Abstract
The ompS1 gene encodes a quiescent porin in Salmonella enterica serovars Typhi and Typhimurium. By using random mariner transposon mutagenesis, mutations that caused derepression of ompS1 expression were isolated, one in S. enterica serovar Typhi and two in S. enterica serovar Typhimurium. All of them mapped in the hns gene in the region coding for the carboxy terminus of the H-NS nucleoid protein. The derepressed ompS1 expression was subject to negative regulation at high osmolarity, both in the presence and in the absence of OmpR. This observation was possible due to the fact that there are two promoters: P1, which is OmpR dependent, and P2, which does not require OmpR for activation (rather, OmpR represses P2). The sequences upstream from position −88, a region previously shown to be involved in the negative regulation of ompS1, can form a static bend, and the integrity of this region was required for function and binding of H-NS and for osmoregulation, as determined with gene reporter fusions of different lengths and with a 31-bp deletion mutant. This is consistent with the notion that this region determines a structure required for repression. Hence, ompS1 shares negative regulation by H-NS with other loci, such as the bgl operon and the ade gene.
Salmonella enterica serovar Typhi is the etiological agent of typhoid fever in humans (24). Its outer membrane proteins (OMPs) have an important role in triggering the host immune response (15, 21), and S. enterica serovar Typhimurium ompC ompF double mutants are attenuated for virulence in mice (2). Thus, aside from their roles in the exchange of small molecules and in participating in preserving the bacterial cell shape, the OMPs are relevant to the interaction between a bacterial pathogen and its host.
S. enterica serovar Typhi synthesizes three major OMPs that are highly abundant upon growth in standard laboratory media: the OmpC and OmpF porins and OmpA, a structural protein (29). In S. enterica serovar Typhi, as in Escherichia coli, expression of OmpC and OmpF is under the control of the ompB (ompR-envZ) locus. The relative levels of expression of OmpC and OmpF in E. coli are modulated by changes in osmolarity, which has been proposed to affect the level of phosphorylated OmpR (9, 20, 26). In S. enterica serovar Typhi, a shift in osmolarity affects only the expression of OmpF, whereas OmpC is not osmoregulated (17, 29).
Besides genes for the major porins, E. coli possesses porin genes that are quiescent under standard laboratory growth conditions, such as those for Lc (1), NmpC, and OmpN (27). In Klebsiella pneumoniae, ompK37 is expressed at low levels under standard laboratory growth conditions but is highly expressed in β-lactam-resistant clinical isolates (6). This gene is 80% identical to S. enterica serovar Typhi ompS2 and to E. coli ompN. Likewise, serovar Typhi ompS1 belongs to this group of low-level-expressed porins (8).
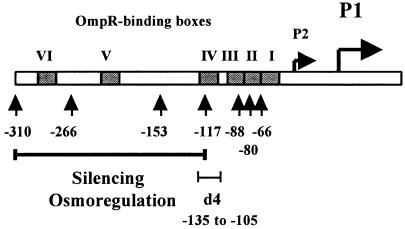
In contrast to the ompC and ompF genes, which contain three and four OmpR-binding sites, respectively, ompS1 possesses six OmpR-binding sites (22) (Fig. 1). cis-acting elements up to −310 bp upstream of the OmpR-dependent P1 transcription start site have been described. There is also a P2 promoter, which does not require OmpR for activation but which, however, is repressed in the presence of OmpR. The −310 region contains both positive and negative regulatory elements. Sequences upstream of position −88 are required for negative regulation, and sequences downstream of −88 are required for positive regulation (Fig. 1) (22). Expression of ompS1 in the S. enterica serovar Typhi wild-type strain IMSS-1 was not affected by a shift in osmolarity or by other stress growth conditions that can affect porin expression, such as changes in pH and temperature (22).
FIG. 1.
The S. enterica serovar Typhi ompS1 5′ upstream regulatory region. The P1 OmpR-dependent promoter, the P2 OmpR-repressed promoter, and the six OmpR-binding boxes (I to VI), as described previously (22), are shown. The number of base pairs upstream of the P1 transcriptional start, as contained in the ompS1-lacZ fusions (22), is indicated throughout the region. The long horizontal bar delimits the region required for silencing and osmoregulation. The d4 bar shows the 31-bp deletion from −135 to −105.
The wide porin repertoire in Salmonella raises new questions regarding their function and regulation. In the present work, we report that the H-NS nucleoid protein is a repressor of ompS1. Previous studies on porin regulation reported that H-NS binds to the intergenic region of the ompC-micF genes, repressing them transcriptionally and interfering with the posttranscriptional negative regulation of ompF translation via micF (37). Moreover, another nucleoid protein, HU, has been found to affect the levels of OmpF (23).
MATERIALS AND METHODS
Bacterial strains, plasmids, and recombinant DNA techniques.
The relevant bacterial strains and plasmids used in this study are listed in Table 1. DNA manipulations were performed according to standard protocols (33). Oligonucleotides used for amplification by PCR were provided by the Oligonucleotide Synthesis Facility at our institute and are listed in Table 2. PCRs were performed with Expand (Boehringer Mannheim Inc.) or with Taq DNA polymerase, according to the instructions by the manufacturer. The one-step mutagenesis procedure described by Datsenko and Wanner (3) for bacterial chromosomal genes was used to generate gene deletions and replacements for antibiotic resistance markers.
TABLE 1.
Relevant bacterial strains and plasmids used
| Strain or plasmid | Genotype and/or relevant markers | Reference or source |
|---|---|---|
| E. coli strains | ||
| BL21(DE3) | F−ompT gal (dcm) (lon) hsdSB(rB− mB−) met(DE3) | Novagen |
| DH5α | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 Δ(lacZYA-argF)U169 φ80ΔlacZΔM15 | Invitrogen |
| SM10λpir | thi thr leu tonA lacY supE recA RP4-2-Tc::Mu λpir | 18 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 Δ(lac-pro) [F′proAB lac1qlacZΔM15 Tn10] | Stratagene |
| S. enterica serovar Typhi and Typhimurium strains | ||
| IMSS-1 | 9, 12, d, Vi serotype; reference clinical strain | 28 |
| IMSS-41 | IMSS-1 ΔompR::Cmr | This study |
| STYhns99 | IMSS-1 hns99::Kmrmariner | This study |
| STY9941 | STYhns99 ΔompR::Cmr | This study |
| ATCC 14028s | Wild-type S. enterica serovar Typhimurium | American Type Culture Collection |
| STMhns114 | ATCC 14028s hns114::Kmrmariner | This study |
| STMhns127 | ATCC 14028s hns127::Kmrmariner | This study |
| Plasmids | ||
| pACYC184 | Vector for cloning of S. enterica serovar Typhi hns, p15A; Tcr Cmr | New England Biolabs |
| pACYC184ΔTc | pACYC184 containing a NruI-EcoRV deletion within the tetracycline resistance gene | This study |
| phnsty184 | Vector pACYC184 carrying S. enterica serovar typhi wild-type hns; Cmr | This study |
| pKD46 | oriR101ts, λ Red recombinase system under paraB promoter; Apr | 3 |
| pKD3 | pANTSγ derivative containing an FRT-flanked Cmr gene from pSC140 | 3 |
| pMC1871 | Vector pBR322 carrying a promoterless E. coli lacZ gene; Tcr | 35 |
| pT6HNS | Vector pMPM-T6Ω carrying the E. coli hns gene fused to His6 under pBAD promoter | V. H. Bustamante et al., unpublished results |
TABLE 2.
