Abstract
Mycothiol (MSH) is the major low-molecular-mass thiol in mycobacteria and is associated with the protection of Mycobacterium tuberculosis from toxic oxidants and antibiotics. The biosynthesis of MSH is a multistep process, with the enzymatic reaction designated MshC being the ligase step in MSH production. A targeted disruption of the native mshC gene in M. tuberculosis Erdman produced no viable clones possessing either a disrupted mshC gene or reduced levels of MSH. However, when a second copy of the mshC gene was incorporated into the chromosome prior to the targeted disruption, multiple clones having the native gene disrupted and the second copy of mshC intact were obtained. These clones produced normal levels of MSH. These results demonstrate that the mshC gene and, more generally, the production of MSH are essential for the growth of M. tuberculosis Erdman under laboratory conditions.
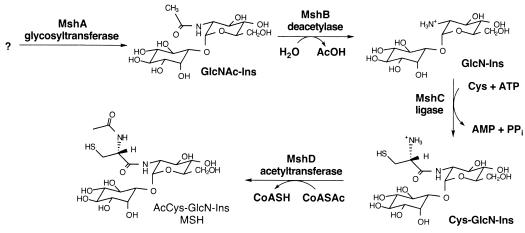
Mycothiol (MSH; AcCys-GlcN-Ins) is a conjugate of N-acetylcysteine (AcCys) with 1-d-myo-inosityl 2-amino-2-deoxy-α-d-glucopyranoside (GlcN-Ins) and is the major low-molecular-mass thiol in most actinomycetes (7). MSH is the functional equivalent of glutathione in mycobacteria (8) and is associated with the protection of Mycobacterium tuberculosis from toxic oxidants and antibiotics (4). The biosynthesis of MSH is a multistep process involving four enzymatic reactions designated MshA, MshB, MshC, and MshD (Fig. 1). An M. tuberculosis mshB mutant demonstrated a heightened sensitivity to the toxic oxidant cumene hydroperoxide and to the antibiotic rifampin even though the mshB mutant produced approximately 20% of the wild-type levels of MSH (4). A compensating deacetylase activity was apparently sufficient for the production of moderate levels of MSH in the mshB mutant. In Mycobacterium smegmatis, mutants have been isolated with mutations in either the mshA gene (9, 10) or the mshC gene (13) that are devoid of detectable levels of MSH. The MshC activity in M. tuberculosis was recently identified as being encoded by open reading frame Rv2130c (14). The mshC gene or Rv2130c had originally been annotated as a cysteinyl-tRNA synthetase gene called cysS2 (5). In order to examine the effect upon M. tuberculosis of very low levels of MSH, the construction of a targeted gene disruption in the mshC gene of M. tuberculosis was attempted.
FIG. 1.
Key enzymes in the biosynthesis of MSH in M. tuberculosis include the glycosyltransferase (MshA, encoded by Rv0486), the GlcNAc-Ins deacetylase (MshB, encoded by Rv1170), the ATP-dependent Cys:GlcN-Ins ligase (MshC, encoded by Rv2130c), and the acetyl-CoA (CoASAc):Cys-GlcN-Ins acetyltransferase (MSH synthase; MshD, encoded by Rv0819).
