Abstract
Adenosine kinase (AK) is a purine salvage enzyme that catalyzes the phosphorylation of adenosine to AMP. In Mycobacterium tuberculosis, AK can also catalyze the phosphorylation of the adenosine analog 2-methyladenosine (methyl-Ado), the first step in the metabolism of this compound to an active form. Purification of AK from M. tuberculosis yielded a 35-kDa protein that existed as a dimer in its native form. Adenosine (Ado) was preferred as a substrate at least 30-fold (Km = 0.8 ± 0.08 μM) over other natural nucleosides, and substrate inhibition was observed when Ado concentrations exceeded 5 μM. M. tuberculosis and human AKs exhibited different affinities for methyl-Ado, with Km values of 79 and 960 μM, respectively, indicating that differences exist between the substrate binding sites of these enzymes. ATP was a good phosphate donor (Km = 1100 ± 140 μM); however, the activity levels observed with dGTP and GTP were 4.7 and 2.5 times the levels observed with ATP, respectively. M. tuberculosis AK activity was dependent on Mg2+, and activity was stimulated by potassium, as reflected by a decrease in the Km and an increase in Vmax for both Ado and methyl-Ado. The N-terminal amino acid sequence of the purified enzyme revealed complete identity with Rv2202c, a protein currently classified as a hypothetical sugar kinase. When an AK-deficient strain of M. tuberculosis (SRICK1) was transformed with this gene, it exhibited a 5,000-fold increase in AK activity compared to extracts from the original mutants. These results verified that the protein that we identified as AK was coded for by Rv2202c. AK is not commonly found in bacteria, and to the best of our knowledge, M. tuberculosis AK is the first bacterial AK to be characterized. The enzyme shows greater sequence homology with ribokinase and fructokinase than it does with other AKs. The multiple differences that exist between M. tuberculosis and human AKs may provide the molecular basis for the development of nucleoside analog compounds with selective activity against M. tuberculosis.
Mycobacterium tuberculosis is a pathogenic mycobacterium that is the causative agent of tuberculosis (TB). TB was once ranked as the second leading cause of death in the United States; however, the mortality rate dropped from 50% to <1% with the advent of effective antibiotic therapy (24, 26). During the 1980s, the simultaneous emergence of multidrug-resistant TB and human immunodeficiency virus-TB coinfection acted synchronously to reverse the downward trends in TB infection and mortality. Between 1985 and 1992, newly reported cases of TB increased by 20% in the United States alone (26). Today, tuberculosis infects one-third of the world's population and ranks as the leading cause of death from an infectious disease, claiming 2 million lives annually (10, 26). The World Health Organization has recognized M. tuberculosis infection as a world health emergency and has raised a call for new treatments for this old pathogen.
Nucleoside analogs are an important class of drugs used in the treatment of viral infections and cancer that may prove to be particularly useful in treating multidrug-resistant TB, since their mechanism of action is likely to be different from those of existing therapies. Nucleoside analogs are prodrugs resembling natural nucleosides that usually must be metabolized intracellularly to active compounds. Purine salvage enzymes can permit the metabolism of nucleoside analogs to active compounds, and the presence of these enzymes in other Mycobacterium spp. has been well documented (30, 31, 32). The analysis of the genomic sequence has permitted identification of several genes that purportedly code for purine salvage enzymes in M. tuberculosis (7), and this information has led to the study of M. tuberculosis purine nucleoside phosphorylase (3). Identification and characterization of other purine salvage enzymes in M. tuberculosis may prove beneficial to the development of novel nucleoside analogs with antitubercular activity; indeed, several nucleoside analogs have demonstrated antitubercular activity in vitro (28).
2-Methyladenosine (methyl-Ado) is a nucleoside analog that has demonstrated promising antitubercular activity, with an in vitro MIC of 3 μg/ml (6). The role that adenosine kinase (AK) plays in methyl-Ado activation was established in previous studies that focused on the in vitro metabolism of methyl-Ado in Mycobacterium smegmatis strain mc2155, a closely related, fast-growing model system for M. tuberculosis (6). These studies revealed that the main intracellular metabolites of methyl-Ado were its phosphorylated products, methyl-AMP, methyl-ADP, and methyl-ATP, with little incorporation into bacterial RNA (6). Furthermore, SRI101, an AK-deficient strain of M. smegmatis, was not sensitive to methyl-Ado, indicating that AK is a requirement for methyl-Ado activity (6).
AK is a purine salvage enzyme that is responsible for the phosphorylation of Ado to AMP. AKs from various mammalian and parasitic sources have been characterized, but the enzyme is not commonly found in bacteria. AKs belong to the PfkB family of carbohydrate and nucleoside kinases, a family of structurally related enzymes that includes ribokinase, fructokinase, and hexokinase among its members (27). Characterization of AK from M. tuberculosis should reveal some insights into the basis for the selective antimycobacterial activity of methyl-Ado. To this end, an effort has been made to compare biochemical characteristics of AKs from M. tuberculosis and human sources, focusing on differences in metabolism that may be responsible for the selective activation of methyl-Ado in M. tuberculosis. The characterization of AK and other enzymes in the purine salvage pathway will hopefully aid in the development of selective antimycobacterial nucleoside analogs like methyl-Ado.
MATERIALS AND METHODS
Reagents.
Radiolabeled nucleosides, including [2,8-3H]Ado (30 Ci/mol), [8-3H]guanosine (6.3 Ci/mmol), [2,8-3H]inosine (14.6 Ci/mmol), [5-3H]cytidine (25.3 Ci/mmol), [methyl-3H]thymidine (60 Ci/mmol), [5-3H]uridine (17.7 Ci/mmol), and [2,8-3H]deoxyadenosine (13 Ci/mmol), were purchased from Moravek Biochemicals, Inc. (Brea, Calif.). methyl-Ado was prepared at the Southern Research Institute, Birmingham, Ala. (6), and radiolabeled by Moravek Biochemicals, Inc., to form [8-3H]methyl-Ado (10.2 Ci/mmol). Bradford dye reagent, sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE) minigels, low-molecular-weight standards, and silver stain reagents were purchased from Bio-Rad Laboratories (Hercules, Calif.). The pVV16 shuttle vector was a gift from Varalakshmi Vissa (Colorado State University). Oligonucleotide primers were purchased from Operon Technologies (Alameda, Calif.). Dynazyme EXT was purchased from Finnzymes (Espoo, Finland). The Qiagen (Valencia, Calif.) miniprep kit was used to purify plasmid DNA.
Cell lines and growth conditions.
