Abstract
The Synechocystis sp. strain PCC 6803, which has a T192H mutation in the D2 protein of photosystem II, is an obligate photoheterotroph due to the lack of assembled photosystem II complexes. A secondary mutant, Rg2, has been selected that retains the T192H mutation but is able to grow photoautotrophically. Restoration of photoautotrophic growth in this mutant was caused by early termination at position 294 in the Slr2013 protein. The T192H mutant with truncated Slr2013 forms fully functional photosystem II reaction centers that differ from wild-type reaction centers only by a 30% higher rate of charge recombination between the primary electron acceptor, QA−, and the donor side and by a reduced stability of the oxidized form of the redox-active Tyr residue, YD, in the D2 protein. This suggests that the T192H mutation itself did not directly affect electron transfer components, but rather affected protein folding and/or stable assembly of photosystem II, and that Slr2013 is involved in the folding of the D2 protein and the assembly of photosystem II. Besides participation in photosystem II assembly, Slr2013 plays a critical role in the cell, because the corresponding gene cannot be deleted completely under conditions in which photosystem II is dispensable. Truncation of Slr2013 by itself does not affect photosynthetic activity of Synechocystis sp. strain PCC 6803. Slr2013 is annotated in CyanoBase as a hypothetical protein and shares a DUF58 family signature with other hypothetical proteins of unknown function. Genes for close homologues of Slr2013 are found in other cyanobacteria (Nostoc punctiforme, Anabaena sp. strain PCC 7120, and Thermosynechococcus elongatus BP-1), and apparent orthologs of this protein are found in Eubacteria and Archaea, but not in eukaryotes. We suggest that Slr2013 regulates functional assembly of photosystem II and has at least one other important function in the cell.
Much information has been gathered regarding the molecular organization and functional role of structural components of the photosynthetic apparatus in microorganisms and plants. However, little is known about mechanisms that regulate the biosynthesis and functional assembly of the photosynthetic machinery. Molecular-genetic, biochemical, and biophysical approaches have generated insight regarding the functional role of most of the components of photosystems I (PS I) and II in the cell and regarding the requirement for structural proteins of the photosystems for the successful assembly and function of these multicomponent systems in photosynthesis (5). In addition, evidence has accumulated for proteins that are important for biosynthesis and/or functional assembly of the photosynthetic machinery in the cell but that are not known to be structural components of the photosynthetic apparatus (6, 11, 13, 25). Much of this evidence was obtained by using cyanobacteria, particularly Synechocystis sp. strain PCC 6803, which was the first photosynthetic organism whose genome was sequenced and which is one of the easiest photosynthetic microorganisms to perform genetic manipulations on.
Several open reading frames (ORFs) coding for proteins that are not structural components of the photosynthetic apparatus but that influence its function have been discovered. Deletion of slr0228, one of four genes in the Synechocystis sp. strain PCC 6803 genome coding for a putative FtsH (a protease of the AAA [ATP-dependent metallopeptidase] superfamily), was found to lead to a reduction in the abundance of functional PS I in the cells. This polypeptide may be involved in PS I biogenesis (13). Slr0171 may be a chaperone for PS I formation, because inactivation of the corresponding gene caused a decrease in the PS I/PS II ratio and an increased phycocyanin/chlorophyll ratio in Synechocystis sp. strain PCC 6803 (25). A similar effect was observed after the deletion of the homologous gene in Chlamydomonas reinhardtii (15). Slr0399 seems to be important for restoration of PS II activity in mutants with impaired QA function, and the protein is considered a chaperone that aids in quinone insertion and protein folding around QA in PS II (6). Slr0286 was shown to be important for the functional assembly and stability of the oxygen-evolving complex in Synechocystis sp. strain PCC 6803. Deletion of this gene caused a decrease in the apparent affinity of PS II complexes for Ca2+ and a delay in the photoactivation of the water-splitting complex (11).
This paper describes the function of Slr2013 in Synechocystis sp. strain PCC 6803 as a chaperone involved in folding of the D2 protein of PS II, thus adding one more protein to the list of factors involved in functional and stable assembly of photosynthetic complexes.
MATERIALS AND METHODS
Growth conditions.
The Synechocystis sp. strain PCC 6803 wild type and mutant strains retaining PS I were grown in liquid BG-11 medium according to the method of Rippka et al. (18) at a light intensity of 40 μmol of photons m−2 s−1 and a temperature of 30°C. Liquid cultures were incubated on a rotatory shaker at 120 rpm. Solid medium was supplemented with 1.5% (wt/vol) agar, 0.3% (wt/vol) sodium thiosulfate, 10 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid]-NaOH buffer (pH 8.2), and, if indicated, 5 mM glucose.
PS I-less strains lacking psaAB coding for the main PS I components were grown as described above, except that a lower light intensity (5 μmol of photons m−2 s−1) was used, and 5 mM glucose was present in the growth medium.
The T192H mutation (Thr192 replaced by His) was introduced into the D2 protein by introducing the corresponding mutation into the psbDI gene of Synechocystis sp. strain PCC 6803 lacking psbDII, which is the second gene copy encoding the D2 protein of PS II (23). The resulting T192H mutant was not capable of photoautotrophic growth and was maintained in the presence of 5 mM glucose. Upon growth on plates, 25 μg of kanamycin ml−1 (the selective marker for the introduction of T192H mutation in psbDI), 25 μg of spectinomycin ml−1 (the selective marker for the psbDII deletion), and 20 μM atrazine [a PS II inhibitor to remove selective pressure for photoautotrophic (pseudo)revertants] were added.
Pseudorevertant selection.
The Rg2 pseudorevertant of the T192H mutant of Synechocystis sp. strain PCC 6803 was selected by plating T192H cultures on solid BG-11 medium from which glucose had been omitted, followed by screening for colonies that were capable of photoautotrophic growth in the presence of the original T192H mutation in psbDI. PCR amplification of this gene and sequencing of the obtained product were used to confirm the desired psbDI sequence.
Introduction of wild-type D2 into the pseudorevertant.
