence related to IS150 and IS3 of Escherichia coli. J. Bacteriol. 171:5531-5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J. E., F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 25.Stock, J. B., A. J. Ninfa, and A. M. Stock. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53:450-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tettelin, H., et al. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 294:498-506. [DOI] [PubMed] [Google Scholar]
- 27.Tettelin, H., et al. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wertman, K. F., A. R. Wyman, and D. Botstein. 1986. Host/vector interactions which affect the viability of recombinant phage lambda clones. Gene 49:253-262. [DOI] [PubMed] [Google Scholar]
- 29.Willett, N., and G. E. Morse. 1966. Long-chain fatty acid inhibition of growth of Streptococcus agalactiae in a chemically defined medium. J. Bacteriol. 91:2245-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yim, H. H., and C. E. Rubens. 1998. Site-specific homologous recombination mutagenesis in group B streptococci. Methods Cell Sci. 20:13-20. [Google Scholar]
- 31.Yim, H. H., and C. E. Rubens. 1997. Use of a dental amalgamator to extract RNA from the gram-positive bacterium Streptococcus agalactiae. BioTechniques 23:229-231. [DOI] [PubMed] [Google Scholar]