M. tuberculosis strain H37Ra (ATCC 25177) and M. smegmatis strain mc2155 (ATCC 700084) were cultured in Middlebrook 7H9 medium supplemented with oleic acid, albumin, dextrose, and catalase (OADC) and 0.05% Tween 80. The AK-deficient strains SRICK1 and SRI101 were used for activity complementation studies. SRICK1 is an AK-deficient spontaneous mutant obtained by growing M. tuberculosis H37Ra on Middlebrook 7H10 medium containing 10 μg of methyl-Ado/ml; mutants occurred with a frequency of ∼1 in 106 cells. SRI101 was created by transposon mutagenesis of M. smegmatis mc2155 (6). AK-deficient strains were grown in Middlebrook 7H9 medium containing OADC and 0.05% Tween 80, supplemented when necessary with 50 μg of hygromycin/ml. Escherichia coli strain DH5α was cultured in Luria-Bertani medium supplemented when necessary with 50 μg of kanamycin/ml. Bacteria were grown at 37°C under either stationary or shaking conditions, as required.
CEM cells (ATCC CCL-119) were used as a source of human AK. The CEM cells were cultured in RPMI 1640 medium supplemented with l-glutamine, 10% fetal bovine serum, and 25 mM HEPES. Cultures were grown at 37°C with 5% CO2.
Activity assays.
M. tuberculosis AK activity was followed throughout purification by a filter disk assay. Phosphorylation of [3H]Ado was quantified by the amount of [3H]AMP bound to a DE-81 cellulose disk following the reaction. The assay conditions consisted of 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 5 mM ATP, 20 μM [3H]Ado (10 μCi/ml), and 10 μM deoxycoformycin, an inhibitor of Ado deaminase. KCl (10 mM) was added to the reaction mixture when appropriate. The reaction was started by the addition of enzyme, incubated for the desired time at 37°C, and stopped by the addition of 10 μl of 0.1 M EDTA. Aliquots of 50 μl of reaction mixture were applied to DEAE cellulose disks, and the disks were batch washed three times with 1 mM ammonium acetate (pH 5.0), rinsed with 95% ethanol, and dried. The filter disks were transferred to scintillation vials with 10 ml of Complete Counting Cocktail (Research Products International, Mount Prospect, Ill.), and radioactivity was detected with a Packard Tri-Carb model 1900 TR liquid scintillation analyzer. Human AK was assayed as described above with the following variations: 50 mM Tris-HCl (pH 7.5), 1 mM MgCl2, 1 mM ATP, 2 μM [3H]Ado (10 μCi/ml), 10 μM deoxycoformycin, and 40 mM KCl.
Protein characterization and substrate studies for M. tuberculosis AK were performed with partially purified AK that had been carried through the first three steps of purification and had been shown to be free of Ado deaminase and AMP kinase activity, as these enzymes can interfere with AK kinetics by depleting the substrate and product, respectively. The presence of Ado deaminase activity was determined by reverse-phase high-performance liquid chromatography analysis of assay products, as previously described (6). Deoxycoformycin was excluded from activity assays when the protein preparation was free of the enzyme. AMP kinase activity was detected by strong anion-exchange high-performance liquid chromatography analysis of assay products (6). All enzyme reactions were linear during the incubation period, and substrate conversions were maintained in the 5 to 10% range.
Preparation of crude protein extracts.
Protein was extracted from CEM cells or M. tuberculosis strain H37Ra as follows. The cells were pelleted by centrifugation, resuspended with an equal volume of PBS, and rinsed twice with 50 mM Tris-HCl (pH 7.5) containing 5 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, 2.3 μg of leupeptin/ml, and 1.1 μg of pepstatin/ml and resuspended in the same buffer. The resuspended M. tuberculosis cells were disrupted with 0.1-mm-diameter glass beads using a Mini bead beater apparatus (Bio Spec Products, Inc.), and the CEM cells were disrupted by sonication. Particulate matter was removed by centrifugation at 3,000 × g for 20 min, followed by centrifugation at 40,000 × g for 1 h at 4°C in a Beckman (Fullerton, Calif.) L-70 ultracentrifuge. The supernatant was filtered through a 0.2-μm-pore-size filter and dialyzed against 50 mM Tris-HCl (pH 7.5) with 10 mM NaCl, 1 mM dithiothreitol (DTT), and 20% glycerol.
Purification of M. tuberculosis AK.
A 4-ml aliquot containing 14 mg of total protein was subjected to stepwise ammonium sulfate precipitation using 40, 60, and 80% ammonium sulfate. The precipitated pellets were resuspended in 50 mM Tris-HCl (pH 7.5) containing 10 mM NaCl and 1 mM DTT (buffer A), and the resuspended protein was dialyzed against the same buffer. AK activity was found in the 40 to 60% ammonium sulfate fraction. Protein was subject to separation by anion-exchange chromatography on a Hi Trap Q column, where separation was achieved by running a linear salt gradient from 100 to 400 mM NaCl in buffer A. Fractions containing AK activity were pooled, dialyzed against buffer A, and then concentrated to a final volume of 200 μl using a Centricon Plus-20 centrifugal filter device (10,000-Da cutoff; Amicon). The concentrated protein was loaded onto a Superose 12 column and subjected to gel filtration chromatography using an isocratic run with buffer A. Fractions containing AK activity were pooled and applied to a Mono Q anion-exchange column, where a linear salt gradient from 100 to 400 mM NaCl in buffer A was used to elute the protein. Protein concentrations were determined at each step by the Bradford method, using bovine serum albumin as a standard (5). The purified protein was subjected to sequencing on a Beckman model PI 2090E amino acid sequencer, using Edman degradation.
Preparation of human AK.
A CEM cell extract was subjected to precipitation with 50% ammonium sulfate and dialyzed against 50 mM Tris (pH 7.5) containing 10 mM NaCl, 1 mM DTT, and 10% glycerol (buffer B). AK activity was found in the >50% ammonium sulfate fraction. Protein was subjected to separation by anion-exchange chromatography on a Hi Trap Q column, followed by cation-exchange chromatography on a Hi Trap SP column. At pH 7.5, human AK did not adhere to either resin; however, >50% of the total protein was removed in each step. Preparation of human AK removed 83% of the total protein and provided fivefold purification, with 75% recovery of AK activity. The partially purified protein preparation was found to be free of AMP kinase, but some Ado deaminase activity remained; therefore, deoxycoformycin was included in all assays involving the human enzyme.
Enzyme kinetics.