Primers used
| Primer | Sequencea |
|---|---|
| LFhns-S | 5′-caaaataaGtcgaCttgcttaaaggc-3′ (SalI site) |
| RRhns-X | 5′-cgttattctAgacgaatatgagtccga-3′ (XbaI site) |
| Mar3-2 | 5′-ccggggacttatcagccaacctgttat-3′ |
| S1B-CAT5 | 5′-ggtcgacggatccggggaat-3′ |
| S1S-CAT3 | 5′-caagcaggtcgacatcttttctgttcat-3′ |
| ompS1-PE | 5′-ttgctgcgcctgccactaataac-3′ |
| ompA-PE | 5′-tttgcgcctcgttatcatccaa-3′ |
| 310b-1 | 5′-tagccttttatcatttattttatc-3′ |
| 310b-2 | 5′-atgagttatgtgttttgatttga-3′ |
| 310b-3 | 5′-caaagcatcaaatacatataaaaa-3′ |
| 310b-4 | 5′-tgtttctatttggttttttataatac-3′ |
| 310b-7 | 5′-attcttagtcacttatatatcctgtattat-3′ |
| 310b-8 | 5′-aatatgtagccacttcaacaaaac-3′ |
Uppercase letter indicate changes in the primer sequence with respect to the wild type, designed to introduce restriction enzyme sites.
Bacterial culture and enzymatic assays.
Bacteria were grown in nutrient broth (low osmolarity) or nutrient broth plus 300 mM NaCl (high osmolarity) at 37°C and collected at mid-logarithmic phase. The culture conditions and microplate protein and β-galactosidase assays were as previously described (22).
Random transposon mutagenesis.
The S. enterica serovar Typhi IMSS-1 and S. enterica serovar Typhimurium 14028 wild-type strains were conjugated with E. coli SM10 λ pir/pFD1 harboring the mariner transposon (Kmr) (32). They were then grown in Luria-Bertani broth (containing, per liter, 10 g of peptone, 5 g of yeast extract, and 10 g of NaCl) or SOC broth (containing, per liter, 20 g of tryptone, 5 g of yeast extract, 0.5544 g of NaCl, 0.1864 g of KCl, 1.2038 g of MgSO4, 0.9522 g of MgCl2 and 3.2 g of glucose). They were then subcultured in fresh medium and incubated at 37°C with shaking until they reached mid-logarithmic growth phase. Cells were collected by centrifugation and washed once with 1× phosphate-buffered saline (containing, per liter, 8.0 g of NaCl, 0.61 g of Na2HPO4, 0.2 g of KH2PO4, and 0.2 g of KCl and adjusted to pH 7.4). They were then resuspended in 1× phosphate-buffered saline and plated onto SOC agar for incubation at 37°C overnight. Mutants derived from this conjugation were scraped and resuspended in 0.9% NaCl, plated on MacConkey agar (containing, per liter, 1.5 g of casein peptone, 17.0 g of gelatin peptone, 1.5g of meat peptone, 10.0 g of lactose, 1.5 g of bile salts, 5.0 g of NaCl, 13.5 g of agar, 0.03 g of neutral red, and 0.001 g of crystal violet [pH 7.2]), and incubated overnight at 37°C. When needed, 12 μg of tetracycline per ml or 25 μg of kanamycin per ml was used.
Localization of the mariner transposon insertion site in the Salmonella mutants.
Bacterial genomic DNA was isolated by using a commercial kit (Aqua Pure genomic DNA isolation kit; Bio-Rad) and digested with either BamHI, KpnI, or SalI. The fragments were cloned into pUC18 or pUC19 vector and selected for Kmr colonies. Clones containing from 1.2 to 4 kb of insert were selected for sequencing. The DNA sequence near the 3′ end of the site of insertion of the mariner transposon was determined with the Thermosequenase kit (United States Biochemicals), according to the instructions of the manufacturer, or at the Sequencing Facility at our institute by using the Mar3-2 primer (Table 2). The sequences obtained were compared with those currently available at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST/) by using N-Blast or T-BLAST-X (National Institutes of Health) with the default parameters. The computer-assisted analysis of nucleotide and amino acid sequences was performed with the GENE WORKS (IntelliGenetics), DNA Strider 1.0 (CEA France), Amplify 1.2 (University of Wisconsin), and OLIGO 4.0 (National Biosciences Inc.) software. Graphics were constructed with Cricket Graph-III version 1.0.1 (Computer Associates International, Inc.), and radioactive gels were developed and analyzed with a PhosphorImager (Molecular Dynamics).
Cloning of the wild-type S. enterica serovar Typhi hns gene.
The wild-type S. enterica serovar Typhi hns gene was amplified by PCR with Expand (Boehringer Mannheim Inc.) and cloned into the pACYC184 vector (New England Biolabs). Briefly, S. enterica serovar Typhi IMSS-1 genomic DNA was used for PCR amplification with LFhns-S (annealing from position −538 to −513 of the serovar Typhi hns translational start site) and RRhns-X (annealing from position +630 to +604 of the serovar Typhi hns translational termination site) primers (Table 2). The 1,584-bp PCR fragment was digested and cloned into the SalI and XbaI sites of pACYC184, giving phnsty184. The identity of the cloned fragment was verified by sequencing.
Purification of H-NS His-tagged protein.