The production of targeted gene disruptions within the chromosome of M. tuberculosis Erdman was carried out via allelic exchange by using the conditionally replicating mycobacteriophage phAE87 (generous gift of J. S. Cox) (2). phAE87 is a temperature-sensitive shuttle phasmid that replicates at 30°C but not at 37°C. The method for specialized transduction to generate targeted gene disruptions in M. tuberculosis was recently described by Bardarov et al. (1). For construction of the mshC knockout phage, an ∼500-bp fragment comprising 102 bp of the N-terminus of the mshC gene plus the adjacent 370 flanking bases was amplified by PCR. A second fragment containing residues 195 to 708 of the mshC gene was also amplified by PCR. A 93-bp section within the mshC gene that encodes the active HLGH region of the MshC protein was not included within the amplified fragments (14). Each PCR fragment included suitable endonuclease sites for directional cloning into pJSC284 (gift of J. S. Cox). pJSC284 is a cosmid containing a PacI site and a res-hyg-res cassette (2) flanked by multiple cloning sites. The mshC fragments were sequentially cloned on either side of the hygromycin resistance cassette in pJSC284 to produce a mutated copy of mshC interrupted by the Hygr gene. This mshC knockout cosmid was digested with PacI and ligated into the PacI site of the specialized transducing phage phAE87 to generate a phasmid capable of replicating as a cosmid in Escherichia coli and as a temperature-sensitive phage in mycobacteria. The ligated DNA was packaged into phage λ with Gigapack III Gold packaging extract (Stratagene), and E. coli HB101 cells grown on maltose were infected with the phage. Colonies were selected for growth on LB plates containing 200 μg of hygromycin/ml. Cosmid DNA was extracted from two of the Hygr colonies, and the DNA was electroporated into M. smegmatis mc2155. The transformation plates were incubated at 30°C until plaques appeared (2 to 3 days later). Several plaques were picked, and a high-titer phage stock (1010 to 1011 PFU/ml) was prepared with M. smegmatis mc2155.
For infection of M. tuberculosis Erdman cells, 10 ml of mid-log to late log phase bacteria was washed with MP buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 2 mM CaCl2, 10 mM MgCl2) and resuspended in 1 ml of MP buffer at 39°C. Phage were added at a multiplicity of infection of 10, and the mixture was incubated at 39°C for 4 h to allow for phage infection. The bacteria were then pelleted by centrifugation, resuspended in 500 μl of MP buffer, and plated on Middlebrook 7H11 plates supplemented with OADC (oleic acid, albumin, dextrose, catalase; BBL) and containing hygromycin (50 μg/ml). Hygromycin-resistant colonies appeared in 3 to 5 weeks. Individual colonies were cultured for analysis by Southern hybridization to identify clones in which allelic exchange had occurred within the mshC gene and to measure MSH content (4).
In our first experiment, few hygromycin-resistant clones grew after transduction of the wild-type M. tuberculosis Erdman cells with the mshC allelic exchange phage. Of the nine clones recovered, none had undergone allelic exchange within the mshC gene, and all had normal levels of MSH (data not shown). In a parallel experiment with a similar allelic exchange phage for the corA gene (which encodes a putative magnesium transport protein), multiple corA mutants were identified. The failure to identify mshC mutants suggested that either the region encompassing the mshC gene in M. tuberculosis is refractive to homologous recombination or that a functional mshC gene is essential to the growth of M. tuberculosis.
In order to determine whether the mshC gene is required for the viability of M. tuberculosis, a second copy of the mshC gene was introduced into the chromosome of M. tuberculosis, generating strain 2X-mshC, prior to transduction with the mshC allelic exchange phage. This strategy had been employed by Parish and Stoker (11) to establish the essential nature of the glnE gene. If allelic exchange within the mshC gene could be demonstrated in the 2X-mshC strain but not in the wild-type M. tuberculosis, this result would strongly suggest that a functional mshC gene is essential for growth of M. tuberculosis.
A second copy of the M. tuberculosis mshC gene and its ribosomal binding site were incorporated into the genome by using integrative vector pCV125. pCV125 had been previously modified to include the streptomycin and spectinomycin resistance genes (3). The mshC open reading frame plus its ribosomal binding site (71 bp upstream of the ATG start codon) were amplified by PCR with genomic M. tuberculosis Erdman DNA. For directional cloning, a forward primer, 5′-TCCCCCGGGACGCGTGGCGCTGAT-3′, containing an SmaI restriction site and a reverse primer, 5′-GGACTAGTCTACAGGTCCACCCCGAGCAG-3′, containing an SpeI restriction site were used. The PCR fragment was cloned into pCR 2.1 (Invitrogen), and the fragment's sequence was confirmed by restriction analysis and sequencing. The SmaI/SpeI fragment containing the mshC gene was then cloned between the NruI and SpeI sites within the aph gene in pCV125. This process resulted in a vector (pCV125::mshC) containing a copy of the mshC gene that is transcribed from the aph promoter. pCV125::mshC DNA was introduced into wild-type M. tuberculosis Erdman by electroporation with selection on Middlebrook 7H11 plates containing streptomycin (30 μg/ml). Streptomycin-resistant colonies were grown, chromosomal DNA was extracted, and the presence of two copies of the mshC gene was confirmed by Southern hybridization. One of the 2X-mshC clones was named Mtb1682 and was used in further experiments.