Michaelis-Menton parameters were determined from linear double-reciprocal plots of 1/velocity versus 1/concentration of the substrate. The best line was determined by linear regression of at least five data points (the regression coefficient for each line was >0.95), and the Km and Vmax values were determined from the intercept of the x and y axes, respectively. The values were expressed as means ± standard errors of the mean for at least three determinations.
Cloning.
The RV2202c gene (accession no. Q10391) (7) was amplified by PCR from M. tuberculosis strain H37Rv chromosomal DNA using the oligonucleotide primers 5′-GGACGGAGATCATATGACGATCGCGGTACC-3′ and 3′-ACGCCGAGCGACTAGACGTCGTGGTGCGAC-5′, containing NdeI and PstI restriction sites, respectively. The PCR product was digested with NdeI and PstI restriction enzymes and cloned into a pVV16 shuttle vector, which imparts hygromycin resistance. After propagation in E. coli strain DH5α, plasmid DNA was purified according to the Qiagen miniprep protocol. DNA sequencing confirmed that cloned Rv2202c had no mutations. The final construct (pVV16/Rv2202c) was transformed by electroporation into AK-deficient M. smegmatis and M. tuberculosis strains, SRI101 and SRICK1, respectively, and transcription was driven by the hsp60 promoter present in pVV16.
Determination of native molecular mass.
The native molecular mass of the enzyme was estimated using size exclusion chromatography. A Superose 12 analytical-grade column (Pharmacia) was calibrated using molecular mass standards ranging from 25 to 669 kDa, and a calibration curve was created based on the elution volumes of these standards. The native molecular mass of AK was extrapolated from the calibration curve based on its elution from the column.
A PD2020A light-scattering detector (Precision Detectors, Franklin, Mass.) was used in conjunction with size exclusion chromatography. As peaks eluted from the column, changes in the refractive index and light-scattering properties were collected using an 810-nm laser with detectors at 15 and 90°. Light-scattering data were analyzed on Discovery 32 software (Precision Detectors).
The native molecular mass of the enzyme was determined by sedimentation velocity and sedimentation equilibrium analytical ultracentrifugation using a Beckman XLA ultracentrifuge. The sedimentation velocity was determined by spinning three protein concentrations at 59,000 rpm for 10 h. Sedimentation velocity data were collected at 250 and 280 nm and then analyzed with Sedfit87 software (National Institutes of Health). Sedimentation equilibrium data were obtained for two protein concentrations at 10,000, 12,500, 15,000, and 18,000 rpm. Data were collected at 250 and 280 nm and then analyzed using Beckman XL-A/XL-1 data analysis software, version 4.0.
Determination of MIC.
The MIC of methyl-Ado was evaluated for M. tuberculosis strains H37Ra, SRICK1, and SRICK1-pVV16/Rv2202c using a colorimetric broth microdilution assay as previously described (29). Dilutions of methyl-Ado, ethambutol, and dimethyl sulfoxide (DMSO) were prepared in Middlebrook 7H9 medium supplemented with 0.2% glycerol and OADC. For each concentration of methyl-Ado, ethambutol, and DMSO, 50 μl of the appropriate dilution of the compound was added to triplicate wells. Two of the three wells were inoculated with ∼5 × 104 CFU/well in 50 μl of Middlebrook 7H9 supplemented with OADC, and 50 μl of Middlebrook 7H9 supplemented with OADC was added to the third well, which served as a color control. For methyl-Ado and ethambutol, seven dilutions ranging from 50 to 0.5 μg/ml were used, while DMSO concentrations ranged from 0.5 to 0.05%. Viability controls and medium control wells were included in each assay. The assay plates were incubated at 37°C. On the sixth day postinoculation, 50 μl containing Alamar blue dye reagent diluted in 0.05%Tween 80 was added to each test well, and the plates were incubated for an additional 18 h. Reduction of Alamar blue dye was measured on an optical microtiter plate reader programmed to subtract the absorbance at 600 nm from that at 570 nm. The MIC was reported as the lowest concentration of drug with a differential absorbance of zero or less.
RESULTS
Purification of M. tuberculosis AK.
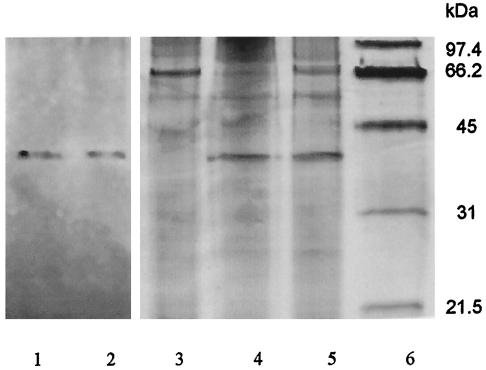
Ammonium sulfate precipitation removed ∼60% of contaminating proteins; however, it did not yield a large purification factor due to the concurrent loss of AK activity. The most efficient step in protein purification was anion-exchange chromatography using the Hi Trap Q column, where AK eluted in fractions containing 250 to 320 mM NaCl. AK eluted from the Superose 12 gel filtration column in a single peak at 136 kDa (data not shown). In the final purification step, enzyme eluted from the Mono Q anion-exchange column at 260 to 320 mM NaCl. This procedure eliminated 99.99% of the protein and produced 100-fold purification (Table 1). Only fractions containing maximum activity were carried over to the next step in order to minimize contamination by other proteins, which is reflected by the low (1%) percent yield. Enzyme preparations were stable when stored at −20°C in 20% glycerol for up to 6 months. The appearance of a single band at 35 kDa on a silver-stained SDS-PAGE gel (Fig. 1) verified that AK was purified to apparent homogeneity.
TABLE 1.
Purification table for AK from M. tuberculosis
| Step | Vol (ml) | Total protein (μg) | Sp act (nmol/mg-min) | Purification factor | Total activity (nmol/min) | Recovery (%) |
|---|---|---|---|---|---|---|
| M. tuberculosis crude | 4.0 | 14,400 | 3.9 | 1 | 56,000 | 100 |
| Ammonium sulfate | 2.3 | 5,300 | 3.9 | 1 | 21,000 | 37 |
| Hi Trap Q | 0.2 | 92 | 53 | 14 | 4,900 | 9 |
| Superose 12 | 1.6 | 14 | 115 | 30 | 1,600 | 3 |
| Mono Q | 0.3 | 14 | 390 | 100 | 550 | 1 |
FIG. 1.