Purification of His6-tagged H-NS protein was performed with Ni-nitrilotriacetic acid resin (QIAExpress; Qiagen) according to the instructions of the manufacturer and the modifications established by V. H. Bustamante et al. (unpublished results). Briefly, E. coli BL21(DE3) carrying the pT6HNS plasmid (Table 1) was grown to mid-logarithmic phase. l(+)-Arabinose (Sigma-Aldrich) was added to a final concentration of 0.1%, and the bacteria were incubated for 3 h at 30°C and 250 rpm. Cells were then pelleted by centrifugation, resuspended in urea buffer (8 M urea, 100 mM NaH2PO4, 10 mM Tris-HCl [pH 8.0]), and disrupted by sonication. The suspension was centrifuged, and the supernatant was filtered through an Ni-nitrilotriacetic acid agarose column (QIAExpress; Qiagen); the column was washed with urea buffer at pH 8.0 and urea buffer at pH 6.0, and finally the bound protein was eluted with urea buffer at pH 4.5. Fractions containing purified His6-tagged H-NS protein were selected after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) and loaded into a Slyde-A-Lyzer 10K cassette (Pierce) for extensive dialysis, first in 50 mM Tris-HCl (pH 7.5)-10 mM MgCl2-20% glycerol-0.5 M NaCl-0.1% Triton X-100-4 M urea and then in the same buffer containing 1 M urea instead of 4 M urea. Finally, dialysis was done in 30 mM Tris-HCl (pH 7.5)-10 mM MgCl2-20% glycerol-240 mM NaCl-0.1% Triton X-100-3 mM EDTA.
Electrophoretic mobility shift assays.
We conducted PCRs that generated ompS1 products encompassing the length of the regulatory region harbored in representative ompS1-lacZ fusions of the pRO series (22), using those plasmids as templates and with oligonucleotide primers S1B-CAT5 (annealing from position 6 to 25 in the pRO plasmid series) and S1S-CAT3 (annealing from position 425 to 398 in pRO310, position 268 to 241 in pRO153, and position 203 to 176 in pRO88). The sites of primer annealing are with respect to the PstI site at the beginning of the lacZ reporter gene. These fragments and the enteropathogenic E. coli (EPEC) ler regulatory and structural gene fragments (100 ng of each), used as positive and negative controls, respectively (Bustamante et al., unpublished), were mixed with increasing concentrations of His-tagged H-NS in the presence of 10 mM Tris-HCl, 1 mM EDTA, 80 mM NaCl, 10 mM 2-mercaptoethanol, 1 mM dithiothreitol, and 10% glycerol. They were incubated 20 min at 37°C and then separated by electrophoresis in 6% polyacrylamide gels in 1× Tris-borate-EDTA buffer. The DNA bands were visualized by staining with ethidium bromide.
Analysis of DNA static curvature.
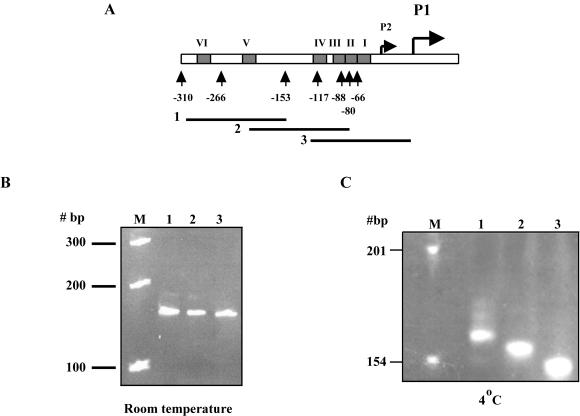
The BEND-IT software, from the International Centre for Genetic Engineering and Biotechnology website (http://www.icgeb.org/dna/bend_it.html), was used to identify regions of potential intrinsic curvature. The curvature is calculated as a vector sum of dinucleotide geometries by using the BEND algorithms of Goodsell and Dickerson (11) and is expressed as degrees per helical turn (10.5o/helical turn = 1o/bp). In addition, the mobility of PCR fragments of 152 or 153 bp was analyzed by 8% PAGE at room temperature and at 4°C in 0.5× Tris-borate-EDTA. The PCR fragments were amplified with primer pairs 310b-1 (annealing from position −303 to −280)-310b-2 (−154 to −176), 310b-3 (−226 to −203)-310b-4 (−76 to −101), and 310b-7 (−124 to −95)-310b-8 (+27 to + 4) (Table 2; see Fig. 8A).
FIG. 8.
The ompS1 5′ upstream region is statically curved. (A) Schematic diagram showing the ompS1 5′ region. Numbers indicate the length of the regulatory region contained in each lacZ fusion. The six OmpR-binding boxes are represented as filled rectangles. Lines 1, 2, and 3, PCR fragments encompassing from −303 to −154, −226 to −76, and −124 to +27, respectively, of the ompS1 5′ region. (B and C) PCR fragments 1, 2, and 3 separated by 8% PAGE at room temperature(B) or at 4°C (C). Lanes M, molecular size markers.
Primer extension analysis.
One microgram of total RNA (for ompA) or 50 μg of total RNA (for ompS1, in two reaction mixtures with 25 μg each), isolated by using commercial kits (RNeasy [Qiagen] or High Pure RNA isolation kit [Roche]) was denatured at 90°C for 3 min and then slowly cooled to 45°C. The RNA was annealed with [γ-32P]ATP-labeled ompA-PE (annealing from position +3 to −24 with respect to the S. enterica serovar Typhi ompA translational start site) or ompS1-PE (annealing from +57 to +35 with respect to the serovar Typhi ompS1 translational start site). The oligonucleotide primers were extended with Moloney murine leukemia virus reverse transcriptase at 37°C for 2 h, and the extended products were collected with a Microcon-30 microconcentrator (Amicon) and analyzed by electrophoresis in urea-8% polyacrylamide gels.
RESULTS
Interruption of the Salmonella hns gene results in derepression of ompS1 expression.
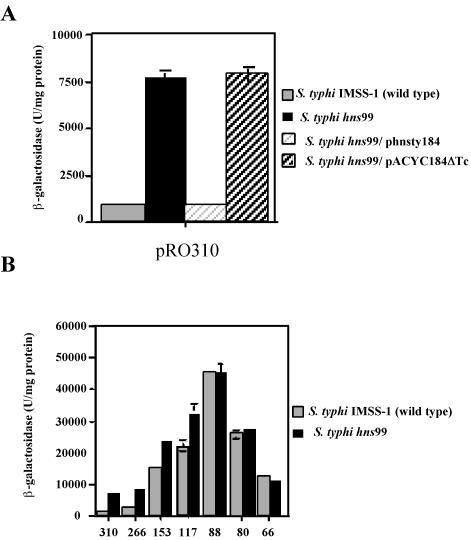
We used random transposon mutagenesis to identify putative negative effectors of ompS1 expression. Briefly, E. coli SM10 λ pir/pFD1 (harboring the mariner transposase; Kmr) was conjugated with the wild-type strain S. enterica serovar Typhi IMSS-1 or S. enterica serovar Typhimurium 14028 transformed with the ompS1-lacZ fusion pRO310. This construct has the longest fused regulatory region (Fig. 1) and had the lowest β-galactosidase activity in both wild-type strains with respect to shorter constructs (22) (Fig. 2B). It produced white colonies on MacConkey indicator plates. From approximately 18 × 103 Kmr serovar Typhi mutants and 80 × 103 Kmr serovar Typhimurium mutants, we were able to isolate one serovar Typhi and two serovar Typhimurium red colonies on MacConkey plates. β-Galactosidase assays on cultured samples of these mutants (Fig. 2A; see Fig. 4A) showed a 10-fold derepression of ompS1 expression compared to that in the wild-type strains.
