Abstract
TolC is a multifunctional outer membrane protein of Escherichia coli that folds into a novel α-β-barrel conformation absent in the other model outer membrane proteins used in assembly studies. The data presented in this work show that the unique folded structure of TolC reflects a unique assembly pathway. During its assembly, the newly translocated nascent TolC monomers are released in the periplasm. Maturation of these nascent monomers, and possibly their oligomerization, in the periplasm precedes their insertion in the outer membrane. The completion of the assembly process is signaled by the development of a characteristic proteinase K-resistant fragment generated by cleavage at a single, periplasmically exposed, protease-sensitive site of the membrane-anchored trimer. None of the assembly steps of TolC is affected by known folding factors, such as SurA, Skp, and lipopolysaccharide, which have profound effects on the assembly of other model trimeric outer membrane proteins. Two assembly-defective TolC mutants were isolated and characterized. One of the mutants (TolCI106N) was defective in the folding of nascent monomers, while the other (TolCS350F) was impaired in steps involving trimerization and membrane insertion of folded monomers.
The β-barrel outer membrane proteins (OMPs) of Escherichia coli have been extensively used in assembly studies (for a review, see reference 12). Their journey to the outer membrane begins with the translocation of OMP precursor molecules through the Sec machinery located in the inner membrane (31). Removal of the signal sequence from precursors during translocation yields mature nascent polypeptides, which are thought to transiently exist in the aqueous environment of the periplasm. One of the challenges these nascent polypeptides face is to remain assembly competent. This is achieved through their interactions with periplasmic folding factors and at least one outer membrane component, lipopolysaccharide (LPS) (1, 22). For trimeric OMPs, the association of folded monomers with the outer membrane presumably triggers their trimerization and insertion in the membrane.
Several periplasmic proteins have been identified that function as chaperones or foldases to assist OMP assembly (12, 30). Skp is thought to be a general chaperone because denatured OmpF, but not the native form, was shown to bind to it in vitro (9). Consistent with this, it was found that the skp null mutant had drastically reduced OMP levels (9). Although the original genetic observation did not withstand further scrutiny, additional biochemical (6, 35) and genetic (33) experiments reiterated the role of Skp as a chaperone.
Among foldases, SurA (23, 34), FkpA (2, 29), and PpiD (13) represent periplasmic proteins that catalyze peptidyl prolyl cis-trans isomerization (PPIase) in vitro. However, despite their in vitro PPIase activity, it has not been demonstrated that these proteins influence OMP assembly by affecting the cis-trans isomerization of proline peptide bonds. In fact, both SurA (4) and FkpA (2) have been shown to assist in OMP assembly independent of their isomerase activities. Moreover, FkpA has been reported to influence protein folding independent of the presence of cis-prolines (5). In light of this, it is conceivable that SurA and FkpA promote OMP assembly by acting as general chaperones. The periplasm also contains proteins that catalyze disulfide bond formation (3). However, since many OMPs, including porins and TolC, the subject of this study, do not contain cysteine residues, they assemble independent of the disulfide isomerase activity.
A periplasmic protease, DegP, has received the most attention in connection with OMP assembly. degP null mutants do not grow at growth temperatures above 39°C (24, 40), presumably because of a greater need for the removal of misfolded proteins at elevated temperatures. However, expression of certain misfolded OMPs renders DegP's presence essential at lower growth temperatures (8, 27). Both biochemical (38) and genetic (33) studies have suggested that DegP can also function as a chaperone.
Not all OMPs follow the same assembly pathway involving common assembly factors. Such exceptions are typified by the outer membrane lipoproteins, whose targeting and assembly rely on five dedicated proteins, LolABCDE (25, 43). This variation most likely reflects the posttranslocational modifications and other structural distinctiveness of lipoproteins. This raises an interesting question of whether other OMPs with a distinct folded structure may also ensue novel targeting and assembly pathways. With this in mind, we examined the assembly of TolC, whose three-dimensional structure and barrel composition are astonishingly different from characteristics of other OMPs used in biogenesis studies.
TolC is a minor but functionally important OMP. It is involved in hemolysin secretion (42, 44), colicin import (10, 14), and antibiotic efflux (16). It is also exploited by a bacteriophage as a cell surface receptor (17). The folded structure of TolC is novel in that it forms an extended α-β-barrel (20), in contrast to the exclusive transmembrane β-barrels of OmpF (11), OmpA (32), and LamB (36). Due to similar three-dimensional structures of these latter proteins, a common folding and assembly principle governs their biogenesis. However, this may not be true for TolC, whose folded structure contains a much higher proportion of α-helices than any other OMP of known structure. The hallmark of the majority of OMPs is the presence of a conserved carboxy-terminal phenylalanine residue, which is thought to influence nascent monomer folding (18, 41). In TolC, however, asparagine occupies this position. Lastly, unlike porins, where three individual monomeric barrels assemble to form a three-barrel trimer, a TolC monomer constitutes only one-third of a single-barrel TolC trimer. These structural and sequence differences may reflect a distinct folding and assembly pathway for TolC. The results obtained in this study indeed support this notion.
MATERIALS AND METHODS
Bacterial strains, growth media, and chemicals.