To assess the effectiveness of pCV125::mshC in providing a functioning MshC protein during MSH biosynthesis, pCV125::mshC was introduced into an MshC chemical mutant of M. smegmatis (strain I64) which produces about 1% of the parental level of MSH (13). Incorporation of the M. tuberculosis mshC gene resulted in the production of 150% of the parental level of MSH in this MSH mutant (results not shown).
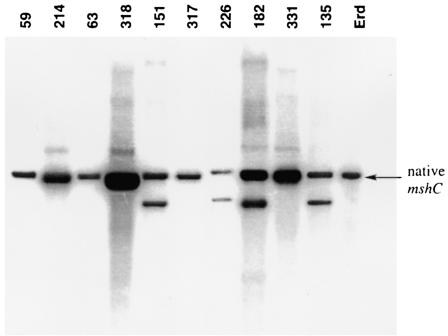
In three separate experiments, the mshC allelic exchange phage was used to infect either wild-type M. tuberculosis Erdman or the 2X-mshC strain Mtb1682. As a positive control for the transduction procedures, wild-type M. tuberculosis Erdman was infected with the corA allelic exchange phage construct. Several hundred hygromycin-resistant clones resulted from these transductions, and each clone was transferred into Middlebrook 7H9 broth supplemented with OADC and containing hygromycin (50 μg/ml) and, for the 2X-mshC strain transductants, also streptomycin (30 μg/ml). Approximately one-third of the hygromycin-resistant clones grew sufficiently for further analysis. These clones were tested for homologous recombination within the mshC gene (by using Southern hybridization after digestion with NcoI- or SacI-digested chromosomal DNA and, as a probe, the PCR fragment from residues 195 to 708 within the mshC gene) and for MSH production. With the wild-type M. tuberculosis Erdman strain used as the transduction recipient, 0 out of 67 hygromycin-resistant clones had undergone homologous recombination within the mshC gene according to Southern analysis (Table 1). The NcoI fragment containing the entire mshC gene is predicted to be 3.0 kb, as found for the native strain (Fig. 2). If homologous recombination had occurred, the predicted NcoI fragmentation pattern would lack the 3.0-kb fragment and include a 2.4-kb fragment generated from NcoI sites at the ends of the allelic exchange substrate. The Southern blots (Fig. 2) revealed that in some of the clones, the mshC knockout substrate had been incorporated in a nonhomologous region of the chromosome, while the native copy of the mshC gene remained unchanged (Fig. 2, clones 151, 226,182, and 135). Other clones contained only a single copy of the mshC gene that was identical to wild-type M. tuberculosis (Fig. 2, clones 59, 214, 63, 318, 317, and 331). Thus, by Southern analysis and by MSH analysis (results not shown), no mshC mutants were produced when only a single copy of the mshC gene was present. In the control for the transduction procedures with the corA gene, Southern hybridization determined that 33% of the 18 tested clones had a disruption in the corA gene (data not shown). While this result demonstrates that the general methods were valid, it does not allow prediction of the success rate for disruption of the mshC gene, which may undergo homologous recombination at a substantially different frequency.
TABLE 1.
Summary of targeted gene disruptions in M. tuberculosis Erdman
| Gene target | Second copy of gene present before mutagenesis | No. of clones screened | No. of clones with disruption in targeted gene | Clones (%) with disruption in targeted gene |
|---|---|---|---|---|
| mshC | No | 67 | 0 | 0 |
| mshC | Yes | 12 | 6 | 50 |
| corA | No | 18 | 6 | 33 |
FIG. 2.