FIG. 2.
Derepression of S. enterica serovar Typhi ompS1 in an hns background. Bars indicate the specific β-galactosidase activity. (A) Expression in the S. enterica serovar Typhi wild-type strain and in the isogenic hns99 derivative. Complementation was with cloned S. enterica serovar Typhi wild-type hns or with the pACYC184ΔTc vector. (B) Expression from reporter fusions of different lengths in the S. enterica serovar Typhi wild-type strain and in the hns99 derivative. Numbers indicate the length of the ompS1 regulatory region fused to lacZ, as depicted in Fig. 1. Results are the averages and standard deviations from three independent experiments. An absence of error bars indicates that the standard deviation was too small to be shown.
FIG. 4.
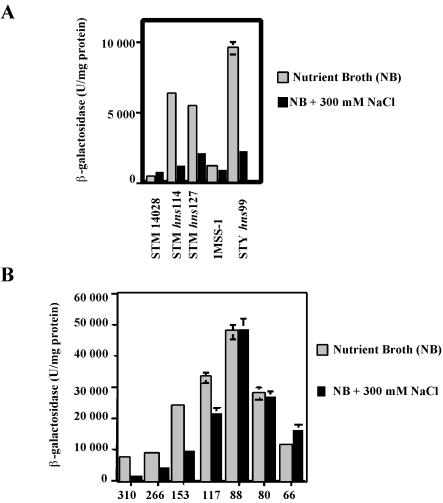
Osmoregulation of ompS1 in Salmonella hns. (A) Expression in S. enterica serovar Typhi (STY) and in S. enterica serovar Typhimurium (STM) wild-type strains and in their isogenic hns derivatives, as assessed with the bp −310 lacZ reporter fusion, at low (NB) and high (NB + 300 mM NaCl) osmolarities. (B) Expression in S. enterica serovar Typhi hns99 of ompS1-lacZ fusions of different lengths (as shown in Fig. 1) at low and high osmolarities. Error bars indicate standard deviations.
The DNA flanking the Kmr marker from the mutants was cloned, and the interrupted gene in all three events was identified as hns. All of the insertions mapped at or near the region coding for the DNA-binding domain (amino acids 89 to 119 of the 137-residue protein) (5, 7). In S. enterica serovar Typhi, the resistance marker inserted into the codon for amino acid 99, and in S. enterica serovar Typhimurium, the insertions were in codons 114 and 127 of H-NS. Thus, these mutations were designated hns99, hns114, and hns127, respectively.
In order to further strengthen the evidence for a role of H-NS in ompS1 expression, we transformed either a plasmid bearing the wild-type S. enterica serovar Typhi hns gene or the pACYC184ΔTc vector plasmid into the mutants. Expression of cloned hns was under the control of ca. 500 bp of its own regulatory region, and the clone does not contain any flanking gene upstream or downstream. Upon complementation, ompS1 expression was restored to low levels close to those observed in the wild-type strain (Fig. 2A). Such complementation was not observed with the pACYC184ΔTc vector plasmid.
We also analyzed the length of the ompS1 regulatory sequence needed for the effect by H-NS. The fusions containing between bp −310 and −117 (Fig. 1) were positively affected by interruption of hns, whereas fusions from −88 and downstream were not affected by the hns interruption (Fig. 2B). Thus, the region upstream of position −88 was required for negative regulation by H-NS.
The shorter the fusions, the higher was the activity in both the wild-type and hns backgrounds, in agreement with the notion that as the fusions are shortened, there is a gradual release of an appropriate structure for repression.
H-NS binds to the upstream regulatory region of ompS1.
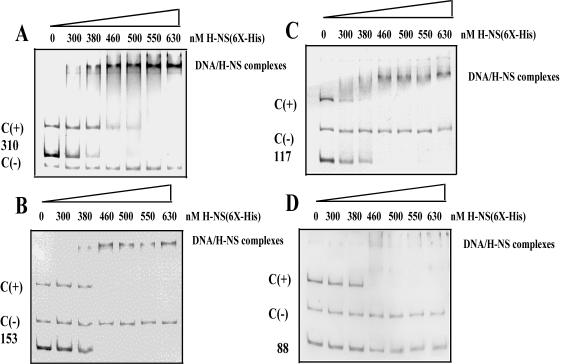
In order to test whether H-NS directly interacted with the ompS1 regulatory region, we conducted electrophoretic mobility shift assays with PCR fragments encompassing the regulatory region contained in four representative ompS1-lacZ fusions (Fig. 3). We were able to detect binding of H-NS to the fragments spanning bp −310, −153, and −117 upstream of the P1 promoter (Fig. 3A to C). In contrast, with a −88 upstream fragment, no shift in mobility was observed (Fig. 3D). All fragments contained 21 bp into the structural gene on their 3′ end. These data were consistent with the results of ompS1 expression, where fusions upstream of bp −88 were derepressed in Salmonella hns strains (Fig. 2B).
FIG. 3.
Binding of H-NS to the ompS1 upstream regulatory region. Electrophoretic mobility shift assays were carried out with the ompS1 regulatory region (100 ng), using His6-tagged H-NS at the indicated concentrations. C(+), is the EPEC ler regulatory region, used as a positive control; C(−), EPEC ler coding region, used as negative control. 310 (A), 153 (B), 117 (C), and 88 (D), fragments of the ompS1 regulatory region encompassing the indicated length as shown in Fig. 1. The complexes were separated on 6% polyacrylamide gels.
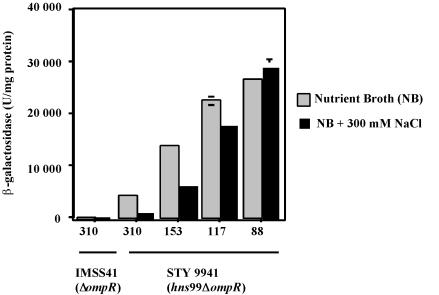
Osmoregulation of ompS1 expression in Salmonella hns mutants.