The E. coli strains and plasmids used in this study are listed in Table 1. Minimal medium (M63) and Luria broth were prepared as previously described by Silhavy et al. (37). When cells were grown on minimal medium, 0.4% glycerol was used as the sole carbon source. A mixture of [35S]methionine-[35S]cysteine was purchased from Perkin-Elmer Life Sciences, Inc. Enzyme-catalyzed fluorescence (ECF) substrate was purchased from Amersham Pharmacia Biotechnologies. Formalin-killed Staphylococcus aureus cells (Pansorbin) were purchased from Calbiochem. All other chemicals were of analytical grade.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Strains | ||
| MC4100 | F−araD139 Δ(argF-lac)U139 rpsL150 flbB5301 ptsF25 deoC1 thi-1 rbsR relA | 7 |
| RAM958 | MC4100 tolC::Tn10-48 | 42 |
| RAM1149 | RAM958/pTrc99A | This study |
| RAM1150 | RAM958/pTrc-tolC+ (BspHI-HindIII clone) | This study |
| RAM1151 | RAM958/pTrc-tolC(TolC1106N) | This study |
| RAM1152 | RAM958/pTrc-tolC(TolCS350F) | This study |
| RAM787 | MC4100 rcsB::Kmr | 22 |
| RAM789 | MC4100 rcsB::Kmr Δrfa-2057 (Cmr) | 22 |
| RAM913 | MC4100 ΦdegP::lacZ+ Tn10::zad-475 | R. Misra |
| RAM914 | MC4100 ΦdegP::lacZ+ Δskp Tn10::zad-475 | R. Misra |
| RAM951 | MC4100 surA::Kmr | R. Misra |
| RAM1116 | RAM958 surA::Kmr/pTrc-tolC+ | This study |
| RAM1117 | RAM958 Δrfa-2057/pTrc-tolC+ | This study |
| XL1-Red | endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac mutD5 mutS mutT::Tn10 | Stratagene |
| Plasmid | ||
| pTrc99A | Apr; expression vector | Pharmacia |
DNA manipulations.
Biogenesis studies with chromosomally expressed TolC were often difficult owing to low protein levels. To circumvent this, the tolC gene was cloned into an expression vector, pTrc99A (Pharmacia). For this, two mutagenic primers were utilized: the forward primer (5′-CAGGAAACAGATCATGAGGAAATTGCTCCC-3′) created a unique BspHI site (underlined), and the reverse primer (5′-GCGGCAGATAACCCGAAGCTTTACGGTTGCC-3′) created a unique HindIII site (underlined). In creating the BspHI site, the second codon of the TolC signal sequence was changed from AAG (lysine) to AGG (arginine). tolC DNA was amplified from the chromosome by PCR, digested with BspHI and HindIII, and ligated into appropriately restricted pTrc99A. In the absence of the inducer isopropyl-β-d-thiogalactopyranoside (IPTG), the level of plasmid-borne TolC was slightly higher (115%) than that of the chromosomally expressed TolC protein. However, when induced for 10 min with 0.01 mM IPTG, plasmid-borne TolC levels increased about three times above the uninduced level.
Radioactive labeling and assembly assays.
Cells were grown overnight at 37°C on minimal medium with glycerol as the carbon source. The following day, cultures were diluted 50-fold with the same medium and grown to mid-log phase (optical density at 600 nm = 0.3 to 0.35). Ten minutes prior to labeling, 0.01 mM IPTG was added to the cultures to induce TolC synthesis. Cells were labeled as described previously (28). Labeled cells were pelleted by spinning tubes in a microcentrifuge at 14,000 × g for 15 min at 4°C. To allow proteinase K to enter the periplasm, the outer membrane was permeabilized by resuspending the cells in a buffer containing 20% sucrose, 20 mM Tris-HCl (pH 8.0), 10 mM EDTA, and 0.5% Triton X-100 and incubated for 10 min at 4°C. Proteinase K was then added to a final concentration of 10 μg/ml, and tubes were further incubated for 10 min at 30°C. The protease inhibitor phenylmethylsulfonyl fluoride (PMSF; 1 mM) was added to inactivate the protease. Membrane proteins were extracted from pelleted cells by the gentle detergent lysis procedure as described previously (26, 28).
Labeled cell extracts were mixed with an immunoprecipitation buffer (1% Triton X-100, 50 mM Tris-HCl; pH 7.5) containing appropriate antibodies and incubated rocking at 4°C for a minimum of 4 h. The immunocomplexes were precipitated with Pansorbin. The immunoprecipitates were washed several times and analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). The gels were dried and exposed to X-ray films at −80°C. Protein bands were scanned and quantified using ImageQuant software (Molecular Dynamics).
Fractionation procedures.
The French press lysis method was used to separate the soluble (cytoplasm and periplasm) and insoluble (inner and outer membranes) fractions (28). To isolate the periplasm, a gentle osmotic shock method was performed (2). Labeled cell pellets were resuspended in a buffer containing 0.5% sucrose, 10 mM Tris-HCl (pH 7.5), 200 μg of lysozyme/ml, and 10 mM EDTA to permeabilize the outer membrane. This mixture was incubated at 4°C for 20 min. The cells were centrifuged at 14,000 × g for 15 min at 4°C. The supernatant, now containing the periplasm, was removed and treated with proteinase K as described above. PMSF (1 mM) was added to inactivate the protease, and immunoprecipitation was carried out using appropriate antibodies.
Western blot analysis.
Whole-cell extracts were analyzed on mini SDS-PAGE gels and transferred onto polyvinylidene difluoride membranes (Immobilon-Millipore). After transfer, the membranes were incubated for 1.5 h with primary antibodies raised against LamB, maltose-binding protein (MBP), or TolC. The membranes were washed, and secondary antibodies (goat anti-rabbit alkaline phosphatase-conjugated immunoglobulin G) were added for 1 h. The membranes were exposed to ECF substrate for 5 min and analyzed using a phosphorimager.