In the Rg2 pseudorevertant, the psbDI/C operon contains the T192H mutation. In order to determine whether the secondary mutation by itself affects the phenotype of the organism, the wild-type psbDI/C operon was reintroduced into the genome. The Rg2 mutant was transformed with a wild-type psbDI/C PCR product followed by growth under photoautotrophic conditions for 14 generations. Single colonies were obtained and sequenced. One of these had Thr in position 192 of the D2 protein but preserved the secondary mutation.
DNA isolation and genetic mapping.
Chromosomal DNA was isolated from photoautotrophic pseudorevertant strains of T192H by a standard method of N-lauryl sarcosine lysis and phenol-chloroform extraction (8, 12).
Mapping of the second-site mutation was performed as described by us in a previous study (11) using digestion of chromosomal DNA from the Rg2 pseudorevertant with either BamHI, BspEI, EcoRV, KpnI, PstI, or SmaI; size fractionation of the restriction products; DNA elution from the gel; transformation of the T192H mutant; and selection for photoautotrophic growth. The DNA elution from the gel and transformation of the T192H strain were conducted under conditions that were optimal for transformation efficiency (10). Colonies of photoautotrophic transformants were visible after 10 to 12 days of incubation at 30°C in the light at an intensity of 40 μmol of photons m−2 s−1.
slr2013 cloning and deletion constructs.
The slr2013 gene and flanking regions in the Synechocystis sp. strain PCC 6803 genome were amplified by PCR; the resulting fragment corresponds to nucleotides 753176 to 755828 (numbering according to CyanoBase). A DraI-StuI fragment of the PCR product, corresponding to nucleotides 753291 to 754965 and containing slr2013 and the flanking regions, was cloned in the HincII site of the pUC19 plasmid, resulting in pslr2013. The pΔslr2013 constructs were created by deleting the middle part of the cloned gene by restriction with BsaBI (278 bp from the start codon of slr2013) and NheI (1,056 bp from the start codon of slr2013) and inserting either a chloramphenicol resistance cassette or kanamycin resistance cassette in both possible orientations in place of the deleted fragment. Transformation with each of the four slr2013 deletion constructs was performed with the wild type and the ΔpsaAB (PS I-less) strain of Synechocystis sp. strain PCC 6803. Segregation of the transformants was attempted under photoautotrophic and photomixotrophic growth conditions. Cells were grown on plates and in liquid cultures at a light intensity of 40 μmol of photons m−2 s−1 (wild type only) or 5 μmol of photons m−2 s−1 or under light-activated photoheterotrophic growth (LAHG) conditions (darkness with 15 min of light per day) (1).
psaAB deletion.
Deletion of psaAB in the Rg2 pseudorevertant of T192H and in wild type was carried out with a ΔpsaAB/Zeor plasmid derived from pBWV (2), which carries the NdeI-SphI fragment of psaAB (926 to 3728 bp from the start codon of psaA). The BsaAI-HpaI fragment (1629 to 2840 bp from the start codon of psaA) in the ΔpsaAB/Zeor plasmid was replaced by the PvuII/Ecl136I fragment of pZeo1 carrying the zeocin resistance gene (ble) (pZeo1 was constructed by cloning the MscI-EcoRI fragment of pSP109 [19] into the SmaI-EcoRI sites of pUC19.)
The resulting Rg2/PS I-less mutant, together with the ΔpsaAB mutant (PS I-less control strain) obtained by using the same plasmid, were used for all experiments in which the functional activity of PS II complexes in the pseudorevertant was tested by monitoring decay of chlorophyll fluorescence and formation and stability of the YDox radical.
Spectroscopy.
Low-temperature fluorescence emission spectra were recorded upon excitation of cell suspensions (chlorophyll concentration, 5 μg ml−1) with 435-nm light by using a FluoroMax spectrofluorometer (Spex Industries, Inc.). The excitation wavelength was 435 nm, and the excitation and emission bandwidths were 4 and 1 nm, respectively.
Fluorescence decay measurements.
The decay kinetics of the chlorophyll fluorescence yield in intact cells after actinic illumination in the presence of 40 μM 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) were obtained with a Walz fluorometer (Walz, Effeltrich, Germany). After illumination of a 400-μl cell suspension (chlorophyll concentration, 5 μg ml−1) with red light (intensity, 50 μmol of photons m−2 s−1) emitted by PAM LEDs for 400 ms, the actinic light was turned off, and the decaying fluorescence yield was monitored as a function of time by using weak measuring light. The intensity of the measuring light was chosen so that it did not have an actinic effect. The data were recorded and analyzed with FIP software (QA-Data, Turku, Finland).
Oxygen evolution.
The steady-state rate of oxygen evolution in intact cells was measured with a Clark-type electrode (Hansatech) at a chlorophyll concentration of 5 μg ml−1 with 0.1 mM 2,5-dimethyl-p-benzoquinone and 0.5 mM K3[Fe(CN)6] as electron acceptors. Light from a xenon arc lamp was filtered through water and passed through an orange cutoff filter so that the cell suspension was illuminated by light with a wavelength above 580 nm and an incident intensity of 1 mmol of photons m−2 s−1.
PS II assembly under different growth conditions.
The influence of light on the assembly of PS II reaction centers was studied upon growth of the wild-type, T192H, and Rg2 strains of Synechocystis sp. strain PCC 6803 under normal light conditions (40 μmol of photons m−2 s−1) or low light intensity (5 μmol of photons m−2 s−1) or under LAHG conditions. The functional efficiency of PS II reaction centers was assessed based on oxygen evolution activity, 77-K fluorescence spectra, and fluorescence induction measurements as described above.
The optical density of liquid cell cultures was determined at 730 nm with a Shimadzu UV-160 spectrophotometer.
EPR measurements.