Molecular mass of AK purified from M. tuberculosis. The monomer molecular mass of purified AK was estimated based on a denaturing SDS-12% PAGE. Lanes 1 and 2 contain Mono-Q fractions with AK activity from purification of native enzyme. This protein was used for amino acid sequencing. Lane 3 contains 2 μg of crude protein extract of SRI101, an AK-deficient strain created from M. smegmatis mc2155. Lanes 4 and 5 each contain 2 μg of different preparations of SRICK1 with cloned AK, and lane 6 contains molecular mass markers.
Amino acid sequence and gene identification.
Protein sequencing of purified M. tuberculosis AK revealed that the 15 N-terminal amino acids were TIAVTGSIATDHLMR. A BLAST search (http://www.ncbi.nlm.nih.gov/BLAST) of the N-terminal sequence against the translated M. tuberculosis genome showed complete identity with the N-terminal sequence of a 324-amino-acid protein with a molecular mass of 34,341 Da. The protein identified, Rv 2202c (accession no. Q10391), is annotated as a hypothetical sugar kinase and is coded for by a gene currently named cbhK. From this point on, this gene will be referred to as adoK.
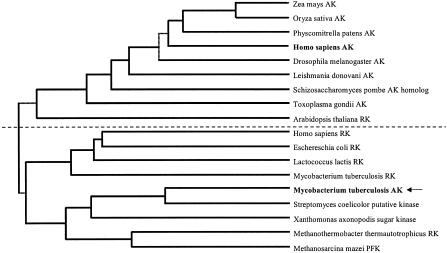
M. tuberculosis AK showed low (24%) overall homology with other known AKs, which prevented the identification of the function of the enzyme based on its amino acid sequence. In light of its low sequence homology with other AKs, a phylogenetic analysis of the enzyme was performed. The amino acid sequence from M. tuberculosis AK was aligned with sequences from related AKs and ribokinases using ClustalW sequence alignment (European Bioinformatics Institute; http://www.ebi.ac.uk/clustalw/index.html), and a phylogenetic tree was created based on this alignment using genebee software (http://www.genebee.msu.su/index.html). The phylogenetic tree indicated that M. tuberculosis AK was more closely related to ribokinases from various sources than to other AKs (Fig. 2).
FIG. 2.
Phylogenetic analysis of M. tuberculosis AK and related enzymes from the PfkB family. Amino acid sequences of several AKs and ribokinases (RK) from various sources were aligned with phosphofructokinase (PFK) from Methanosarcina mazei, an S. coelicolor putative kinase, and sugar kinase from Xanthomonas axonopodis. All proteins were members of the PfkB family of carbohydrate and nucleoside kinases. The phylogenetic tree was created as described in Results and is displayed as a cluster phylogram. The dashed line bisecting the tree roughly separates AKs from RK.
Amino acid sequence alignment with other AKs permitted the identification of some regions in the M. tuberculosis enzyme that are highly conserved with other AKs. Analysis of the aligned sequences revealed that there is one region from D250 to L263 of M. tuberculosis AK that is homologous with the region that interacts with the γ-phosphate of ATP in the human enzyme (19). In this region, 9 of 14 residues were identical in the human and M. tuberculosis AKs, with overall homology between the sequences comprising 11 of 14 residues. Unlike the amino acids that interact with ATP, those that interact with Ado are mostly scattered in the N-terminal half of the sequence and do not form a highly conserved linear motif. Sequence alignment with human AK (19) revealed several conserved amino acid residues that interact with Ado in the active site. The amino acid sequence also contained an NXXE motif that is highly conserved in the PfkB family of carbohydrate kinases and that has been identified as being important for binding pentavalent ions and Mg2+ in the active site (17).
The amino acid sequence of M. tuberculosis AK showed at least 50% sequence homology with predicted proteins from Streptomyces coelicolor, Streptomyces fradiae, Burkholderia mallei, Burkholderia pseudomallei, Desulfovibrio vulgaris, Methylococcus capsulatus, Thiobacillus ferrooxidans, Bordetella parapertussis, Bordetella bronchiseptica, Xanthomonas campestris, and other Mycobacterium spp. These proteins either have unspecified functions or predicted functions as carbohydrate or sugar kinases.
Characterization of cloned AK.
To confirm that Rv2202c encoded an AK, the gene was cloned and expressed in AK-deficient strains of M. smegmatis and M. tuberculosis, SRI101 and SRICK1, respectively. Following transformation with pVV16/Rv2202c, AK activity was restored in both AK-deficient bacterial strains to a level at least 900 times higher than those of the original mutants (Table 2).
TABLE 2.
AK activities and MICs for methyl-Ado in different strains of M. tuberculosis and M. smegmatisa
| Strain | Properties | AK activity (nmol/mg-min) | MIC for methyl-Ado (μg/ml)b |
|---|---|---|---|
| M. tuberculosis H37Ra | Wild type | 5 | 5 |
| M. tuberculosis SRICK1 | AK deficient | <0.02 | >50 |
| M. tuberculosis SRICK1-pVV16/Rv2202c | SRICK1 transformed with Rv2202c | 100 | 1 |
| M. smegmatis mc2155 | Wild type | 15 | ND |
| M. smegmatis SRI101 | AK deficient | 0.4 | ND |
| M. smegmatis SRI101-pVV16/Rv2202c | SRI101 transformed with Rv2202c | 360 | ND |
AK activity was measured in crude protein extracts of the indicated bacterial strains.
ND, not determined.
While M. tuberculosis strain H37Ra is sensitive to methyl-Ado, the AK-deficient mutant, SRICK1, is methyl-Ado resistant, presumably because it lacks AK activity. Transformation of SRICK1 with pvv16/Rv2202c plasmid DNA restored its sensitivity to methyl-Ado (Table 2). The sensitivity of each M. tuberculosis strain to methyl-Ado corresponded to the amount of AK activity detectable in protein extracts from each M. tuberculosis strain (Table 2). Denaturing SDS-PAGE of crude protein extracts from complemented bacterial strains revealed a prominent protein band at 35 kDa (Fig. 1), while size exclusion chromatography revealed that cloned Rv2202c eluted around 135 kDa, the same as native AK (data not shown). Mass spectrometry of cloned AK demonstrated that the protein was 34,337 Da, confirming the fidelity of the construct (data not shown). Cloned AK purified in exactly the same manner as the native enzyme, and kinetic studies with cloned AK demonstrated that the Km values for Ado and methyl-Ado were the same as those of the native enzyme (data not shown). Restoration of AK activity and methyl-Ado sensitivity to SRICK1 and SRI101 cells that had been complemented with Rv2202c confirmed that Rv2202c is the gene that codes for AK in M. tuberculosis.
Characterization of native AK.