RESULTS
In vivo assembly assay.
Fully assembled, membrane-bound TolC shows a characteristic protease sensitivity pattern. It was discovered that the treatment of membrane-anchored TolC with proteinase K yields a stable membrane-bound fragment of 46 kDa that is truncated from its carboxyl end (21). Unlike the membrane-bound species, TolC solubilized from the membrane with nonionic detergents is rapidly degraded by the protease. Thus, the appearance of the 46-kDa TolC fragment from cells withdrawn at various chase points should not only reflect a conformational transition but also its membrane insertion (Fig. 1A).
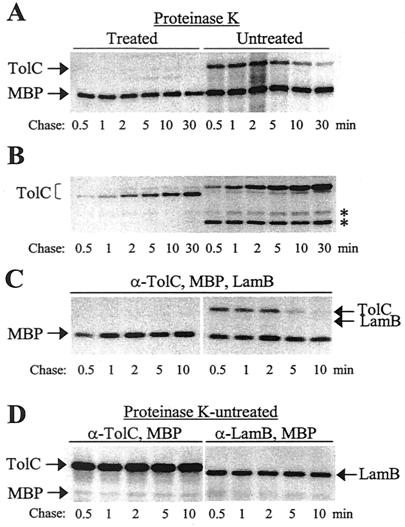
FIG. 1.
TolC assembly assay. (A) Cartoon showing the TolC assembly assay. The treatment of outer membrane (OM)-permeabilized cells with proteinase K (PK) results in the complete degradation of soluble TolC assembly intermediates, while assembled TolC molecules produce an OM-anchored 46-kDa fragment. The C-terminal 5-kDa fragment is either completely degraded or not detected by SDS-PAGE. (B) An autoradiograph of an SDS-polyacrylamide gel carrying immunoprecipitated samples from pulse-chase labeling experiments. Cells expressing wild-type TolC were labeled with [35S]methionine-[35S]cysteine as described in Materials and Methods. Half of the labeled samples was treated with proteinase K. TolC and MBP were immunoprecipitaed from protein extracts and analyzed by SDS-PAGE. Positions of TolC and MBP are shown. Note that TolC from proteinase K-treated samples runs faster (46 kDa) than that from untreated samples (51 kDa). Chase times are shown below the gel. (C) TolC levels, relative to MBP levels, were quantified and graphed. Total TolC (♦), proteinase K-resistant TolC (▪), and proteinase K-sensitive TolC (▴) results are shown.
To monitor this event in vivo, we combined two techniques in which whole-cell proteolysis is followed by trimer extraction using a gentle lysis procedure (26, 28) that preserves TolC's native trimeric state. As the proteinase K-sensitive site in TolC is accessible only from the periplasm, whole-cell proteolysis was carried out after permeabilizing the outer membrane. TolC trimers were extracted by the SDS-lysis procedure. TolC and MBP were immunoprecipitated from supernatants, and immunoprecipitated samples were analyzed by SDS-PAGE (Fig. 1B and C).
The results presented in Fig. 1B and C showed that although some TolC molecules had already attained the characteristic protease-resistant state at the earliest chase point, the majority of TolC molecules present at the first chase point were protease sensitive. Gradually, all TolC molecules matured into a form that produced the characteristic membrane-anchored 46-kDa fragments. These results showed that TolC progresses through a protease-sensitive state to a protease-resistant state during assembly.
Localization of assembly intermediates.
An element concerning TolC's assembly involves identifying various assembly intermediates based on their cellular location. Of course, the fully assembled product of TolC is localized in the outer membrane, but nothing is known about the localization of assembly intermediates. This is important to determine, because it will shed light on the cellular pathway TolC takes to get to the outer membrane. Two popular models of OMP assembly evoke that either nascent polypeptides of OMPs are dumped into a soluble periplasmic environment or they remain associated, albeit weakly, with the inner membrane and reach the outer membrane through membrane contact sites. As the physiological significance of such contact sites remains unclear, the discovery of several periplasmic OMP assembly factors has given greater impetus to the notion of soluble OMP assembly intermediates.
Our fractionation analyses entailed two methods. The first method separated the soluble cytoplasmic and periplasmic fractions from the insoluble fractions of inner and outer membranes. The second method aided in the isolation of the soluble periplasmic fraction. To assist in the isolation of soluble and insoluble contents, [35S]methionine-cysteine-labeled cells (∼2 × 108) withdrawn at various chase points were mixed with unlabeled TolC̄ cells (∼1 × 109). The addition of cold cells compensates for sample loss during the fractionation procedure and aids in monitoring the cross-contamination of various fractions by Western blot analysis of unlabeled proteins. Membranes were separated from soluble fractions by the high-speed centrifugation of cell lysates obtained by a French press cell. The soluble periplasmic fraction was isolated using methods involving a mild spheroplast formation that does not release the membrane and cytoplasmic fractions (see Materials and Methods for details). TolC was detected from various fractions by immunoprecipitation reactions using TolC-specific antibodies (Fig. 2).
FIG. 2.