Formation and stability of the YD radical were studied by electron paramagnetic resonance (EPR). Thylakoid membranes were isolated from cells of PS I-less and Rg2/PS I-less strains of Synechocystis sp. strain PCC 6803, washed in buffer (20 mM Mes-NaOH [pH 6.5], 5 mM MgCl2, 5 mM CaCl2, 10% [vol/vol] glycerol), and resuspended in the same buffer containing 1 mM ɛ-aminocaproic acid and 1 mM benzamidine (protease inhibitors). The cell pellet was resuspended, mixed with an equal volume of glass beads, and broken in a prechilled Braun homogenizer (Bronwill Scientific, Inc., Rochester, N.Y.) chamber (2 cycles of 45 s each with a 2-min incubation on ice between them). After removal of glass beads and intact cells by centrifugation (1,000 × g, 3 min), thylakoids were pelleted (7,000 × g, 20 min). Thylakoids were washed with a 50 mM Mes-NaOH-20 mM pyrophosphate buffer (pH 6.5) to remove phycobilisomes, and thylakoids were resuspended in thylakoid buffer without protease inhibitors to a concentration of 0.1 mg of chlorophyll per ml, frozen, and stored in liquid nitrogen. After thawing, thylakoids were treated with 4 mM hydroxylamine (pH 7.0) for 30 min on ice to inactivate the water-splitting apparatus. Hydroxylamine was washed out by three consecutive washes in the thylakoid buffer. To oxidize the acceptor side of PS II and facilitate the formation of the YDox under illumination, potassium ferricyanide (fourfold molar excess over the concentration of PS II) was then added to thylakoids that were resuspended in a small volume of phycobilisome wash buffer. Thylakoids were then pelleted and resuspended in a small volume of thylakoid buffer to a chlorophyll concentration of 0.10 to 0.15 mg/ml and illuminated with red light (light intensity, 2 mmol of photons m−2 s−1) for 10 min with constant stirring to prevent thylakoid sedimentation. At the end of illumination, 0.3 ml of the thylakoid suspension was withdrawn, placed in an EPR tube, frozen immediately during illumination in a dry ice-ethanol bath, and then stored in liquid nitrogen. The rest of the thylakoid suspension was placed in a microcooler set at 4°C, and 0.3 ml of the suspension was withdrawn at different time intervals, frozen in darkness, and stored as described above. EPR spectra were recorded at 125 K with a Bruker ESP 550E spectrometer. The EPR parameters were as follows: microwave power, 5 mW; microwave frequency, 9.4 GHz; modulation amplitude, 0.4 mT; modulation frequency, 100 kHz.
Electron microscopy.
Transmission electron microscopy was used to analyze the cell ultrastructure of the Synechocystis sp. strain PCC 6803 strains employed in this study. Samples were prepared for electron microscopy as follows. Liquid cell cultures (optical density at 730 nm of 0.4 to 0.6) in BG-11 medium supplemented with 5 mM glucose were harvested, and the cell pellet was placed promptly between two interlocking planchets (3) and immediately frozen under high pressure (9) with a Bal-Tec HPM 010 high-pressure freezing system (Bal-Tec Products, Inc., Middlebury, Conn.). Freeze substitution and fixation with 2% glutaraldehyde, 0.1% tannic acid, and 0.2% osmium tetroxide in anhydrous acetone were performed with an FSU 010 freeze substitution unit (Bal-Tec Products, Inc.). The conditions of freeze substitution were as follows: −90°C for 24 h, −60°C for 4 h, −20°C for 4 h, 0°C for 2 h, and room temperature for 1 h. Freeze substitution was followed by slow infiltration with hard Eponate 12 resin (Ted Pella, Inc., Redding, Calif.) in a series of acetone-resin mixtures with an increasing content of resin at room temperature for 72 h. Fully infiltrated samples were flat embedded in PELCO resin molds (Ted Pella, Inc.) and polymerized for 48 h at 60°C. Ultrathin sections were cut on a Leica Ultracut R ultramicrotome (Leica Mikrosysteme GmbH, Vienna, Austria) and collected on mesh copper grids. Staining in 2% uranyl acetate in 50% ethanol for 5 min was followed by lead citrate staining as described by Reynolds (17). Prepared sections were viewed with a Philips CM12 electron microscope (Philips Electronic Instruments Co., Mahwah, N.J.) at an accelerating voltage of 80 kV and recorded with a Gatan Digital Micrograph 3.4 imaging system (Gatan, Inc., Pleasanton, Calif.).
Protein sequence analysis.
The primary sequence of Slr2013 was obtained from CyanoBase (http://www.kazusa.or.jp/cyanobase). Analysis for a possible protein targeting sequence was performed by using the SignalP V2.0 program (16), and prediction of protein topology was performed with the DAS program (4). The search for protein family signatures was done by using InterProScan (26). For functional motif prediction, relevant programs on the EXPASY web site (http://www.expasy.ch) were used. Alignment of multiple protein sequences and calculation of the percentage of identity and similarity were performed with CLUSTALW (21).
RNA isolation and RT-PCR.
Total RNA isolation from wild-type cells of Synechocystis sp. strain PCC 6803 was performed as described earlier (14). The reverse transcription (RT) reaction step was conducted at 47°C for 1 h. The PCR step consisted of 30 cycles, each of which included incubation at 94°C for 30 s, 62°C for 30 s, and 72°C for 45 s. For quantitative analysis, 10-μl aliquots were withdrawn from the RT-PCR tubes after a certain numbers of PCR cycles. The amount of PCR product was determined by ethidium bromide staining after separation on a 1.2% (wt/vol) agarose gel. Standard size markers were obtained from Gibco BRL. The transcription level of slr2013 compared to that of psbDI was determined by RT-PCR with primers described in Table 1. The priming efficiency of primers used for RT-PCR of these two genes was compared by PCR under the conditions described above and using equal amounts of genomic DNA from wild-type cells as template.
TABLE 1.
Sequence of primers used for RT-PCR in this study and their positions in the genome of Synechocystis sp. strain PCC 6803
| Primer | Sequence | Positiona |
|---|---|---|
| 5′ slr2013 | 5′GCG GTG GAG CTA TGG TGG TAG CTC 3′ | 753470-753493 |
| 3′ slr2013 | 5′CCT ACC GTC CAT TGT TGC CAA GTC 3′ | 753881-753858 |
| 5′ psbDI | 5′CTT CGG CCA TTC CCT CCT GTT CCT 3′ | 1349647-1349624 |
| 3′ psbDI | 5′GTG TTT TCC ACC GTG GCA CCG TGG 3′ | 1349237-1349260 |
Numbering according to CyanoBase.