Kinase activity was evaluated with 20 μM Ado, guanosine, inosine, deoxyadenosine, cytidine, uridine, or thymidine. Of the nucleosides tested, Ado was the only nucleoside that served as a substrate for this kinase. In the presence of other nucleosides, the kinase activity was below the detection limit (3% of the activity seen with Ado [data not shown]), which indicated that Ado was at least 30-fold better than other nucleosides as a substrate for this enzyme. Because deoxyadenosine is a substrate for human AK with 7% of the activity of Ado at 25 μM (34), deoxyadenosine was further examined as a substrate for M. tuberculosis AK. The assay conditions were altered by increasing the protein concentration so that the detection limit was 0.002% of the activity of Ado; under these conditions, deoxyadenosine exhibited 0.014 ± 0.004% of the activity of Ado (data not shown), which indicated that it was at least 7,000-fold worse than Ado as a substrate.
Potential phosphate donors, including ATP, GTP, UTP, CTP, dTTP, dATP, dGTP, and dCTP, were tested at 5 mM each. GTP and dGTP were the best phosphate donors, with 2.5 and 4.7 times the activity observed with ATP, respectively (data not shown). UTP, dATP, and dTTP exhibited ∼33% of the activity observed with ATP, while CTP and dCTP were the worst phosphate donors, with 1.7 and 0.2% of the activity observed with ATP, respectively (data not shown).
In order to eliminate the possibility that the AK activity that we detected was an artifact of a related sugar kinase, ribose, glucose, fructose, and fructose-6-phosphate were evaluated as inhibitors in the presence of 20 μM Ado. None of the sugars assayed were inhibitors at 1,000 μM (data not shown), which indicated that these compounds were at best poor substrates and suggested that this protein is a specific AK.
Iodotubercidin is an Ado analog that is a potent competitive inhibitor of human AK, with a Ki between 3 and 30 nM (8, 22). Double-reciprocal plots of AK activity in the presence of iodotubercidin were constructed with five data points for each concentration of iodotubercidin (the regression coefficients were at least 0.94). These showed that iodotubercidin was a competitive inhibitor of M. tuberculosis AK. The Ki was determined to be 210 ± 100 nM (n = 3) from replots of 1/slope of the double-reciprocal plot versus the concentration of iodotubercidin (the regression coefficients were at least 0.99).
Substrate inhibition is a common characteristic of AKs from various sources. The M. tuberculosis AK reaction velocity was evaluated in the presence of 0.05 to 100 μM Ado. The reaction velocity increased linearly up to 0.5 μM and reached a maximum at 5 μM Ado, then decreased with increasing concentrations of Ado. By 25 μM, the reaction rate had decreased to 75% of the maximum rate, and the reaction velocity plateaued at 70% of the maximum velocity from 50 to 100 μM Ado (data not shown). This substrate inhibition is similar to that reported for human AK (23).
M. tuberculosis AK was dependent on the presence of Mg2+ for activity. Optimum AK activity occurred in the presence of 10 to 50 mM MgCl2; however, MgCl2 concentrations of >50 mM were inhibitory (data not shown). A 50% loss of AK activity occurred with 125 mM MgCl2, corresponding to an ionic strength of 375 mM. A similar decrease in activity did not occur until 750 mM NaCl in the presence of 10 mM MgCl2, corresponding to an ionic strength of 780 mM. Therefore, the loss of activity appears to be a specific effect of Mg2+ and not an effect of ionic strength. The ATP/Mg ratio plays a critical role in regulating activity for AKs from various sources (21, 23, 25, 34), and M. tuberculosis AK assays were typically performed with an ATP/Mg ratio of 1:2.
M. tuberculosis AK activity was assessed in the presence of the monovalent salts LiCl, NaCl, and KCl, and the enzyme exhibited stimulation in the presence of KCl that has not been reported for other AKs. In the presence of 10 mM KCl, the Km for Ado decreased from 3.4 to 0.8 μM (P < 0.05) and the Vmax increased from 60 to 180 nmol/mg-min (P < 0.005). A profound effect was also observed for methyl-Ado in the presence of 10 mM KCl. The Km decreased from 709 to 79 μM (P < 0.005), while the Vmax increased from 2.3 to 72 nmol/mg-min (P < 0.005). The effect of KCl on the phosphorylation of Ado and methyl-Ado resulted in 12- and 300-fold increases in Vmax/Km, respectively, which indicated that AK phosphorylated these substrates more efficiently in the presence of KCl. The stimulatory effects were observed in the presence of KCl but not in the presence of NaCl or LiCl (data not shown). Therefore, we attributed this stimulation of activity to a specific effect of K+.
Methyl-Ado was evaluated as a substrate for human AK in the presence of 40 mM KCl, a concentration that was representative of most human AK assays. With human AK, the Km for methyl-Ado was 960 μM, and the Vmax was 0.64 nmol/mg-min (Table 3), which are similar to the values seen for M. tuberculosis AK in the absence of KCl.
TABLE 3.
Comparative properties of M. tuberculosis and human AK
| Property | Value
|
Referencea | |
|---|---|---|---|
| M. tuberculosis AK | Human AK | ||
| Molecular mass (kDa) | 34.3 | 38.7 | 2, 27 |
| Stoke's radius (Å) | 49.4 | 26.4 | 2 |
| Quaternary structure | Dimer | Monomer | 2, 34 |
| pH optimum | 8-11 | 5.5, 7.5-8.5 | 34 |
| pI | 4.5 | 5.9 | 2 |
| Ki of iodotubercidin (nM) | 210 ± 100 | 3-30 | 8, 14 |
| Km of Ado (μM) | 0.80 ± 0.08 | 0.15-0.4 | 23, 34, 35 |
| Km of ATP (μM) | 1,100 ± 140 | 75 | 23, 34 |
| Km of mAdo (μM) | 79 ± 26 | 961 ± 385 | |
| Vmax of mAdo (nmol/mg-min) | 72 ± 3.9 | 0.64 ± 0.21 | |
| Catalytic efficiency of mAdo | 0.91 | 0.00067 | |
| Phosphate donors | dGTP > GTP > ATP > dATP, UTP, dTTP >> CTP, dCTP | GTP, dGTP > ATP, dATP > CTP, UTP > dTTP, dCTP | 18 |
| Substrates | Adenosine | Adenosine >> deoxyadenosine | 18 |
Reference for properties of human AK. Properties of M. tuberculosis AK were generated as a result of the present work.