In the S. enterica serovar Typhi IMSS-1 wild-type strain, the low level of ompS1 expression was not affected by changes in osmolarity (22). Nevertheless, we decided to test the effect on the derepressed ompS1 activity in the hns background in response to osmolarity. Indeed, the derepressed ompS1 expression was osmoregulated, as it decreased at increasing osmolarity in both S. enterica serovar Typhimurium and S. enterica serovar Typhi (Fig. 4A). This osmoregulation was gradually abolished upon deletion of the region upstream of position −88. The decrease in expression varied, as it ranged from nearly fourfold in the −310 fusion (ca. 8,000 U of β-galactosidase in low-osmolarity medium versus ca. 1,900 U in high-osmolarity medium) to less than twofold in the −117 fusion (from ca. 32,000 U to ca. 20, 000 U), with no effect in the −88 and shorter fusions (Fig. 4B). This dependence on the length of the regulatory region was similar to that observed for the derepressing effect in the hns background (Fig. 2B).
Since OmpR is required for osmoregulation of the ompC and ompF major porin genes (26), we tested whether osmoregulation of ompS1 in the hns mutants was dependent on OmpR: this was possible because there is a P2 promoter that does not require OmpR for activation (22) (Fig. 1). Expression of ompS1 was derepressed in S. enterica serovar Typhi 9941 (hns99 ΔompR double mutant) (Fig. 5) with respect to the ompR strain, and it also decreased at high osmolarity, i.e., even in the absence of OmpR. Fusions containing from bp −310 and up to bp −117 diminished their expression at high osmolarity, whereas no decrease in expression was seen with the −88 construct (Fig. 5). The pattern of expression was remarkably similar to that obtained in the presence of OmpR (Fig. 4B). Indeed, the decrease in expression ranged from almost fourfold in the −310 fusion (ca. 4,100 U of β-galactosidase in low-osmolarity medium versus ca. 1,100 U in high-osmolarity medium) to less than twofold in the −117 fusion (from ca. 22,000 to 17,000 U) (Fig. 5). Hence, the region upstream of bp −88 was also determinant for osmoregulation in the absence of OmpR, as it was for H-NS binding and silencing (Fig. 2 to 4). Again, these data are consistent with the proposal that the upstream region determines an appropriate structure for repression, which occurs at high osmolarity in the absence of H-NS or at both low and high osmolarity in the presence of H-NS.
FIG. 5.
Osmoregulation in S. enterica serovar Typhi 9941 (hns99 ΔompR). Expression of the ompS1-lacZ fusions constructs of different lengths in S. enterica serovar Typhi IMSS-41 ΔompR and STY9941 hns99 ΔompR at low (NB) and high (NB + 300 mM NaCl) osmolarities is shown. The number below each bar indicates the length of the ompS1 regulatory region fused to lacZ. Error bars indicate standard deviations.
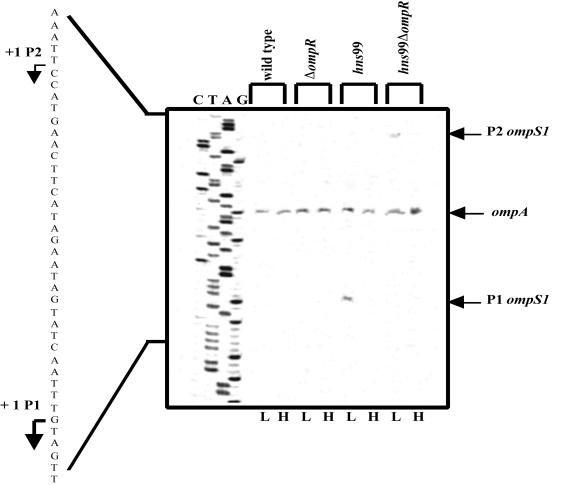
The identity of the promoters affected by the hns interruption was addressed by primer extension analysis (Fig. 6). The expression of chromosomal ompS1 was not detected in the S. enterica serovar Typhi wild-type strain, and transcription from the P1 OmpR-dependent promoter was indeed derepressed in the hns99 mutant. This activity was lower at high osmolarity. In the double hns99 ΔompR mutant, the OmpR activation-independent P2 promoter was derepressed, as no activity was detected in a ΔompR mutant, and its activity was lower at high osmolarity. Furthermore, ompA transcriptional activity was not affected either by the genetic background or by changes in osmolarity. It is worth noting that the promoter strength for P1 was less than 2% the level for ompA, even under derepression in the hns background. This was based on the densitometric scan of the autoradiogram and taking into account that the total amount of RNA used to detect the transcriptional start sites for ompS1 was 50 μg, whereas for ompA it was only 1 μg. Moreover, P2 showed one-half of the promoter strength of P1 under derepression in the hns99 ΔompR background (Fig. 6). These results on the activity of the native chromosomal genes reflect the differences in activity observed with the lacZ fusion studies (Fig. 4B and 5).
FIG. 6.
ompS1 chromosomal promoter activity at low (L) and high (H) osmolarities in S. enterica serovar Typhi strains, as assessed by primer extension. IMSS-1 is the wild-type strain, IMSS-41 is the isogenic ΔompR derivative, hns99 is the isogenic transposon mutant, and STY9941 (hns99 ΔompR) is the isogenic double mutant. The DNA ladder sequence for ompS1 is shown on the left. The bands corresponding to the transcriptional start sites of the OmpR-dependent P1 promoter, the P2 OmpR-repressed promoter, and the ompA gene are marked. The amounts of total RNA used were 50 μg for the ompS1 transcriptional start site and 1 μg for ompA.
Deletion of a region from bp −135 to −105 results in loss of osmoregulation in Salmonella hns.
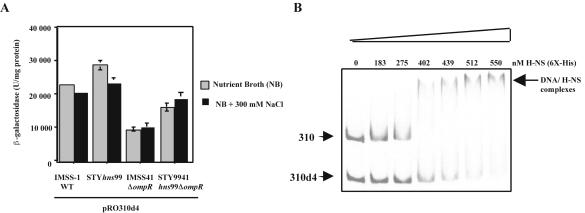
In order to further verify the role of the ompS1 upstream regulatory region in negative regulation, we analyzed the expression of a lacZ fusion containing the bp −310 region with a deletion of 31 bp (from −135 to −105), the pRO310d4 fusion (Fig. 1) (22). We measured expression of the pRO310d4 fusion in S. enterica serovar Typhi IMSS-1 (wild type) and in its isogenic ΔompR (IMSS-41), hns (hns99), and hns99 ΔompR (STY9941) mutants (Fig. 7A). As observed previously (22), the mere deletion of this region results in a 20-fold increase in activity in the wild type. Moreover, no osmoregulation was observed in the hns99 or hns99 ΔompR background, in contrast to that seen with pRO310 (Fig. 4 and 5). Even though some increase was observed in the hns strains, this effect was much lower than the 10-fold observed for pRO310 (Fig. 2A and 5).