RESULTS
Characteristics of T192H.
This study focuses on an obligate photoheterotrophic mutant of Synechocystis sp. strain PCC 6803, T192H, with a corresponding mutation in the D2 protein, and Rg2, a photoautotrophic strain that retains the T192H mutation and carries a secondary mutation. The phenotype of the T192H strain will be presented first.
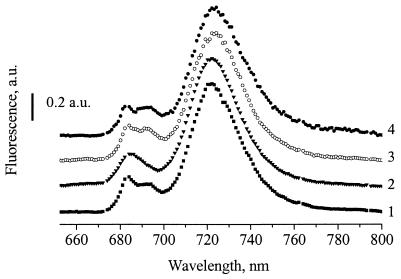
The low-temperature fluorescence emission spectrum of the T192H mutant lacks a peak at approximately 695 nm (Fig. 1), which is characteristic of functional PS II (7). Therefore, the absence of photoautotrophic capacity in the T192H mutant is due to its depletion in assembled PS II centers. No 695-nm fluorescence emission peak was observed in the T192H mutant even when the strain was grown at low light intensity (5 μmol of photons m−2 s−1) or under LAHG conditions (1; data not shown), essentially ruling out light sensitivity of PS II in the T192H mutant.
FIG. 1.
Fluorescence emission spectra (77 K) of intact cells. Shown are spectra of the T192H mutant of Synechocystis sp. strain PCC 6803 (line 2), the Rg2 pseudorevertant (line 3), the T192T/Y294Stop mutant (line 4), and the wild type (line 1). Excitation was at 435 nm. Cells were grown at a light intensity of 40 μmol of photons m−2 s−1. Fluorescence intensities were normalized to the fluorescence yield at 725 nm. a.u., arbitrary units.
Thr192 in the D2 protein is surrounded by functionally important components of PS II: it is close to P680 and is three residues away from His189, the primary proton acceptor for oxidized YD (20, 22). In close proximity to Thr192 is the accessory chlorophyll of the inactive branch of PS II (P. Fromme, personal communication). However, there is no indication that Thr192 directly interacts with any of these PS II components.
Cells of the T192H strain of Synechocystis sp. strain PCC 6803 do not demonstrate oxygen-evolving activity (Table 2), further supporting the absence of functionally active PS II centers in this strain. Their growth rate in the presence of glucose is similar to the rate observed for wild-type cultures (Table 2), suggesting that the effects of the T192H mutation are limited to PS II. Upon transformation of the T192H strain with PCR-amplified wild-type psbDI, photoautotrophic transformants were obtained (data not shown), implying that the reason for the lack of photoautotrophic growth of the T192H strain is limited to the mutation introduced in psbDI.
TABLE 2.
Growth rate and oxygen evolution of the strains in this study under photoautotrophic and photomixotrophic conditionsa
| Strain | Doubling time (h) during growth
|
Oxygen evolution rate (μmol of O2 mg of chlorophyll−1 h−1) | |
|---|---|---|---|
| Photoautotrophic | Photomixotrophic | ||
| Wild type | 12.8 ± 0.4 | 12.7 ± 0.6 | 256 ± 28 |
| T192H | NGb | 13.5 ± 0.8 | 0 |
| Rg2 | 13.3 ± 0.5 | 13.8 ± 0.7 | 275 ± 35 |
| T192T/Y294Stop | 13.4 ± 0.7 | 13.5 ± 0.6 | 248 ± 32 |
| PS I-less | NG | 22.3 ± 1.1 | —c |
| Rg2/PS I-less | NG | 24.1 ± 0.9 | — |
Numbers represent the mean calculated from three independent experiments ± the standard deviation.
NG, no growth (strain does not grow under these conditions).
—, not measured.
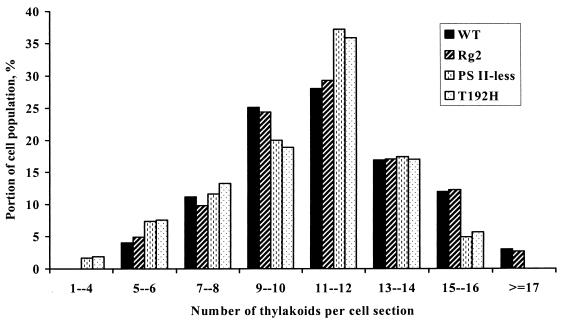
Transmission electron microscopy showed that cells of the T192H mutant and the PS II-less (ΔpsbDIC ΔpsbDII) strain on average have fewer thylakoids per section than wild-type cells (Fig. 2). The proportion of cells with 15 or more thylakoids per section is smaller in the T192H and PS II-less strains, and, in contrast to the wild type, cells with more than 17 thylakoids per section were not observed in the preparations of these strains. On the other hand, the proportion of cells with less than seven thylakoids per section is larger in T192H and the PS II-less strain than in the wild type, and cells with less than four thylakoids per section are present in the PS II-less strain and T192H but are absent from the wild type (Fig. 2).
FIG. 2.
Distribution of the number of thylakoids per section of Synechocystis sp. strain PCC 6803 cells. The distribution was calculated on the basis of transmission electron microscopy images of about 200 cell sections for each strain. WT, wild type.
Properties of the Rg2 pseudorevertant.
The Rg2 pseudorevertant of the T192H mutant was obtained by selection of T192H for photoautotrophic growth. Rg2 retained the T192H mutation (data not shown) and was capable of photoautotrophic growth due to restoration of assembled PS II reaction centers; the latter is evident from the 77-K fluorescence emission spectra (Fig. 1), oxygen-evolution capacity and growth rates (Table 2), and ultrastructural characteristics of the Rg2 cells (Fig. 2).