Localization of TolC assembly products. Bacterial cells from pulse-chase experiments were fractionated into soluble cytoplasm and periplasm (A) and insoluble inner and outer membranes (B). One half of the protein sample was treated with proteinase K, while the other half was left untreated. TolC and MBP were immunoprecipitated and analyzed by SDS-PAGE. Gels were dried and autoradiographed. (C) Purified periplasmic fraction from a separate pulse-chase experiment analyzed as described for panels A and B. Note that unlike in panels A and B, a cocktail of antibodies specific to TolC, MBP, and LamB was used in the immunoprecipitation reactions. (D) Immunoprecipitation of TolC, MBP, and LamB from periplasm-free, shocked cell pellets. Prior to immunoprecipitation, protein samples were divided into halves; antibodies specific to TolC and MBP were used in one half, while those specific to LamB and MBP were used in the other half of immunoprecipitation reactions. Positions of TolC, MBP, and LamB are shown. Identities of two protein bands (*) are currently unknown. Chase time points are shown below the gel.
The results showed the accumulation of a soluble TolC species during early chase points whose levels gradually declined during later chase points (Fig. 2A). In contrast, only a small population of TolC was present in the membrane fraction during early chase points, but its level rose steadily during later chase points (Fig. 2B). This depletion of a soluble TolC species with a concomitant increase in insoluble TolC levels during chase reflected a typical precursor-product relationship. The soluble TolC species showed almost total sensitivity to proteinase K (Fig. 2A), while insoluble TolC produced the characteristic 46-kDa fragments (Fig. 2B). Further separation of envelopes into outer and inner membranes by sucrose density gradients showed that the 46-kDa fragment was generated from TolC localized with the outer membrane (data not shown).
The result presented in Fig. 2C showed that during the early steps of biogenesis, TolC was present in the periplasm but then disappeared around 10 min postchase. As observed for the soluble fraction in Fig. 2A, TolC present in the periplasmic fraction was sensitive to proteinase K. Since cultures were induced with maltose prior to pulse-chase analysis, LamB was also examined as a control protein. No soluble assembly intermediates of LamB were detected (Fig. 2C). Unlike the periplasmic fraction, LamB was present in periplasm-free shocked cell pellets (Fig. 2D). It is important to emphasize that the purity of the periplasmic fraction is critical to our claim for the existence of TolC assembly intermediates in the periplasm. Results presented in Fig. 2C showed that periplasmic fractions from all five chase samples were entirely free of any contamination from the assembled (protease-resistant) forms of TolC and LamB and, thus, insoluble membrane fractions. These results unequivocally demonstrate that prior to assembling in the outer membrane, the proteinase K-sensitive TolC intermediates first emerge in the periplasm.
The role of known assembly factors in TolC assembly.
Several assembly factors have been identified through studies involving model OMPs (12, 30). These include SurA, a periplasmic peptidyl prolyl cis-trans isomerase (23, 34), Skp, a presumed general chaperone (6, 9, 35), DegP, a periplasmic protease (24, 40) that is also proposed to possess a general chaperone activity (38), and LPS, which is thought to be involved in the assembly of all trimeric OMPs (22). We examined the effect of null (surA, skp, and degP) or mutant (LPS) alleles of these factors on the assembly of TolC. The initial examination involved Western blot analysis in which the steady-state levels of TolC, LamB, and MBP were examined (Fig. 3A and B). The latter two proteins served as outer membrane (LamB) and periplasmic (MBP) protein controls. As expected, the level of LamB was reduced in mutant surA and deep-rough LPS backgrounds, whereas MBP levels remained unaltered (Fig. 3A). In contrast, no reduction in TolC level was observed in either mutant background (Fig. 3B). We failed to see any reduction in LamB and TolC levels in an skp null background (Fig. 3A and B). Similarly, the level of TolC, like that of LamB and MBP, was unaffected in a degP null background (data not shown).
FIG. 3.
Western blot analysis of cell extracts obtained from TolC−, TolC+, TolC+ SurA−, TolC+ Tn10::zad475, TolC+ Δskp Tn10::zad475, TolC+ rcsB::Kmr, and TolC+ rcsB::Kmr Δrfa2057 (lanes 1 to 7, respectively). Membrane filters were treated with either LamB and MBP antibodies (A) or TolC and MBP antibodies (B). Positions of LamB, MBP, and TolC are shown. (C and D) Effects of surA::Kmr (C) and Δrfa2057 (D) on TolC assembly. Assembly assays were carried out as described in Materials and Methods and in the legend for Fig. 1. Levels of assembled (proteinase K-resistant) TolC, relative to MBP levels, were quantified and graphed.
Since the examination of the steady-state protein level does not reflect the kinetics of assembly, we studied this through pulse-chase labeling experiments. These experiments involved only mutant surA and LPS backgrounds, as they produced the most severe effect on porin assembly. The results presented in Fig. 3C and D showed that neither the absence of SurA nor the expression of the deep-rough LPS had any significant effect on TolC assembly. These results showed that the known assembly factors (particularly SurA and LPS) that profoundly influence the assembly of most OMPs are not involved in TolC's assembly.
Isolation and characterization of assembly-defective TolC mutants.
OMP mutants defective in assembly have been extremely valuable in (i) dissecting the assembly steps (26, 28), (ii) identifying individual residues that play an important role in assembly (26), and (iii) revealing the cellular factors involved in the assembly (15, 19, 26). We anticipate that this class of TolC mutant would be equally valuable in providing insights into its assembly pathway.
The basic premise behind isolating assembly-defective TolC mutants is that such mutants may also display a generally compromised TolC phenotype due to reduced TolC levels. In an attempt to isolate assembly-defective TolC mutants, we exploited a strategy principally designed to isolate TolC efflux mutants. As noted previously (16), TolC is the outer membrane component of an efflux pump consisting of AcrA and AcrB proteins; null mutations in any one of the three genes display a hypersensitivity phenotype. If we were to mutagenize plasmids carrying the tolC gene, only those mutations mapping in tolC would contribute to a hypersensitive phenotype in a genetic background lacking the chromosomal tolC gene.