FIG. 7.
Effect of the deletion in the ompS1 regulatory region of 31 bp, from −135 to −105. (A) Expression of the pRO310d4 fusion in S. enterica serovar Typhi IMSS-1 (wild type), IMSS-41 ΔompR, hns99, and STY9941 hns99 ΔompR at low (NB) and high (NB + 300 mM NaCl) osmolarities. Error bars indicate standard deviations. (B) Comparison of the binding of H-NS to PCR fragments representing the −310 and −310d4 fusions. The nanomolar final concentrations of His6-tagged H-NS are indicated at the top of the gel.
The notion that the integrity of the −310 region is needed for full H-NS binding and thus derepression in an hns background was also shown by electrophoretic mobility shift assays with H-NS. As can be seen in Fig. 7B, the deletion from position −135 to −105 resulted in a lowering of the affinity of H-NS compared to the full −310 region.
The 5′region of ompS1 is intrinsically curved.
It has been proposed that H-NS preferentially binds to curved DNA (16) and that negative osmoregulation of ompF involves a DNA loop structure (14). Hence, the possibility that the 5′ upstream region of ompS1 might acquire a curved configuration was tested. Three circularly permuted DNA fragments encompassing the upstream regulatory region, present in pRO310, were analyzed by PAGE at 4°C, where curved fragments migrate anomalously compared to at room temperature. The more centered the curvature on a given fragment, the slower the migration (39). As can be seen in Fig. 8C, at 4°C fragment 1 (position −303 to −154) migrated the slowest, fragment 2 (−226 to −76) had an intermediate migration, and fragment 3 (−124 to +27) had the fastest migration. In contrast, all three fragments had the same migration at room temperature (Fig. 8B). This is consistent with the presence of a curved region at the center of fragment 1 and close to the center of fragment 2.
The curvatures predicted by analysis in silico were more prominent at positions −230 and −149, having values of 13 and 11°/10.5 bp of helical turn, respectively. This is consistent with the electrophoretic data. The same in silico analysis on the E. coli ompF 5′ regulatory region rendered values of 11 and 12, near those for OmpR-binding box F3 and the integration host factor-binding sites, where regions of bent DNA have been postulated (14).
DISCUSSION
The Salmonella ompS1 gene belongs to the porin superfamily. Its function and the in vitro signals that lead to the induction of its expression remain unknown so far. Nevertheless, it appears to have a role in the Salmonella life cycle, since mutations causing defects in swarming motility and biofilm formation have been found to map in S. enterica serovar Typhimurium ompS1 (19, 38). Moreover, the fact that a serovar Typhimurium ompS1 mutant is less virulent in mice (O. Rodríguez-Morales et al., unpublished results) points towards a role in pathogenicity. In contrast to the case for the K. pneumoniae minor porin OmpK37, which is expressed in the absence of the major porins (6), OmpS1 expression was not enhanced in an S. enterica serovar Typhi ompC ompF double mutant grown in standard laboratory media (data not shown).
The regulation of ompS1 shares several features with that of the quiescent E. coli bgl operon, which codes for the uptake and utilization of β-glucosides, and with that of a quiescent adenine deaminase gene, yicP (ade), recently discovered in E. coli. First, the mechanisms for their induction in a wild-type background, and thus their putative role in nature, have remained elusive. Second, their expression is negatively affected by the H-NS nucleoid protein; i.e., they are activated when hns is mutated. Moreover, derepression of bgl has also been observed by deletions in the upstream silencer. Accordingly, integration of insertion elements in the upstream silencer regions has contributed to the derepression of both the bgl and ade loci (4, 25, 31, 34, 36).
Previously we showed that ompS1 contains two cis-acting regions: one from bp −310 to −88 upstream of the transcriptional start site of the P1 promoter, which is involved in silencing; and other downstream from bp −88, which is necessary for the positive control of ompS1 expression (22) (Fig. 1). In order to elucidate the mechanism that maintains a low level of expression of ompS1, random transposon mutagenesis was performed in S. enterica serovar Typhi and S. enterica serovar Typhimurium. We found mutations in hns that resulted in the derepression of ompS1 (Fig. 2A and 4A). All of the transposon insertions mapped to the coding region for the carboxy terminus, at or near the DNA-binding domain. Consistently, in another study, a TnphoA-generated mutation in S. enterica serovar Typhimurium strain C5 was also found to map at the 3′end of hns (12).
In S. enterica serovar Typhi, fusions containing sequences upstream of position −88 showed higher levels of expression in the hns background with respect to the wild type, whereas those at and downstream of −88 were not affected (Fig. 2B). Moreover, H-NS bound to promoter fragments including sequences up to −117 and further upstream but not to a promoter fragment which includes DNA up to position −88 (Fig. 3). The result that derepression of the −310 construct in hns99 did not reach the levels attained by the −88 construct and that gradual derepression was observed as the constructs were shortened (Fig. 2B) suggests that the sequences upstream of this position could form a DNA structure that promotes silencing.
In the S. enterica serovar Typhi IMSS-1 wild-type strain, ompS1 expression was not affected by a shift in osmolarity, leading to the proposal that ompS1 was a nonosmoregulated porin gene (22). The lack of osmoregulation was also observed for ompS1 with the lacZ fusions in wild-type S. enterica serovar Typhimurium 14028 (Fig. 4A). However, when ompS1 expression was derepressed in the hns mutants, in either serovar Typhi or serovar Typhimurium, it was negatively regulated in high-osmolarity medium (Fig. 4A). The analysis of ompS1-lacZ fusions encompassing different lengths of the regulatory region showed that the negative regulatory region needed for H-NS repression, i.e., upstream of bp −88 (Fig. 2B), was also required for osmoregulation (Fig. 4B). Furthermore, osmoregulation occurred in the absence of OmpR (Fig. 5) and also required this sequence. The ratio of ompS1 expression at low osmolarity to that at high osmolarity in the hns99 and hns99 ΔompR strains was almost identical for all of the fusions (Figs. 4B and 5). Hence, both ompS1 promoters share a common mechanism that leads to lower expression at high osmolarity in the absence of H-NS function. It appears that a repressing structure is formed under these conditions, which is also promoted in the presence of H-NS.