Pi has been reported to have a stimulatory effect on various AKs, with a concurrent depression of the Km for Ado and ATP and an increase in Vmax for these reactants (12, 13, 16, 17). The effect of Pi on M. tuberculosis AK was evaluated, using up to 50 mM sodium phosphate with Ado concentrations both above and below the Km for Ado, and little or no stimulation of activity was observed (data not shown).
M. tuberculosis AK activity was dependent on pH. The optimum pH range for AK activity was from pH 8 to 11 (data not shown), with activity decreasing rapidly at values above pH 11 and below pH 8. AK was inactive at its calculated pI of 4.3, and activity was not restored by dialyzing the enzyme back to pH 8. Although the enzyme is fairly stable, it cannot tolerate dramatic reductions in pH.
AK eluted from a Superose 12 size exclusion column as a single peak near the aldolase standard (158 kDa), and absorbance at 280 nm correlated with AK activity. The molecular mass of native AK was determined to be 136 kDa, with a Stoke's radius of 49.4 Å from the calibration curve (R = 0.99).
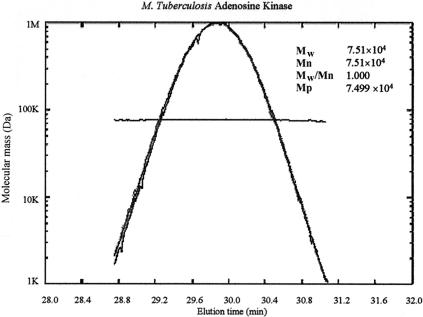
Light-scattering analysis of the peak of AK activity as it eluted from the column indicated that the molecular mass of the protein was 74,840 ± 710 Da (n = 4) (Fig. 3); this molecular mass is approximately that of a dimer. The molecular mass distribution across the width of the peak was uniform, as indicated by the line traversing the peak of the refractive index and 15 and 90° light scatter (Fig. 3). Sedimentation velocity and sedimentation equilibrium analytical ultracentrifugation confirmed that the native form of the enzyme was a dimer with a molecular mass of ∼70 kDa and a sedimentation coefficient of 4.47. The dissociation constant for the dimer was 2.3 × 10−7 M, and weight-fraction plots demonstrated that >90% of the enzyme was a dimer at 1 mg/ml (∼30 μM) (data not shown).
FIG. 3.
Native molecular mass of AK as determined by light scattering. Refractive indexes and 15 and 90° light-scattering data were acquired for AK as it eluted from a Superdex 200 size exclusion column. The scale on the ordinate axis is a reference for the molecular mass distribution line; the refractive index and 15 and 90° light-scattering traces are scaled to the maximum height of the plot. The plot of elution time versus molecular mass shows the distribution of molecular masses across the width of the AK peak. Mw is the weighted-average molecular mass, Mn is the average molecular mass, and Mp is the molecular mass associated with the maximum peak height.
DISCUSSION
M. tuberculosis AK was studied in an attempt to characterize the enzymes involved in the metabolism of methyl-Ado. AK catalyzes the phosphorylation of Ado to AMP, an important step in the purine salvage pathway. M. tuberculosis AK activity was dependent on Mg2+, exhibited substrate inhibition, and demonstrated affinities for Ado and ATP that were similar to those of other AKs. However, the AK that we identified exhibited some characteristics that are unique among this group of enzymes, including its pH optimum profile, pI, quaternary structure, and lack of stimulation by Pi, which were markedly different from those of other AKs (Table 3) (2, 9, 13, 35).
As for the physical attributes of M. tuberculosis AK, those of the native form of the enzyme proved to be the most challenging to discern. M. tuberculosis AK was initially thought to be a tetramer based on its elution from a gel filtration column, but subsequent analysis by light-scattering photometry and analytical ultracentrifugation revealed that the native form of the enzyme was a dimer. Gel filtration chromatography separates proteins based on their sizes (Stoke's radius), and molecular weight is extrapolated from elution volume using the migration of globular proteins of known molecular weight as standards. Since it is not a direct measurement of molecular mass, molecular weight determination by gel filtration is prone to error. Light-scattering photometry, sedimentation velocity, and sedimentation equilibrium analytical ultracentrifugation are considered to be direct measurements of molecular mass, and their results are weighted accordingly. Even with buffer conditions similar to those for gel filtration, these methods uniformly indicated that the native form of the enzyme was a dimer. Therefore, we concluded that the native form of M. tuberculosis AK is a dimer.
The physical parameters of AKs from M. tuberculosis and human sources differed, and perhaps more interestingly, the substrate and phosphate donor specificities also differed. M. tuberculosis and human AKs are different enough to permit selective activation of Ado analogs like methyl-Ado. The MIC of methyl-Ado for M. tuberculosis (3 μg/ml) is 50 to 70 times lower than the concentration of methyl-Ado that inhibits 99% of growth in CEM cells (150 to 200 μg/ml). Studies of methyl-Ado phosphorylation by M. tuberculosis and human AK helped to explain the basis for the selective activation of methyl-Ado in M. tuberculosis. The Km values for methyl-Ado demonstrated that methyl-Ado is a better substrate for M. tuberculosis AK than the human homolog (Table 3). These results suggest that the rate of phosphorylation of methyl-Ado by M. tuberculosis AK is at least partially responsible for the selectivity of the compound.
Initially, the difference in methyl-Ado metabolism was not appreciated because assays for M. tuberculosis AK were performed without added KCl; under these conditions, the Km for methyl-Ado in M. tuberculosis was 710 μM, which was similar to the 960 μM Km for methyl-Ado in human cells. After it was observed that K+ had a stimulatory effect on Ado metabolism, the Km for methyl-Ado was determined in the presence of KCl and was found to have been significantly reduced, to 79 μM. In the presence of KCl, the Vmax for methyl-Ado increased from 60 to 180 nmol/mg-min. Overall, methyl-Ado phosphorylation was stimulated 300-fold in the presence of K+. Since K+ exists at millimolar levels within cells, stimulation by K+ is physiologically relevant, and Km and Vmax values obtained in the presence of K+ more closely reflect the true physiological state of the enzyme than values obtained in the absence of K+.
The influence of K+ on M. tuberculosis AK activity could not have been predicted based on its effects on other AKs, although it is a characteristic of ribokinase (1). There are conflicting reports regarding the effect of K+ on Ado phosphorylation by mammalian AK (11, 13, 15, 20); however, the human enzyme requires K+ in order to phosphorylate deoxyadenosine (14, 15). Studies with AK from Leishmania donovani have shown that KCl stimulates phosphorylation of formycin A but not Ado (4), and emission spectral analysis of AK from rabbit liver indicates that 0.9 mol of K+ is bound per mol of AK (20). Taken in context with the data from human and mammalian AK studies, it appears that K+ may be bound to AKs from various sources with little effect on the phosphorylation of Ado. However, phosphorylation of other nucleosides and nucleoside analogs, such as deoxyadenosine and formycin A, may be stimulated in the presence of K+. Unlike the other AKs, in the M. tuberculosis enzyme, Ado phosphorylation was also stimulated in the presence of K+.