A plasmid tolC clone was constructed that produces TolC at a level similar to that produced from the chromosomal allele. Random tolC mutations were generated by allowing the replication of the tolC plasmid in a mutator strain (mutT, mutS, or mutD). Mutagenized plasmid DNA was transformed into a genetic background lacking the chromosomal tolC allele. Transformed bacterial colonies were screened by replica plating for their ability to grow on medium containing low or high concentrations of novobiocin, an antibiotic that is normally extruded by the TolC-AcrA/B pump. We concentrated on colonies that grew on low (5 μg/ml) but not on high (50 μg/ml) concentrations of novobiocin, because both the desired class of assembly mutants as well as efflux mutants are expected to be present among them. Colonies that fail to grow on plates containing 5 μg of novobiocin/ml are likely to contain tolC null mutations. On the other hand, colonies that are able to grow on a medium containing a high (50 μg/ml) novobiocin concentration presumably synthesize TolC with normal assembly and efflux activity.
The above genetic strategy led to the isolation of two TolC mutants that had reduced TolC levels. DNA sequence analysis of the plasmid tolC gene revealed the presence of point mutations, resulting in the I106N or S350F substitution in the mature portion of the protein. Low TolC levels in these mutants indicated that either the assembly intermediates or assembled molecules are unstable. We carried out assembly assays to test whether the two mutants carrying an I106N or S350F substitution are assembly defective.
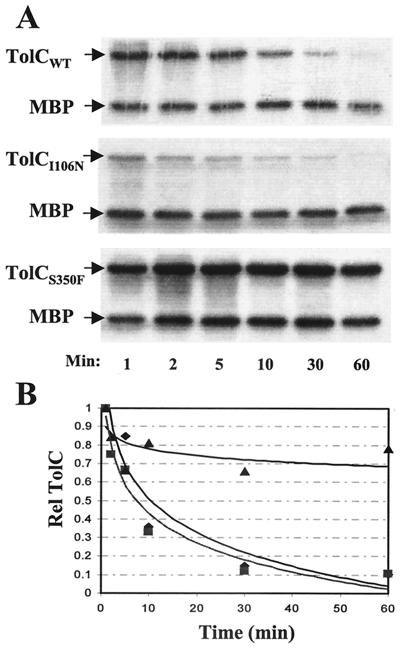
The results presented in Fig. 4 reveal two interesting observations concerning mutant TolC's assembly: first, unlike wild-type TolC (Fig. 4A) where the level of trimer antibody-recognizable forms reaches a plateau around 10 min postchase, in the mutants a plateau was not reached even after 60 min of chase (Fig. 4B and C). Secondly, in the case of wild-type TolC, biogenesis of the proteinase K-resistant form closely followed that of total TolC, and by 60 min virtually all TolC molecules had adopted the proteinase K-resistant conformation. In contrast, the mutant proteins assumed the proteinase K-resistant conformation extremely slowly and remained largely proteinase K sensitive even after 60 min of chase.
FIG. 4.
Assembly of wild-type TolC, TolCI106N, and TolCS350F. Assembly assays were carried out as described in Materials and Methods and in the legend for Fig. 1. Levels of untreated and proteinase K-treated TolC (+ PK), relative to MBP, were quantified and graphed.
Next, we examined the status of the periplasmic assembly intermediates through pulse-chase labeling experiments (Fig. 5). The results showed that the pool of soluble intermediates of TolCI106N disappeared at a rate similar to that observed for the wild-type protein (Fig. 5B). However, unlike the wild-type protein, the depletion of TolCI106N molecules did not result in the concomitant accumulation of the proteinase K-resistant assembly product (Fig. 4B). This suggested that the majority of the soluble intermediates was degraded prior to adopting a protease-resistant conformation.
FIG. 5.
Examination of periplasmic TolC intermediates from strains expressing wild-type TolC (TolCwt), TolCI106N, and TolCS350F. TolC and MBP were immunoprecipitated from purified periplasmic fractions obtained from pulse-chase experiments. (A) Autoradiographs of SDS-polyacrylamide gels carrying immunoprecipitated samples. (B) TolC levels, relative to MBP levels, were quantified and graphed. ♦, TolCwt; ▪, TolCI106N; ▴, TolCS350F.
The mutant TolC protein with an S350F substitution produced results quite distinct from the wild-type and TolCI106N proteins. We not only observed a much larger pool of the soluble periplasmic intermediates than that present in the other two proteins, but these intermediates persisted in the soluble state even after 60 min of chase (Fig. 5). These results showed that TolCS350F is severely defective in a step involving the conversion of soluble, protease-sensitive periplasmic intermediates into membrane-bound, protease-resistant assembly products. The persistence of the soluble TolCS350F species suggests that it is conformationally distinct from that produced by TolCI106N.
DISCUSSION
TolC assembly pathway.
Data presented in this work showed that TolC follows an assembly pathway distinct from that ensued by other thoroughly studied model trimeric OMPs. We developed an in vivo assay to study the assembly of TolC, a structurally unique OMP of E. coli. The basic premise behind this assay entailed differential proteinase K sensitivity profiles of assembly intermediates and assembled molecules. The proteinase K treatment of permeabilized cells, in which the protease could reach the periplasmic side of the outer membrane, resulted in the cleavage of the fully assembled TolC molecules of 51 kDa into outer membrane-anchored 46-kDa fragments. Pulse-chase experiments demonstrated a clear transition of a TolC population from a state that was completely sensitive to proteinase K to a state that displayed the distinctive proteinase K sensitivity pattern of fully assembled TolC molecules. Our assays also revealed that the development of this final state in TolC assembly is a relatively slow process, taking almost 60 min to fully assemble all of the TolC molecules synthesized during the 30-s pulse period.