Southern blot of chromosomal DNA digested with NcoI and probed with a 500-bp PCR fragment containing residues 195 to 708 from within the mshC gene. Erd, M. tuberculosis Erdman. Clones 59, 214, 63, 318, 317, and 331 have a single copy of mshC identical to that of the wild-type strain. Clones 151, 226, 182, and 135 have the native mshC intact and a component of the knockout substrate incorporated by nonhomologous recombination.
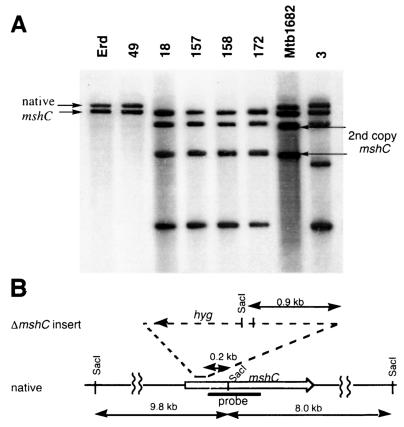
In contrast to the results with wild-type M. tuberculosis Erdman, multiple clones containing a disruption in the mshC gene were identified when the 2X-mshC strain Mtb1682 was used. Although fewer clones were cultured successfully from these transductions, 6 of 12 clones, or 50%, contained a mutated mshC gene (Table 1). The disruption of the mshC gene was most easily observed in Southern blotting with SacI-digested chromosomal DNA and, as the probe, the PCR fragment from residues 195 to 708 within the mshC gene (Fig. 3). SacI cuts within the mshC gene, producing two hybridizing pieces from wild-type DNA and four hybridizing fragments from the 2X-mshC strain Mtb1682. The predicted SacI fragments from the M. tuberculosis H37Rv genome are 9.8 and 8.0 kb (Fig. 3B), as observed in M. tuberculosis Erdman (Fig. 3A). The predicted SacI fragmentation pattern for the mshC knockout includes a 1.1-kb fragment generated from the SacI site within the hygromycin resistance gene which replaces the 9.8-kb fragment (Fig. 3B). The smaller 8.0-kb SacI fragment is unaffected by the knockout insertion and remains unchanged (Fig. 3AB). In clones 18, 157, 158, and 172, the larger fragment from the native copy of mshC is absent and has been replaced by a smaller mshC hybridizing fragment (Fig. 3A). In these clones, the two fragments from the second copy of mshC remained unchanged. In one instance, the second copy of the mshC gene was disrupted, while the native copy of mshC remained unchanged (Fig. 3A, clone 3). Clone 49 contains only a nondisrupted copy of mshC and resulted from a transduction with wild-type M. tuberculosis; it presumably represents an example of spontaneous hygromycin resistance.
FIG. 3.
(A) Southern blot of chromosomal DNA digested with SacI and probed with a 500-bp fragment containing residues 195 to 708 from within the mshC gene. Erd, M. tuberculosis Erdman. Clone 49 is derived from wild-type M. tuberculosis Erdman, and clones 3, 18, 157, 158, and 172 are derived from Mtb1682 (2X-mshC). (B) Diagram showing location of SacI sites within the M. tuberculosis H37Rv genome near mshC and within the insert of the mshC knockout.
The levels of MSH and two precursor molecules in the MSH biosynthetic pathway were analyzed by high-pressure liquid chromatography (4). The MSH contents of all the clones containing a disruption in one of their mshC genes were very similar to the value in wild-type M. tuberculosis Erdman, 13.7 nmol/109 cells (Table 2). This level is in contrast to the reduced level of MSH of 2.9 nmol/109 cells previously observed in the mshB mutant (4). The levels of two precursor molecules, GlcNAc-Ins and GlcN-Ins, exhibited a wider variation but were generally of the same magnitude as found for the wild-type strain. Interestingly, the MSH level of Mtb1682 (2X-mshC) was nearly twice the level of the wild type, and the level of the immediate substrate of MshC, GlcN-Ins, was reduced by 64% from the wild-type level. This result may reflect the presence of two copies of mshC in Mtb1682 and is consistent with ligase substrate depletion due to a higher level of cellular MshC activity.
TABLE 2.