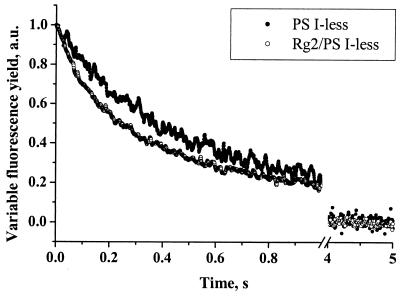
The properties of the PS II complex in Rg2 were somewhat altered. The kinetics of charge recombination between QA and the donor side of PS II, reflecting midpoint redox potentials of contributing cofactors, in Rg2 were somewhat faster than those observed for the wild type (Fig. 3). The half-time of fluorescence decay observed for the Rg2/PS I-less strain of Synechocystis sp. strain PCC 6803 in the presence of DCMU (320 ± 30 ms) was about 30% shorter than what was measured with PS I-less cells (440 ± 42 ms).
FIG. 3.
Decay kinetics of the variable chlorophyll fluorescence yield of intact cells of the PS I-less and Rg2/PS I-less strains of Synechocystis sp. strain PCC 6803. Measurements were conducted in the presence of 40 μM DCMU. Actinic light was turned off at time zero. a.u., arbitrary units.
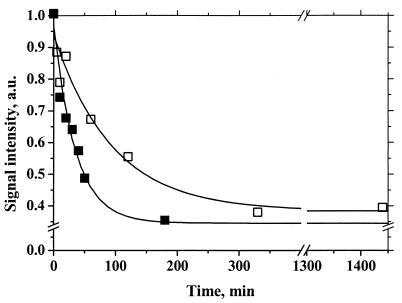
Because Thr192 is located in close proximity to His189, the primary proton acceptor for YD (20, 22), properties of YDox were studied. The shape of the YDox EPR signal was normal (data not shown), but the decay kinetics of YDox were about 2.5 times faster for thylakoids isolated from the Rg2/PS I-less strain than those from the PS I-less strain (Fig. 4). This may indicate that the T192H mutation caused structural changes in the vicinity of His189 leading to a higher rate of reduction of YDox.
FIG. 4.
Decay kinetics of the YDox EPR signal in thylakoids. Thylakoids were isolated from the PS I-less strain (open squares) and the Rg2/PS I-less strain (solid squares) of Synechocystis sp. strain PCC 6803. Illumination was turned off at time zero. The EPR signal intensities were similar in both strains and were normalized to 1.0 at the end of illumination. a.u., arbitrary units.
Mapping of the site of pseudoreversion.
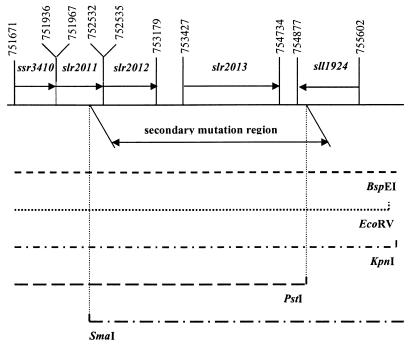
The Rg2 pseudorevertant carries the original T192H mutation and lacks psbDII, the second copy of the gene coding for the D2 protein. Attempts to identify the secondary mutation in some of the single-copy genes coding for PS II proteins (psbB, psbC, and psbDI) by PCR, functional complementation, and sequencing were not successful, indicating that the secondary mutation was not in these genes. The secondary mutation was mapped by functional complementation of the T192H mutant to photoautotrophic growth by using restriction fragments of known size obtained from Rg2 genomic DNA. As indicated in Table 3, five restriction digestions (all except BamHI) yielded one DNA size fraction that was able to functionally complement the T192H mutant, thus indicating that these fractions contained the fragment with the secondary mutation. In a comparison of the size fragments with a genomic restriction map of Synechocystis sp. strain PCC 6803 (24), a unique match was found: the restriction pattern of the genome region carrying the secondary mutation (Table 3) indicates that this mutation is located between nucleotides 752366 (a SmaI site) and 754915 (a PstI site), according to the numbering in CyanoBase. This region of the genome includes four ORFs (slr2011, slr2012, slr2013, and sll1924), three of which are possibly cotranscribed (Fig. 5).
TABLE 3.
Comparison of experimental pseudorevertant restriction fragment sizes that complemented the T192H mutant to photoautotrophic growth, with the theoretical restriction pattern of the sole region of the Synechocystis sp. strain PCC 6803 genome that fits these criteria
| Enzyme | DNA fragment size in fraction that complements photoautotrophic growth (kb) | Actual size of the fragment (bp) | Position of DNA fragment that includes secondary mutation sitea |
|---|---|---|---|
| BamHI | —b | 1,831 | 754522-756352 |
| BspEI | 12.0-20.0 | 17,772 | 745500-763271 |
| EcoRV | 12.0-20.0 | 19,133 | 737521-756653 |
| KpnI | 10.0-15.0 | 12,143 | 747099-759241 |
| PstI | 10.0-15.0 | 14,878 | 740038-754915 |
| SmaI | 12.0-17.0 | 16,525 | 752366-768890 |
Numbering according to CyanoBase.
—, this fragment did not complement T192H to photoautotrophic growth, because one of the restriction sites for this enzyme, 754521 is close to the secondary mutation site, 754308.
FIG. 5.
Location of the secondary mutation site in the genome of the Rg2 photoautotrophic pseudorevertant of the T192H mutant of Synechocystis sp. strain PCC 6803. The position of the secondary mutation region was deduced based on its restriction pattern (Table 3) and the genomic restriction map of Synechocystis sp. strain PCC 6803. M, marker lane.
When this region was amplified by PCR using pseudorevertant DNA, the amplified fragment complemented the original T192H mutant to photoautotrophic growth, thus confirming the accuracy of mapping. Sequencing of this region revealed a mutation of slr2013 codon 294 (coding for Tyr) to a stop codon. The original T192H mutant retained the Tyr294 codon, confirming the accuracy of genomic sequencing in this region.
Attempts to delete slr2013.
To determine whether the secondary mutation caused inactivation of slr2013, a plasmid carrying a deletion of this gene was created (see Materials and Methods for details) and used for the transformation of the wild-type strain. The resulting transformants did not segregate, and all attempts to fully delete slr2013 from the Synechocystis sp. strain PCC 6803 genome have failed, regardless of the antibiotic resistance gene introduced at the site of the slr2013 deletion, the direction of this gene relatively to slr2013, and the growth conditions of the transformants. This demonstrates that (i) the mutation of a Tyr to a stop codon at position 294 of slr2013 did not fully inactivate the corresponding protein, and (ii) Slr2013 in Synechocystis sp. strain PCC 6803 appears to be crucial for cell survival under the conditions explored in this study.