Fractionation studies revealed two topologically distinct TolC populations. The first population primarily appeared during the early chase period and was comprised of soluble molecules present in the periplasm. This population presumably represented a mixture of nascent mature polypeptides and unassembled TolC molecules. The second population predominantly appeared during the later chase period and was membrane bound. This species likely corresponded to fully assembled TolC molecules, since TolC from steady-state cultures also behaves this way. These topologically distinct populations could also be differentiated based on their protease sensitivity patterns. The soluble periplasmic population was fully sensitive to proteinase K, while the membrane-bound TolC produced the distinct proteinase K-sensitive band of fully assembled TolC. Based on these results, we summarize that the assembly of TolC begins with the emergence of soluble, protease-sensitive intermediates in the periplasm. These soluble intermediates may represent multiple assembly ensembles, including nascent mature monomers, folded monomers, and soluble trimers. The insertion of soluble trimers into the outer membrane, perhaps followed by some additional conformational changes as seen in the metastable-to-stable trimer transition of OmpF (26) and LamB (28), completes the assembly process.
It is interesting that, unlike TolC, no soluble LamB assembly intermediates were detected. The existence of soluble LamB intermediates has been hypothesized from genetic data showing that LamB assembly is severely affected in the absence of SurA, a periplasmic foldase (34). The lack of a detectable soluble LamB assembly intermediate may reflect its rapid conversion into membrane-bound forms (28). On the other hand, membrane insertion may be a rate-limiting step in TolC assembly where, unlike LamB, trimerization most likely precedes insertion into the outer membrane. Soluble assembly intermediates of OmpF and PhoE have been reported (for a review, see reference 12).
Known assembly factors do not affect TolC assembly.
Several periplasmic (e.g., SurA and Skp) and outer membrane (LPS) factors have been implicated in the assembly of model OMPs, particularly that of porins and LamB (12). However, in a sharp contrast to these OMPs, we show here that the in vivo assembly of TolC appeared to be unaffected in a genetic background lacking either SurA or Skp or producing a truncated LPS core. This would suggest that (i) TolC assembly intermediates fold in a unique manner, and (ii) novel assembly factors may be involved in TolC assembly. It is worth emphasizing that while we observed no obvious effect of SurA or Skp on TolC assembly, it could be due to the existence of redundant cellular activities. For examples, there are at least two other periplasmic peptidyl prolyl cis-trans isomerases (2, 13, 29) that may compensate for the loss of SurA. Curiously, however, such compensation has not been seen in the case of OmpF and LamB, suggesting a more direct role for SurA, perhaps as a chaperone (4) rather than as a peptidyl prolyl cis-trans isomerase, on OMP assembly. Interestingly, while TolC assembly is independent of the LPS core, certain TolC-dependent activities, such as phage infection (17) and hemolysin secretion (39), are drastically affected by the composition of the LPS core.
Insight on TolC assembly through mutant analysis.
Mutants that influence various assembly steps can be powerful tools in dissecting the assembly pathway. With this in mind, we sought TolC assembly mutants through exploiting TolC's role in antibiotic efflux. We surmised that reduced TolC levels would negatively affect the cell's ability to pump out antibiotics, thus leading to a hypersensitivity phenotype. A TolC mutant-screening method was devised that eliminated null mutants and differentiated isolates with reduced efflux activity from those that maintained full activity. Two TolC mutants bearing I106N and S350F substitutions had an intermediate efflux activity and were defective in assembly, respectively.
Assembly and fractionation analyses revealed that while both mutant proteins were severely impaired in acquiring the membrane-bound conformation of fully assembled molecules, they were defective in different stages of assembly. Even though the periplasmic species of TolCI106N was depleted at the same rate as the wild-type TolC protein, its failure to assume the characteristic proteinase K-resistant pattern suggested rapid turnover of the soluble assembly intermediates. In contrast to TolCI106N, the soluble, periplasmic species of TolCS350F was surprisingly impervious to degradation, and over 70% of it persisted even after 60 min of chase. The perseverance of the soluble TolCS350F species showed that its conformation is significantly different from that of the highly labile TolCI106N species.
The crystal structure of TolC revealed that I106 is part of a hydrophobic cluster that stabilizes neighboring helices within a monomer (20). Assuming that the formation of this hydrophobic cluster and helix stabilization occurs during the commencement of the folding process, their destabilization would negatively impact the initial stages of assembly. Thus, the substitution of I106 with N most likely interferes with the folding of nascent monomers. This is consistent with the rapid turnover of soluble TolCI106N molecules, in which the unfolded monomers presumably constitute the largest fraction.
The S350F substitution likely interferes with the stabilization of subunit interactions, since S350 forms hydrogen bonds with D162 of the neighboring subunit (20). Moreover, due to its location in the folded molecule, the S350F substitution is less likely to interfere with the early folding events involving nascent polypeptide chains. Again, this is consistent with our observation that the accumulated periplasmic species of TolCS350F is highly stable, suggesting that monomers have assumed a folded conformation that renders them resistant to degradation by periplasmic proteases. Based on this, we propose that a defect in the oligomerization event blocks the insertion of a soluble but folded species of TolCS350F in the outer membrane. Thus, unlike TolCI106N, the assembly of TolCS350F is defective at a later stage of assembly involving trimerization and membrane insertion.