Another appreciable difference between human and M. tuberculosis AKs lies in their respective preferences for phosphate donors. M. tuberculosis AK has demonstrated a preference for dGTP and GTP as phosphate donors, with 4.7 and 2.5 times the activity of ATP, respectively. Human AK has also demonstrated a preference for GTP and dGTP, although the preference is not as profound as that for M. tuberculosis AK. In the presence of GTP and dGTP, human AK exhibited at most 1.5 times the activity seen with ATP (23). Since dGTP, GTP, and ATP exist at different intracellular concentrations, it is not yet possible to determine which of these phosphate donors is used intracellularly or what affects the phosphate donors will have on Ado and methyl-Ado metabolism. Differences in the metabolism of methyl-Ado and a preference for dGTP and GTP as phosphate donors suggest that the active sites in M. tuberculosis and human AKs may contain differences that can be exploited for drug development.
The amino acid sequence of M. tuberculosis AK was highly conserved among Mycobacterium spp., with 86 to 100% homology; however, it was different enough from other AKs that no specific function could originally be assigned. In light of the identification of the function of the Rv2202c gene product as AK, we propose that the gene be renamed adoK in order to be consistent with the nomenclature of homologous genes. Human AK shows ≥50% sequence homology with AKs from diverse sources, while it showed only 24% homology with M. tuberculosis AK. Based on primary structure, the M. tuberculosis enzyme could not be identified as AK but was annotated as a member of the PfkB family of carbohydrate and purine nucleoside kinases. Although the percent homology is still low (∼35%), M. tuberculosis AK shows as much or more sequence homology with bacterial ribokinase and fructokinase as other AKs.
According to amino acid sequence homology, M. tuberculosis AK appears to be evolutionarily more closely related to other members of the PfkB family than other AKs, indicating that AK activity may have arisen in M. tuberculosis as a result of convergent evolution. Indeed, phylogenetic analysis has indicated that M. tuberculosis AK is more closely related to ribokinases than other AKs (Fig. 2). It is possible that M. tuberculosis AK represents a new class of bacterial AKs within the PfkB family that is distinguished from other AKs by its unique primary structure, quaternary structure, preference for reactants, and regulatory mechanisms. This work describes the first bacterial AK to be characterized; however, several other bacteria contain genes that show >50% sequence homology with AK from M. tuberculosis that have not yet been assigned specific functions. Based on this homology, it is possible that AKs will be found in these other bacteria.
At this time, it is unclear why M. tuberculosis should contain AK while many other intracellular bacteria do not. AK-deficient strains of M. tuberculosis and M. smegmatis survive well in vitro, suggesting that AK is not essential under these conditions. Mycobacterium spp. contain both de novo synthesis and salvage pathways for nucleotide metabolism (18, 30, 31, 32). It has been suggested that a limiting factor in the growth of M. tuberculosis is the low rate of nucleic acid biosynthesis (33). Therefore, it is possible that the purine salvage pathway in these bacteria has been specialized to allow the bacteria to take full advantage of nutrients available in their intracellular environment. Very small quantities of Ado and other nucleosides are available exogenously; however, nucleotides are readily available. M. leprae has demonstrated the ability to salvage purine nucleosides from nucleotides by using phosphatase adsorbed from its host (30), and it is possible that M. tuberculosis utilizes a similar mechanism for the salvage of exogenous nucleotides.
The purine salvage pathway in M. tuberculosis is an avenue for drug development that has not yet been exploited, and drugs with novel mechanisms of action are needed to combat emerging drug-resistant strains of M. tuberculosis. The selective activity of methyl-Ado against M. tuberculosis has provided proof of concept for the use of nucleoside analogs as antimycobacterial agents. An understanding of the mechanism of action of methyl-Ado may reveal novel drug targets in the purine salvage pathway and pave the way for development of other nucleoside analogs.
Acknowledgments
We thank Rongbao Li, Southern Research Institute, Birmingham, Ala., for his analysis of the primary structure of M. tuberculosis AK; William Suling, Southern Research Institute, for advice regarding the use of the colorimeteric broth microdilution technique for determination of MIC; Peter Prevelige, Department of Microbiology, University of Alabama at Birmingham, for his assistance with light-scattering and mass spectrometry; and Stephen Barnes, Department of Pharmacology and Toxocology, University of Alabama at Birmingham, for his assistance with the phylogenetic analysis.
This work was supported by NIH research grant AI43241. Peptide sequencing was carried out by the Protein Analysis Shared Facility of the Cancer Center at the University of Alabama at Birmingham with support from the National Institutes of Health, Bethesda, Md. (CA13148).