Since the deletion of slr2013 in Synechocystis sp. strain PCC 6803 did not allow full segregation, our analysis of functional effects of slr2013 on the photosynthetic apparatus had to be limited to the study of Synechocystis sp. strain PCC 6803 strains carrying an early termination in slr2013.
Slr2013 truncation in a wild-type background.
Because the function of Slr2013 could not be studied by gene deletion, the effects of truncation of the Slr2013 polypeptide in the absence of D2 mutations were determined. For this purpose, the wild-type sequence of the psbDI/C operon was restored in the Rg2 mutant while retaining the truncation in Slr2013. The resulting T192T/Y294Stop mutant grew normally under both photoautotrophic and photomixotrophic conditions (Table 2). The oxygen-evolving activity of this strain was similar to that observed for the wild type and Rg2 mutant (Table 2). This strain had normally assembled PS II complexes, because the low-temperature fluorescence emission spectrum demonstrated a normal 695-nm peak (Fig. 1) similar to the wild type and the Rg2 strain. The T192T/Y294Stop strain had kinetics of fluorescence decay similar to that of the wild type (data not shown). Therefore, there is no indication that the Slr2013 truncation affects growth and photosynthetic activity of Synechocystis sp. strain PCC 6803 unless the PS II function is compromised by the introduction of T192H mutation in the D2 protein.
Slr2013 truncation in other D2 protein mutants.
The lack of photosynthetic activity of the T192H mutant of the D2 protein can be rescued by a spontaneous Y294Stop mutation in Slr2013 of Synechocystis sp. strain PCC 6803. In order to determine whether this mutation can also rescue other D2 mutants, the PCR product of the mutated slr2013 was used to transform other obligate photoheterotrophic D2 mutants with site-directed mutations introduced on the donor (E69Q) or acceptor (V247M/A249T) sides of PS II. The resulting transformants were not capable to photoautotrophic growth. Apparently, the Slr2013 truncation specifically rescues the T192H mutation and is not a universal means to ameliorate impaired PS II function in Synechocystis sp. strain PCC 6803.
slr2013 transcript abundance.
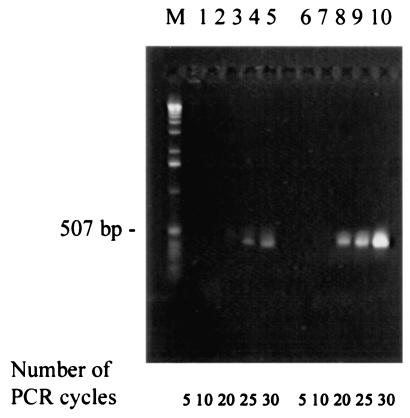
To obtain information on the relative abundance of slr2013 transcripts in comparison with that of transcripts for PS II components, RT-PCR analyses of slr2013 and psbDI, a gene coding for the D2 protein, were performed with wild-type Synechocystis sp. strain PCC 6803. The number of PCR cycles required for visible transcript amplification was increased by at least 10 for slr2013 relative to psbDI (Fig. 6). Therefore, the level of slr2013 transcript is several orders of magnitude lower than the level of psbDI transcript. The differences observed here are due primarily to a different amount of template: PCR analysis of priming efficiency indicated that both sets of primers demonstrate practically identical efficiencies when genomic DNA is used as template (data not shown). The amount of slr2013 transcript suggests that Slr2013 may not be an abundant protein in the cell. A similarly low transcript level was found for slr0286, which codes for a protein that is involved in PS II assembly but that is not a stoichiometric component of the PS II complex (11).
FIG. 6.
RT-PCR analysis of transcript levels of slr2013 and psbDI. Total RNA isolated from the Synechocystis sp. strain PCC 6803 wild type was used as a template. Primers for slr2013 (lanes 1 to 5) and psbDI (lanes 6 to 10) amplification are described in Table 3. The numbers of PCR cycles to which the samples have been exposed have been indicated. Molecular size markers (M) are in the lane to the left.
DISCUSSION
Identification and structural analysis of Slr2013.
A Thr-to-His mutation at position 192 of D2, located in the lumenal CD loop in this protein, caused a complete functional inactivation of PS II complexes, resulting in an obligate photoheterotrophic phenotype of the Synechocystis sp. strain PCC 6803 mutant. The T192H mutant had virtually no active PS II centers, as judged by chlorophyll induction/decay traces (not shown), 77-K fluorescence emission spectra (Fig. 1), and oxygen evolution parameters (Table 2). The absence of assembled PS II centers in the T192H mutant as well as in the PS II-less mutant of Synechocystis sp. strain PCC 6803 correlated with a smaller number of thylakoids per cell section (Fig. 2).
A spontaneous mutation in ORF slr2013 leading to early termination at codon 294 (UAU mutated to UAA) restored photoautotrophic capacity of the T192H mutant. At least part of the N-terminal 293 residues of Slr2013 appeared essential, because transformants with deletion constructs of this gene did not segregate completely. Therefore, the early termination of Slr2013 at residue 294 presumably leads to the production of a truncated polypeptide with an essential function in the cell.
Computer analysis of the Slr2013 primary structure suggests one or two putative transmembrane domains in addition to a cleavable N-terminal transmembrane targeting sequence (Fig. 7). According to the DAS program (4), in the mature protein, a clear transmembrane segment is located between residues 30 and 47, with another possible transmembrane segment around residue 260 (numbering according to the unprocessed translated sequence). A short hydrophobic stretch around residue 333 is too short to be a transmembrane span. Therefore, Slr2013 is likely to be an integral membrane protein with the N terminus of the mature protein on the periplasmic or lumenal side of the cytoplasmic or thylakoid membrane, respectively. Because PS II is in thylakoids, a localization of Slr2013 in thylakoids appears to be most logical. In this case, and provided that the mature protein has two transmembrane helices, the C-terminal part of Slr2013 carrying Tyr294 is exposed to the thylakoid lumen, where it can interact with the Thr192 region of the D2 protein.