The TolC barrel structure is unique in that it is made up of three monomers. This suggests that at least the β-barrel portion must assemble in the periplasm prior to inserting in the outer membrane. We assert this because the monomer interface has large exposed hydrophilic surfaces that would make it energetically unfavorable for the β-strand regions of TolC to be exposed to the hydrophobic environment of the outer membrane. However, if the monomers associate with other monomers to first form a trimer in the periplasm, the assembled β-barrel trimer would assume a conformation suitable for membrane insertion. The TolCS350F protein, which appears to be defective in the formation of stable trimers, is also defective in membrane insertion, which lends further support to the notion that the trimerization of TolC monomers precedes their insertion in the outer membrane.
We have previously reported that the expression of assembly-defective OMPs in a genetic background lacking a major periplasmic protease, DegP, confers lethality (8, 27). Interestingly, we failed to see such lethality when the assembly-defective TolC proteins were expressed, even though the absence of DegP increased the level of mutant proteins, suggesting that DegP is one of the proteases responsible for the degradation of mutant TolC (data not shown).
The work presented in this study showed that cellular factors that drastically influence the assembly of other model OMPs have no effect on the assembly of TolC. Perhaps this is reflective of the novel structure of TolC, which involves an unusually large, periplasmically exposed, α-helical domain that is absent from other model OMPs. This raises the possibility that a novel folding pathway and assembly factors are involved in the biogenesis of TolC. Our present efforts are directed at further dissecting the assembly steps and revealing cellular factors that are involved in the assembly of TolC.
Acknowledgments
We thank Matthew Humbard for purifying TolC used in antibody production. We are grateful to Leanne Misra for critically reading the manuscript and the W. M. Keck Bioimaging Laboratory for the use of a phosphorimager.
J.W. is the recipient of a graduate assistantship from the Molecular and Cellular Graduate Program. A.M.A. was sponsored by the BREU program. This work was supported by National Institutes of Health grant GM48167 to R.M.
REFERENCES
- 1.Ames, G. F.-L., E. N. Spudich, and H. Nikaido. 1974. Protein composition of the outer membrane of Salmonella typhimurium: effect of lipopolysaccharide mutations. J. Bacteriol. 117:406-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arié, J.-P., N. Sassoon, and J.-M. Betton. 2001. Chaperone function of FkpA, a heat shock prolyl isomerase, in the periplasm of Escherichia coli. Mol. Microbiol. 39:199-210. [DOI] [PubMed] [Google Scholar]
- 3.Bardwell, J. C. A., K. McGovern, and J. Beckwith. 1991. Identification of a protein required for disulfide bond formation. Cell 67:581-589. [DOI] [PubMed] [Google Scholar]
- 4.Behrens, S., R. Maier, H. de Cock, F. X. Schmid, and C. A. Gross. 2001. The SurA periplasmic PPIase lacking its parvulin domains functions in vivo and has chaperone activity. EMBO J. 20:285-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bothmann, H., and A. Pluckthun. 2000. The periplasmic Escherichia coli peptidylprolyl cis, trans-isomerase FkpA. Increased functional expression of antibody fragments with and without cis-prolines. J. Biol. Chem. 275:17100-17105. [DOI] [PubMed] [Google Scholar]
- 6.Bulieris, P. V., S. Behrens, O. Host, and J. H. Kleinschmidt. 2003. Folding and insertion of the outer membrane protein OmpA is assisted by the chaperones Skp and lipopolysaccharide. J. Biol. Chem. 278:9092-9099. [DOI] [PubMed] [Google Scholar]
- 7.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 141:541-555. [DOI] [PubMed] [Google Scholar]
- 8.CastilloKeller, M., and R. Misra. 2003. Protease-deficient DegP suppresses lethal effects of a mutant OmpC protein by its capture. J. Bacteriol. 185:148-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, R., and U. Henning. 1996. A periplasmic protein (Skp) of Escherichia coli selectively binds a class of outer membrane proteins. Mol. Microbiol. 19:1287-1294. [DOI] [PubMed] [Google Scholar]
- 10.Clowes, R. C. 1965. Transmission and elimination of colicin factors and some aspects of immunity to colicin E1 in Escherichia coli. Zentralbl. Bakteriol. Parasitenk. D 196:152-160. [Google Scholar]
- 11.Cowen, S. W., T. Schirmer, G. Rummel, M. Steiert, R. Ghose, R. A. Pauptit, J. N. Jansonius, and J. P. Rosenbusch. 1992. Crystal structures explain functional properties of two E. coli porins. Nature 358:727-733. [DOI] [PubMed] [Google Scholar]
- 12.Danese, P. N., and T. J. Silhavy. 1999. Targeting and assembly of periplasmic and outer membrane proteins in Escherichia coli. Annu. Rev. Genet. 32:59-94. [DOI] [PubMed] [Google Scholar]
- 13.Dartigalongue, C., and S. Raina. 1998. A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J. 17:3968-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies, J. K., and P. R. Reeves. 1975. Genetics of resistance to colicins in Escherichia coli K-12: cross-resistance among colicins of group A. J. Bacteriol. 123:102-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng, M., and R. Misra. 1996. Characterization of AsmA, a protein that influences the assembly of Escherichia coli outer membrane proteins. Mol. Microbiol. 21:605-612. [DOI] [PubMed] [Google Scholar]
- 16.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.German, G., and R. Misra. 2001. The TolC protein of Escherichia coli servers as a cell-surface receptor for the newly characterized TLS bacteriophage. J. Mol. Biol. 301:579-585. [DOI] [PubMed] [Google Scholar]