As observed with ompF, ompS1 expression decreased at high osmolarity when derepressed in an hns strain (Fig. 4B and 5). In this respect, the ompF 5′ upstream regulatory region has a role in osmoregulation (14, 30). Using the pRO310d4 fusion, which contains the entire bp −310 regulatory region with a 31-bp deletion (from −135 to −105) (Fig. 1), we observed that ompS1 expression was derepressed 20-fold and was not significantly affected in either the hns99 or the hns99 ΔompR mutant; that is, it no longer responded to changes in osmolarity independently of the presence or absence of OmpR (Fig. 7A). Moreover, the binding affinity of H-NS to the 310d4 regulatory region was lower than that observed for the whole −310 region, although it was still higher than that for the −88 region (Fig. 3A and D and 7B). Thus, the integrity of the −310 regulatory region was required for full H-NS repression and osmoregulation. In this regard, the region upstream of bp −88 has an intrinsic curvature, as assessed both in silico and by DNA bending electrophoretic assays (Fig. 8C). Such curvature could determine a structure needed for repression in the presence of H-NS or at high osmolarity in the hns background.
Because of the fact that ompS1 expression in the Salmonella hns mutants did not reach the level attained in the shortened, most active pRO88 fusion, the presence of additional negative effectors aside from H-NS cannot be excluded. Thus, we can envision at least three scenarios to explain the negative regulation of ompS1 expression by high osmolarity. First, as mentioned above, changes in the DNA structure of the regulatory region could hinder expression, possibly by blocking access of the RNA polymerase. Another possibility would occur if H-NS is produced as a stable, truncated protein in our Salmonella mutants. In this regard, the participation of the nucleoid protein StpA as an adaptor for truncated H-NS in repressing the expression of the proU gene and the bgl locus in E. coli has been illustrated (10). Hence, the binding of a putative complex formed by truncated H-NS and StpA to the ompS1 upstream regulatory region could be enhanced at high osmolarity, leading to the observed decrease in ompS1 expression. Finally, the action of another putative negative effector could be increased at high osmolarity.
Thus, the characterization of the state of the H-NS protein in our mutants, analysis of ompS1 expression in an hns stpA double mutant at both low and high osmolarity, and a search for other negative effectors by mutagenesis in hns mutants are experiments that should help in further defining the mechanism involved in the negative regulation of ompS1 expression.
Hence, our present model to explain the strict negative regulation of ompS1 implies the initial binding of H-NS at or near the −135 to −105 region, thus allowing the formation of a repressing loop by nucleation of H-NS to the rest of the regulatory region. Such a repressing loop would involve the entire regulatory region, not only hindering the access of OmpR and thus preventing activation of P1 but also impeding transcription from P2, which otherwise would have been activated once OmpR could not activate P1. At high osmolarity, in the absence of H-NS function, the repressing loop again would be formed by local changes in DNA structure.
It is worth noting that the promoter strength of the native ompS1 gene on the chromosome is relatively low compared to that of a highly expressed outer membrane protein gene such as ompA (Fig. 6). This likely reflects the low abundance of the OmpS1 porin even under derepression in the hns strains, an idea that is further confirmed by the lack of detectable OmpS1 in outer membrane protein preparations separated by PAGE and stained with Coomassie brilliant blue (data not shown).
The function of nucleoid-associated proteins, such as H-NS, remains elusive despite extensive efforts towards understanding their physiological role. Moreover, whether osmolarity is a true regulatory signal for ompS1 in nature or whether it mimics another signal in the environment is an open question. ompS1 belongs to the pleiotropic H-NS regulatory circuit, where nearly one-third of the genes affected by H-NS are involved in cell envelope physiology and structure, consistent with the notion that genes regulated by H-NS are generally associated with bacterial adaptations to environmental stress (13).
Acknowledgments
We acknowledge Jeannette Barba-León and Víctor Bustamante for helping us with purification of His6-tagged H-NS and gel mobility shift assays, Rob Edwards for helpful discussions, Marcos Fernández-Mora for constructing the S. enterica serovar Typhi IMSS-41 ΔompR mutant, Eugenio López-Bustos for oligonucleotide synthesis, Ricardo Oropeza and Linda J. Kenney for helpful discussions and critical reading of the manuscript, Alejandra Vázquez for constructing the hns99 ΔompR mutant, and Miryam Villalba for technical assistance.
M.A.F.-V. was supported by a Ph.D. fellowship (113352) from the Consejo Nacional de Ciencia y Tecnología and by the PAEP (203336) and DGEP programs of the Universidad Nacional Autónoma de México. J.L.P. is a Howard Hughes International Research Scholar. This work was supported by grants to E.C. from the Universidad Nacional Autónoma de México (DGAPA IN229001) and from the Consejo Nacional de Ciencia y Tecnología, Mexico (CONACyT 37738-N).
REFERENCES
- 1.Blasband, A. J., W. R. Marcotte, Jr., and C. A. Schnaitman. 1986. Structure of the lc and nmpC outer membrane porin protein genes of lambdoid bacteriophage. J. Biol. Chem. 261:12723-12732. [PubMed] [Google Scholar]
- 2.Chatfield, S. N., C. J. Dorman, C. Hayward, and G. Dougan. 1991. Role of ompR-dependent genes in Salmonella typhimurium virulence: mutants deficient in both OmpC and OmpF are attenuated in vivo. Infect. Immun. 59:449-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Defez, R., and M. De Felice. 1981. Cryptic operon for beta-glucoside metabolism in Escherichia coli K-12: genetic evidence for a regulatory protein. Genetics 97:11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dersch, P., K. Schmidt, and E. Bremer. 1993. Synthesis of the Escherichia coli K-12 nucleoid-associated DNA-binding protein H-NS is subjected to growth-phase control and autoregulation. Mol. Microbiol. 8:875-889. [DOI] [PubMed] [Google Scholar]
- 6.Doménech-Sánchez, A., S. Hernández-Alles, L. Martínez-Martínez, B. J. Benedi, and S. Alberti. 1999. Identification and characterization of a new porin gene of Klebsiella pneumoniae: its role in beta-lactam antibiotic resistance. J. Bacteriol. 181:2726-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorman, C. J., J. C. D. Hinton, and A. Free. 1999. Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 7:124-128. [DOI] [PubMed] [Google Scholar]