FIG. 7.
Slr2013 homologues among other cyanobacterial proteins. Sequence alignment between Slr2013 and homologous proteins from N. punctiforme, Anabaena sp. strain PCC 7120 (Alr5035), and T. elongatus BP-1 (Tlr2082). Residues conserved in at least three proteins are in boldface. Asterisks below the alignment indicate amino acids conserved in all four proteins. Amino acids conserved among DUF58 family members are marked “+” above the alignment. Regions of predicted transmembrane spans are marked with a line above the alignment. An arrow indicates the predicted cleavage site of the signal peptide. The site of the Slr2013 truncation (Tyr294) in the Rg2 mutant is marked “X” above the alignment.
Genes for proteins similar to Slr2013 were found in the genomes of some other cyanobacteria, such as Nostoc punctiforme (contig 362, nucleotides 767 to 2089), Anabaena sp. strain PCC 7120 (alr5035), and Thermosynechococcus elongatus BP-1 (tlr2082) (Fig. 7) but were absent in marine cyanobacteria, such as Synechococcus sp. strain WH8102 or Prochlorococcus marinus. Protein sequence analysis software does not identify specific, known functional motifs in Slr2013 or homologues in other cyanobacteria, but all of them have a DUF58 protein family signature; however, this signature does not have an assigned function. The predicted proteins are fairly conserved among these species of cyanobacteria (50% identity and 79% similarity between Synechocystis sp. strain PCC 6803, N. punctiforme, and Anabaena sp. PCC 7120; 35% identity and 69% similarity between these three proteins and Tlr2082 of T. elongatus BP-1). In addition, genes coding for proteins that are less similar to Slr2013 (20 to 30% identity) are present in a wide range of other prokaryotes. These genes code for proteins that belong to COG (cluster of orthologous groups of proteins) 1721. The proteins of this group share a conserved region that corresponds to the residues 193 to 217 of Slr2013 that is present in the truncated version of this polypeptide in the Rg2 strain of Synechocystis sp. strain PCC 6803. The closest homologue of Slr2013 in this COG is the hypothetical protein NP_253013 (National Center for Biotechnology Information database entry) of Pseudomonas aeruginosa PA01 (encoded by NC_002516.1: 4852313 to 4853644). These proteins have about 30% identity over the entire length of the sequence. Similar proteins were found in Bacillus halodurans (BH0732) and in some other Eubacteria and Archaea. However, significant homologues appear to be absent from eukaryotes.
Each of the cyanobacterial genomes with Slr2013 appears to have a second slr2013-like gene. In Synechocystis sp. strain PCC 6803, Slr1927 is 26% identical to Slr2013 over the N-terminal half of the protein. Similar results are found for Alr4064 (31% identity over residues 81 to 238) in Anabaena sp. strain PCC 7120 and for Tll2086 (34% identity over residues 143 to 267) in T. elongatus BP-1. This suggests a gene duplication event early in the evolution of slr2013-like genes in cyanobacteria. The homologues appear to have diverged functionally, because the lack of segregation of an slr2013 deletion indicates that Slr1927 cannot functionally complement Slr2013.
Function of Slr2013.
In Synechocystis sp. strain PCC 6803, slr2013 could not be deleted under the conditions probed regardless of whether the strain had functionally active PS I or PS II. Therefore, the presence of Slr2013 is required for cell survival for reasons other than regulation of assembly and/or function of the photosynthetic apparatus. Nonetheless, Slr2013 appears to have an effect on assembly and functional activity of PS II. With wild-type Slr2013, the T192H mutation leads to a loss of PS II, while with a truncated Slr2013 protein, photoautotrophic growth is restored in the T192H mutant. Our interpretation of these data is that Slr2013 is involved in assembly of PS II, possibly serving as a chaperone. In the case of full-length Slr2013, PS II centers in the T192H mutant do not appear to assemble, whereas truncated Slr2013 allows formation of fully functional PS II reaction centers.
These observations may be interpreted in several ways. For example, the C-terminal part of Slr2013 may have protease activity towards abnormally folded D2. An alternative explanation is that Slr2013 has chaperone activity (altered upon truncation of Slr2013), helping to fold PS II as well as other proteins, and that the T192H mutant does not fold correctly in the presence of full-length Slr2013. A similar explanation was provided for Slr0399, apparently a chaperone for quinone binding (6). This may explain why the T192H mutant does not have active PS II centers, whereas the Rg2 pseudorevertant with a T192H mutation in the D2 protein and truncated Slr2013 is able to assemble functionally active PS II.
The presence of the T192H mutation is not harmful for the function of the D2 protein and PS II if Slr2013 is truncated. This implies that Thr192 is not in direct contact with one of the PS II electron transfer components, in line with the PS II crystal structure. A primary effect of a mutation on protein folding rather than directly on function may be quite common: For example, a similar situation may exist for the E69Q mutation in the D2 protein, which can be rescued by a mutation in Slr0286 (11), and for the V247M/A249T mutation in the QA-binding domain of the D2 protein, which can be functionally complemented by the partial deletion of slr0399 (6).
Truncation of slr2013 is not a universal tool for Synechocystis to deal with the D2 protein mutations: it does not facilitate restoration of the photosynthetic capacity of the D2 protein mutants with mutations on donor (E69Q) or acceptor (V247 M/A249T) sides of PS II. Similarly, other D2 mutations can be functionally complemented by specific secondary mutations in the genes outside of psbDI (6, 11). Apparently, folding of the D2 protein and PS II assembly in vivo are impaired in some of the D2 mutants and may be affected by many different proteins.
Generation and analysis of pseudorevertants (or second-site revertants) has implicated a new protein in PS II assembly and/or function. Gene knockout approaches would not have been able to provide useful information in this respect, because a complete deletion could not be obtained. A combination of several genetic approaches, including pseudorevertant analysis, clearly is the best way to identify the role of the many conserved and widespread proteins of yet unknown function that have been detected in genomic sequencing projects.