- 18.Kloser, A. W., J. T. Reading, T. McDermott, R. Stidham, and R. Misra. 2001. Intragenic suppressors of an OmpF assembly mutant and assessment of the role of various OmpF residues in assembly through informational suppressors. J. Bacteriol. 183:264-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kloser, A. W., M. W. Laird, and R. Misra. 1996. asmB, a suppressor locus for assembly-defective outer membrane proteins of Escherichia coli, is allelic to envA (lpxC). J. Bacteriol. 178:5138-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koronakis, V., A. Sharff, E. Koronakis, B. Luisi, and C. Hughes. 2000. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature 405:914-919. [DOI] [PubMed] [Google Scholar]
- 21.Koronakis, V., J. Li, E. Koronakis, and K. Stauffer. 1997. Structure of TolC, the outer membrane component of the bacterial type I efflux system, derived from two-dimensional crystals. Mol. Microbiol. 23:617-626. [DOI] [PubMed] [Google Scholar]
- 22.Laird, M. W., A. Kloser, and R. Misra. 1994. Assembly of LamB and OmpF in deep rough lipopolysaccharide mutants of Escherichia coli K-12. J. Bacteriol. 176:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazar, S. W., and R. Kolter. 1996. SurA assists the folding of Escherichia coli outer membrane proteins. J. Bacteriol. 178:1770-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipinska, B., O. Fayet, L. Baird, and C. Georgopoulos. 1989. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for growth only at elevated temperatures. J. Bacteriol. 171:1574-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masuda, K., S. Matsuyama, and H. Takuda. 2002. Elucidation of the function of lipoprotein-sorting signals that determine membrane localization. Proc. Natl. Acad. Sci. USA 99:7390-7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Misra, R. 1993. OmpF assembly mutants of Escherichia coli K-12: isolation, characterization, and suppressor analysis. J. Bacteriol. 175:5049-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Misra, R., M. Castillokeller, and M. Deng. 2000. Overexpression of protease-deficient DegPS210A rescues the lethal phenotype of Escherichia coli OmpF assembly mutants in a degP minus background. J. Bacteriol. 182:4882-4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Misra, R., A. Peterson, T. Ferenci, and T. J. Silhavy. 1991. A genetic approach for analyzing the pathway of LamB assembly into the outer membrane of Escherichia coli. J. Biol. Chem. 266:13592-13597. [PubMed] [Google Scholar]
- 29.Missiakas, D., J.-M. Betton, and S. Raina. 1996. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol. Microbiol. 21:871-884. [DOI] [PubMed] [Google Scholar]
- 30.Missiakas, D., and S. Raina. 1997. Protein folding in the bacterial periplasm. J. Bacteriol. 179:2465-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori, H., and K. Ito. 2001. The Sec protein-translocation pathway. Trends Microbiol. 9:494-500. [DOI] [PubMed] [Google Scholar]
- 32.Pautsch, A., and G. E. Schulz. 1998. Structure of the outer membrane protein A transmembrane domain. Nat. Struct. Biol. 5:1013-1017. [DOI] [PubMed] [Google Scholar]
- 33.Rizzitello, A. E., J. R. Harper, and T. J. Silhavy. 2001. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J. Bacteriol. 183:6794-6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rouvière, P. E., and C. A. Gross. 1996. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 10:3170-3182. [DOI] [PubMed] [Google Scholar]
- 35.Schäfer, U., K. Beck, and M. Müller. 1999. Skp, a molecular chaperone of gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J. Biol. Chem. 274:24567-24574. [DOI] [PubMed] [Google Scholar]
- 36.Schirmer, T., T. A. Keller, Y.-F. Wang, and J. P. Rosenbusch. 1995. Structural basis for sugar translocation through maltoporin channels at 3.1 Å resolution. Science 267:512-514. [DOI] [PubMed] [Google Scholar]
- 37.Silhavy, T. J., M. L. Berman, and L. W. Enquist. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Spiess, C., A. Bell, and M. Ehrmann. 1999. A temperature-dependent switch from chaperones to protease in a widely conserved heat shock protein. Cell 97:339-347. [DOI] [PubMed] [Google Scholar]
- 39.Stanley, P. L., P. Diaz, M. J. Bailey, D. Gygi, A. Juarez, and C. Hughes. 1993. Loss of activity in the secreted form of Escherichia coli haemolysis caused by an rfaP lesion in core lipopolysaccharide assembly. Mol. Microbiol. 10:781-787. [DOI] [PubMed] [Google Scholar]
- 40.Strauch, K. L., K. Johnson, and J. Beckwith. 1989. Characterization of degP, a gene required for proteolysis in the cell envelope and essential for growth of Escherichia coli at high temperature. J. Bacteriol. 171:2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Struyvé, M., M. Moons, and J. Tommassen. 1991. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 218:141-148. [DOI] [PubMed] [Google Scholar]
- 42.Vakharia, H., G. J. German, and R. Misra. 2001. Isolation and characterization of Escherichia coli tolC mutants defective in secreting enzymatically active α-hemolysin. J. Bacteriol. 183:6908-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yokota, N., T. Kuroda, S. Matsuyama, and H. Tokuda. 1999. Characterization of the LolA-LolB system as the general lipoprotein localization mechanism of Escherichia coli. J. Biol. Chem. 274:30995-30999. [DOI] [PubMed] [Google Scholar]
- 44.Wandersman, C., and P. Delepelaire. 1990. TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc. Natl. Acad. Sci. USA 87:4776-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]