- 8.Fernández-Mora, M., R. Oropeza, J. L. Puente, and E. Calva. 1995. Isolation and characterization of ompS1, a novel Salmonella typhi outer membrane protein-encoding gene. Gene 158:67-72. [DOI] [PubMed] [Google Scholar]
- 9.Forst, S., and M. Inouye. 1988. Environmentally regulated gene expression for outer membrane proteins in Escherichia coli. Annu. Rev. Cell Biol. 4:21-42. [DOI] [PubMed] [Google Scholar]
- 10.Free, A., M. E. Porter, P. Deighan, and C. J. Dorman. 2001. Requirement for the molecular adapter function of StpA at the Escherichia coli bgl promoter depends upon the level of truncated H-NS protein. Mol. Microbiol. 42:903-917. [DOI] [PubMed] [Google Scholar]
- 11.Goodsell, D. S., and R. E. Dickerson. 1994. Bending and curvature calculations in B-DNA. Nucleic Acids Res. 2:5497-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison, J. A., D. Pickard, C. F. Higgins, A. Khan, S. N. Chatfield, T. Ali, C. J. Dorman, C. E. Hormaeche, and G. Dougan. 1994. Role of hns in the virulence phenotype of pathogenic salmonellae. Mol. Microbiol. 13:133-140. [DOI] [PubMed] [Google Scholar]
- 13.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J. P. LeCaer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40:20-36. [DOI] [PubMed] [Google Scholar]
- 14.Huang, K. J., J. L. Schieberi, and M. M. Igo. 1994. A distant upstream site involved in the negative regulation of the Escherichia coli ompF gene. J. Bacteriol. 176:1309-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isibasi, A., V. Ortíz, M. Vargas, J. Paniagua, C. González, and J. Moreno. 1988. Protection against Salmonella typhi infection in mice after immunization with outer membrane proteins isolated from Salmonella typhi 9, 12, d, Vi. Infect. Immun. 56:2953-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jordi, B. J. A. M., A. E. Fielder, C. M. Burns, J. C. D. Hinton, N. Dover, D. W. Ussery, and C. F. Higgins. 1997. DNA binding is not sufficient for H-NS-mediated repression of proU expression. J. Biol. Chem. 272:12083-12090. [DOI] [PubMed] [Google Scholar]
- 17.Martínez-Flores, I., R. Cano, V. H. Bustamante, E. Calva, and J. L. Puente. 1999. The ompB operon partially determines differential expression of OmpC in Salmonella typhi and Escherichia coli. J. Bacteriol. 181:556-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in the construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mireles, J. R., II, A. Toguchi, and R. M. Harshey. 2001. Salmonella enterica serovar Typhimurium swarming mutants with altered biofilm-forming abilities: surfactin inhibits biofilm formation. J. Bacteriol. 183:5848-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizuno, T., and S. Mizushima. 1990. Signal transduction and gene regulation through the phosphorylation of two regulatory components: the molecular basis for the osmotic regulation of the porin genes. Mol. Microbiol. 4:1077-1082. [DOI] [PubMed] [Google Scholar]
- 21.Muthukummar, S., and V. R. Muthukkaruppan. 1993. Mechanism of protective immunity induced by porin-lipopolysaccharide against murine salmonellosis. Infect. Immun. 61:3017-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oropeza, R., C. L. Sampieri, J. L. Puente, and E. Calva. 1999. Negative and positive regulation of the non-osmoregulated ompS1 porin gene in Salmonella typhi: a novel regulatory mechanism that involves OmpR. Mol. Microbiol. 32:243-252. [DOI] [PubMed] [Google Scholar]
- 23.Painbeni, E., M. Caroff, and J. Rouviere-Yaniv. 1997. Alterations of the outer membrane composition in Escherichia coli lacking the histone-like protein HU. Proc. Natl. Acad. Sci. USA 94:6712-6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pang, T., Z., A. Bhutta, B. B. Finlay, and M. Altwegg. 1995. Typhoid fever and other salmonellosis: a continuing challenge. Trends Microbiol. 3:53-255. [DOI] [PubMed] [Google Scholar]
- 25.Petersen, C., L. B. Moller, and P. Valentin-Hansen. 2002. The cryptic adenine deaminase gene of Escherichia coli. J. Biol. Chem. 277:31373-31380. [DOI] [PubMed] [Google Scholar]
- 26.Pratt, L. A., W. Hsing, K. E. Gibson, and T. J. Silhavy. 1996. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20:911-917. [DOI] [PubMed] [Google Scholar]
- 27.Prilipov, A., P. S. Phale, R. Koebnik, C. Widmer, and J. P. Rosenbusch. 1998. Identification and characterization of two quiescent porin genes, nmpC and ompN, in Escherichia coli BE. J. Bacteriol. 180:3388-3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puente, J. L., V. Alvarez-Scherer, G. Gosset, and E. Calva. 1989. Comparative analysis of the Salmonella typhi and Escherichia coli ompC genes. Gene 83:197-206. [DOI] [PubMed] [Google Scholar]
- 29.Puente, J. L., A. Verdugo-Rodríguez, and E. Calva. 1991. Expression of Salmonella typhi and Escherichia coli OmpC is influenced differently by medium osmolarity: dependence on Escherichia coli OmpR. Mol. Microbiol. 5:1205-1210. [DOI] [PubMed] [Google Scholar]
- 30.Rampersaud, A., S. L. Harlocker, and M. Inouye. 1994. The OmpR protein of Escherichia coli binds to sites in the ompF promoter region in a hierarchical manner determined by its degree of phosphorylation. J. Biol. Chem. 269:12559-12566. [PubMed] [Google Scholar]
- 31.Reynolds, A. E., J. Felton, and A. Wright. 1981. Insertion of DNA activates the cryptic bgl operon in E. coli K-12. Nature 293:625-629. [DOI] [PubMed] [Google Scholar]
- 32.Rubin, A. J., B. J. Akerley, V. M. Novik, D. J. Lampe, R. N. Husson, and J. J. Mekalanos. 1999. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. USA 96:1645-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Schnetz, K. 1995. Silencing of Escherichia coli bgl promoter by flanking sequence elements. EMBO J. 14:2545-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shapira, S. K., J. Chou, F. V. Richaud, and M. J. Casadaban. 1983. New versatile plasmid vectors of hybrid proteins code by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of β-galactosidase. Gene 25:71-82. [DOI] [PubMed] [Google Scholar]
- 36.Singh, J., M. Mukerji, and S. Mahadevan. 1995. Transcriptional activation of the Escherichia coli bgl operon: negative regulation by DNA structural elements near the promoter. Mol. Microbiol. 17:1085-1092. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki, T., C. Ueguchi, and T. Mizuno. 1996. H-NS regulates OmpF expression through micF antisense RNA in Escherichia coli. J. Bacteriol. 178:3650-3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toguchi, A., M. Siano, M. Burkart, and R. M. Harshey. 2000. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J. Bacteriol. 182:6308-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valle, F. 1994. PCR-based method to map the bending locus of DNA molecules. PCR Methods Appl. 4:44-45. [DOI] [PubMed] [Google Scholar]