Acknowledgments
We are grateful to Robert Roberson and William Sharp for assistance with electron microscopy studies and to Russell LoBrutto for help with EPR measurements. Useful discussion with Petra Fromme is greatly appreciated.
The work was supported by a grant from the National Science Foundation (MCB-0111058).
REFERENCES
- 1.Anderson, S. L., and L. McIntosh. 1991. Light-activated heterotrophic growth of the cyanobacterium Synechocystis sp. strain PCC 6803: a blue-light-requiring process. J. Bacteriol. 173:2761-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boussiba, S., and W. F. J. Vermaas. 1992. Creation of a mutant with an enriched photosystem II/pigment ratio in the cyanobacterium Synechocystis sp. PCC6803, p. 429-432. In N. Murata (ed.), Research in photosynthesis, vol. III. Kluwer, Dordrecht, The Netherlands.
- 3.Craig, S., J. C. Gilkey, and L. A. Staehelin. 1987. Improved specimen support cups and auxiliary devices for the Balzers high pressure freezing apparatus. J. Microsc. 148:103-106. [DOI] [PubMed] [Google Scholar]
- 4.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Prot. Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 5.Diner, B. A., and F. Rappaport. 2002. Structure, dynamics, and energetics of the primary photochemistry of photosystem II of oxygenic photosynthesis. Annu. Rev. Plant Biol. 53:551-580. [DOI] [PubMed] [Google Scholar]
- 6.Ermakova-Gerdes, S., and W. Vermaas. 1999. Inactivation of the open reading frame slr0399 in Synechocystis sp. PCC 6803 functionally complements mutations near the QA niche of photosystem II: a possible role of Slr0399 as a chaperone for quinone binding. J. Biol. Chem. 43:30540-30549. [DOI] [PubMed] [Google Scholar]
- 7.Haag, E., J. J. Eaton-Rye, G. Renger, and W. F. J. Vermaas. 1993. Functionally important domains of the large hydrophilic loop of CP47 as probed by oligonucleotide-directed mutagenesis in Synechocystis sp. PCC 6803. Biochemistry 32:4444-4454. [DOI] [PubMed] [Google Scholar]
- 8.Joset, F. 1988. Transformation in Synechocystis PCC 6714 and PCC 6803: preparation of chromosomal DNA. Methods Enzymol. 167:712-714. [DOI] [PubMed] [Google Scholar]
- 9.Kiss, J. Z., T. H. Giddings, Jr., L. A. Staehelin, and F. D. Sack. 1990. Comparison of the ultrastructure of conventionally fixed and high-pressure frozen freeze substituted root-tips of Nicotiana and Arabidopsis. Protoplasma 157:64-74. [DOI] [PubMed] [Google Scholar]
- 10.Kufryk, G. I., M. Sachet, G. Schmetterer, and W. F. J. Vermaas. 2002. Transformation of the cyanobacterium Synechocystis sp. PCC 6803 as a tool for genetic mapping: optimization of efficiency. FEMS Microbiol. Lett. 206:215-219. [DOI] [PubMed] [Google Scholar]
- 11.Kufryk, G. I., and W. F. J. Vermaas. 2001. A novel protein involved in the functional assembly of the oxygen-evolving complex of photosystem II in Synechocystis sp. PCC 6803. Biochemistry 40:9247-9255. [DOI] [PubMed] [Google Scholar]
- 12.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Mann, N. H., N. Novac, C. W. Mullineaux, J. Newman, S. Bailey, and C. Robinson. 2000. Involvement of an FtsH homologue in the assembly of functional photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 479:72-77. [DOI] [PubMed] [Google Scholar]
- 14.Mohamed, A., and C. Jansson. 1989. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol. Biol. 13:693-700. [DOI] [PubMed] [Google Scholar]
- 15.Naver, H., E. Bodreau, and J.-D. Rochaix. 2001. Functional studies of Ycf3: its role in assembly of photosystem I and interactions with some of its subunits. Plant Cell 13:2731-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Prot. Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 17.Reynolds, E. S. 1963. The use of lead citrate at high pH as an electron opaque stain in electron microscopy. J. Cell Biol. 17:208-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 19.Stevens, D. R., J.-D. Rochaix, and S. Purton. 1996. The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol. Gen. Genet. 251:23-30. [DOI] [PubMed] [Google Scholar]
- 20.Tang, X.-S., D. A. Chisholm, G. C. Dismukes, G. W. Brudvig, and B. A. Diner. 1993. Spectroscopic evidence from site-directed mutants of Synechocystis PCC6803 in favor of a close interaction between histidine-189 and redox-active tyrosine-160 both of polypeptide-D2 of the photosystem-II reaction center. Biochemistry 32:13742-13748. [DOI] [PubMed] [Google Scholar]
- 21.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tommos, C., L. Davidsson, B. Svensson, C. Madsen, W. Vermaas, and S. Styring. 1993. Modified EPR-spectra of the tyrosine(D) radical in photosystem II in site-directed mutants of Synechocystis sp. PCC 6803: identification of side-chains in the immediate vicinity of tyrosineD on the D2 protein. Biochemistry 32:5436-5441. [DOI] [PubMed] [Google Scholar]
- 23.Vermaas, W., J. Charité, and B. Eggers. 1990. System for site-directed mutagenesis in the psbDI/C operon of Synechocystis sp. PCC 6803, p. 231-238. In M. Baltscheffsky (ed.), Current research in photosynthesis, vol. I. Kluwer, Dordrecht, The Netherlands.
- 24.Vermaas, W. F. J. 1998. Gene modifications and mutation mapping to study the function of photosystem II. Methods Enzymol. 297:293-310. [DOI] [PubMed] [Google Scholar]
- 25.Wilde, A., K. Lünser, F. Ossenbühl, J. Nickelsen, and T. Börner. 2001. Characterization of the cyanobacterial ycf37: mutation decreases the photosystem I content. Biochem. J. 357:211-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zdobnov, E. M., and R. Apweiler. 2001. InterProScan: an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847-848. [DOI] [PubMed] [Google Scholar]