REFERENCES
- 1.Andersson, C. E., and S. L. Mowbray. 2002. Activation of ribokinase by monovalent cations. J. Mol. Biol. 315:409-419. [DOI] [PubMed] [Google Scholar]
- 2.Andres, C. M., and I. H. Fox. 1979. Purification and properties of human placental adenosine kinase. J. Biol. Chem. 254:11388-11393. [PubMed] [Google Scholar]
- 3.Basso, L. A., D. S. Santos, W. Shi, R. H. Furneaux, P. C. Tyler, V. L. Schramm, and J. S. Blanchard. 2001. Purine nucleoside phosphorylase from Mycobacterium tuberculosis. Analysis of inhibition by a transition-state analog and dissection by parts. Biochemistry 40:8196-8203. [DOI] [PubMed] [Google Scholar]
- 4.Bhaumik, D., and A. K. Datta. 1989. Immunochemical and catalytic characteristics of adenosine kinase from Leishmania donovani. J. Biol. Chem. 264:4356-4361. [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C.-K., E. W. Barrow, P. W. Allan, N. Bansal, J. A. Maddry, W. J. Suling, W. W. Barrow, and W. B. Parker. 2002. The metabolism of 2-methyladenosine in Mycobacterium smegmatis. Microbiology 148:289-295. [DOI] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, S. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rojers, S. Rutter, K. Seeger, S. Skelton, S. Squares, R. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Cottam, H. B., D. B. Wasson, H. C. Shih, A. Raychaudhuri, G. Di Pasquale, and D. A. Carson. 1993. New adenosine kinase inhibitors with oral anti-inflammatory activity: synthesis and biological function. J. Med. Chem. 36:3424-3430. [DOI] [PubMed] [Google Scholar]
- 9.Datta, A. K., D. Bhaumik, and R. Chatterjee. 1987. Isolation and characterization of adenosine kinase from Leishmania donovani. J. Biol. Chem. 262:5515-5521. [PubMed] [Google Scholar]
- 10.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 11.Fisher, M. N., and E. A. Newsholme. 1984. Properties of rat heart adenosine kinase. Biochem. J. 221:521-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta, R. S. 1996. Adenosine-AMP exchange activity is an integral part of the mammalian adenosine kinase. Biochem. Mol. Biol. Int. 39:493-502. [DOI] [PubMed] [Google Scholar]
- 13.Hao, W., and R. S. Gupta. 1996. Pentavalent ions dependency of mammalian adenosine kinase. Biochem. Mol. Biol. Int. 38:889-899. [PubMed] [Google Scholar]
- 14.Hurley, M. C., B. Lin, and I. H. Fox. 1985. Regulation of deoxyadenosine and nucleoside analog phosphorylation by human placental adenosine kinase. J. Biol. Chem. 260:15675-15681. [PubMed] [Google Scholar]
- 15.Hurley, M. C., T. D. Pallela, and I. H. Fox. 1983. Human placental deoxyadenosine and deoxyguanosine phosphorylating activity. J. Biol. Chem. 258:15021-15027. [PubMed] [Google Scholar]
- 16.Maj, M., B. Singh, and R. S. Gupta. 2000. The influence of inorganic phosphate on the activity of adenosine kinase. Biochim. Biophys. Acta 1476:33-42. [DOI] [PubMed] [Google Scholar]
- 17.Maj, M. C., B. Singh, and R. S. Gupta. 2002. Pentavalent ions dependency is a conserved property of adenosine kinase from diverse sources: identification of a novel motif implicated in phosphate and magnesium ion binding and substrate inhibition. Biochemistry 41:4059-4069. [DOI] [PubMed] [Google Scholar]
- 18.Malathi, V. G., and T. R. Ramakrishnan. 1966. Biosynthesis of nucleic acid purines in Mycobacterium tuberculosis H37Rv. Biochem. J. 98:594-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathews, I. I., M. D. Erion, and S. E. Ealick. 1998. Structure of human adenosine kinase at 1.5 Å resolution. Biochemistry 37:15607-15620. [DOI] [PubMed] [Google Scholar]
- 20.Miller, R. L., D. L. Adamczyk, and W. H. Miller. 1979. Adenosine kinase from rabbit liver. I. Purification by affinity chromatography and properties. J. Biol. Chem. 254:2339-2345. [PubMed] [Google Scholar]
- 21.Miller, R. L., D. L. Adamczyk, J. L. Rideout, and T. A. Krenitsky. 1982. Purification, characterization, substrate and inhibitor specificity of adenosine kinase from several Eimeria species. Mol. Biochem. Parasitol. 6:209-223. [DOI] [PubMed] [Google Scholar]
- 22.Newby, A. C. 1981. The interactions of inhibitors with adenosine metabolizing enzymes in intact isolated cells. Biochem. Pharmacol. 30:2611-2615. [DOI] [PubMed] [Google Scholar]
- 23.Palella, T. D., C. M. Andres, and I. H. Fox. 1980. Human placental adenosine kinase. Kinetic mechanism and inhibition. J. Biol. Chem. 255:5264-5269. [PubMed] [Google Scholar]
- 24.Raviglione, M. C., D. E. Snider, and A. Kochi. 1995. Global epidemiology of tuberculosis. Morbidity and mortality of a worldwide epidemic. JAMA 273:220-226. [PubMed] [Google Scholar]
- 25.Rotllan, P., and M. T. Miras Portugal. 1985. Adenosine kinase from bovine adrenal medulla. Eur. J. Biochem. 151:365-371. [DOI] [PubMed] [Google Scholar]
- 26.Snider, D. E., M. Raviglione, and A. Kochi. 1994. Global burden of tuberculosis, p. 3-11. In B. R. Bloom (ed.), Tuberculosis pathogenesis, protection, and control. ASM Press, Washington, D.C.
- 27.Spychala, J., N. S. Datta, K. Takabayashi, M. Datta, I. H. Fox, T. Gribbin, and B. S. Mitchell. 1996. Cloning of human adenosine kinase cDNA: sequence similarity to microbial ribokinases and fructokinases. Proc. Natl. Acad. Sci. USA 93:1232-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suhadolnik, R. J. 1970. Nucleoside antibiotics. Wiley-Interscience, New York, N.Y.
- 29.Suling, W. J., L. E. Seitz, V. Pathak, L. Westbrook, E. W. Barrow, S. Zywno-Van-Ginkel, R. E. Reynolds, J. R. Piper, and W. W. Barrow. 2000. Antimicrobial activities of 2,4-diamino-5-deazapteridine derivatives and effects on mycobacterial dihydrofolate reductase. Antimicrob. Agents Chemother. 44:2784-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wheeler, P. R. 1987. Biosynthesis and scavenging of purines by pathogenic mycobacteria including Mycobacterium leprae. J. Gen. Microbiol. 133:2999-3011. [DOI] [PubMed] [Google Scholar]
- 31.Wheeler, P. R. 1987. Enzymes for purine synthesis and scavenging in pathogenic mycobacteria and their distribution in Mycobacterium leprae. J. Gen. Microbiol. 133:3013-3018. [DOI] [PubMed] [Google Scholar]
- 32.Wheeler, P. R. 1990. Biosynthesis and scavenging of pyrimidines by pathogenic mycobacteria. J. Gen. Microbiol. 136:189-201. [DOI] [PubMed] [Google Scholar]
- 33.Wheeler, P. R., and C. Ratledge. 1994. Metabolism of Mycobacterium tuberculosis, p. 353-380. In B. R. Bloom (ed.), Tuberculosis pathogenesis, protection, and control. ASM Press, Washington, D.C.
- 34.Yamada, Y., H. Goto, and N. Ogasawara. 1981. Adenosine kinase from human liver. Biochim. Biophys. Acta 660:36-43. [DOI] [PubMed] [Google Scholar]
- 35.Yamada, Y., H. Goto, and N. Ogasawara. 1982. Differences of adenosine kinases from various mammalian tissues. Comp. Biochem. Physiol. 71B:367-372. [DOI] [PubMed] [Google Scholar]