Levels of MSH and precursors in M. tuberculosis mshC disruption clones
| Strain | Optical density (600 nm) | Cellular content (nmol/109 cells)a
|
||
|---|---|---|---|---|
| GlcNAc-Ins | GlcN-Ins | MSH | ||
| Wild-type M. tuberculosis Erdman | 0.50 | 1.7 ± 0.7 | 8.9 ± 0.2 | 13.7 ± 0.2 |
| Mtb1682 (2X-mshC) | 0.85 | 0.9 ± 0.1 | 3.2 ± 0.1 | 26.2 ± 1.2 |
| Mtb1682 transformant | ||||
| Clone 3 | 0.41 | ≤0.71 | 7.3 ± 0.1 | 12.1 ± 1.3 |
| Clone 14 | 0.36 | ≤1.4 | 14.1 ± 0.2 | 12 ± 0.3 |
| Clone 16 | 0.37 | ≤0.6 | 5.7 ± 0.3 | 14.6 ± 0.1 |
| Clone 18 | 0.43 | ≤1.2 | 12 ± 1 | 10.2 ± 0.1 |
| Clone 157 | 0.58 | 1.5 ± 0.7 | 12 ± 1.4 | 10.2 ± 0.1 |
| Clone 158 | 0.52 | 1.7 ± 0.4 | 15 ± 1 | 11.8 ± 0.1 |
| Clone 172 | 0.49 | 1.0 ± 0.5 | 8.5 ± 0.2 | 11.3 ± 0.2 |
Results are expressed as the means and ranges of duplicate samples.
The only plausible explanation for the failure to detect any mshC gene knockouts among 67 clones from the transduction of native M. tuberculosis is that the mshC gene is essential. The rates of production of spontaneous hygromycin-resistant mutants and of illegitimate recombination mutants in the native strain and in the 2X-mshC strain are expected to be the same. For the 2X-mshC strain, the results show that the ratio of native mshC gene knockouts to spontaneous resistance plus illegitimate recombination mutants is 6:5, with one additional mutant (Fig. 3, clone 3) representing a disruption of the second copy of mshC. We would therefore expect that about half (6/11) of the clones examined from the transduction of native M. tuberculosis would have the mshC gene disrupted. The probability of finding 0 such mutants among the 67 clones tested is ∼0.567, or about 1 in 1020. Thus, we are forced to conclude that no mshC knockouts were found because such knockouts were unable to produce MSH and were thus prevented from replicating sufficiently to produce detectible colonies.
In studies with M. smegmatis, mutants in the mshA, mshC, or mshD genes produce ≤1% of wild-type levels of MSH during exponential growth (6, 9, 10, 13). In the late stationary phase, the Tn5::mshD mutant in M. smegmatis produces low levels of MSH and thus may have a compensatory acetyltransferase activity for MshD (T. Koledin, G. L. Newton, and R. C. Fahey, unpublished results). Thus, we consider mshA and mshC to be the primary genes for testing the essential nature of MSH in M. tuberculosis. The successful identification of a disruption in the mshC gene only when a second copy of mshC was present demonstrates that mshC and, by inference, MSH are essential for the growth of M. tuberculosis.
The absolute requirement for MSH by M. tuberculosis contrasts with observations from experiments with M. smegmatis, where we have isolated mutants that lack any detectable MSH. Although the MSH null mutants of M. smegmatis often grow poorly, these combined observations indicate that M. tuberculosis relies to a greater extent than M. smegmatis upon the detoxification activity of MSH for growth during normal metabolism. One key difference between these organisms is their growth rate, the doubling time for M. tuberculosis being ca. sixfold longer than that for M. smegmatis. It is conceivable that the rate of lethal oxidative damage is such that most M. smegmatis cells can replicate without incurring lethal damage sufficient to prevent normal growth but that for M. tuberculosis the ca. sixfold increase in damage per cell division is sufficient to prevent growth. Alternatively, M. smegmatis may have additional protective mechanisms, absent in M. tuberculosis, which supplement the role played by MSH. When the fully annotated genome for M. smegmatis becomes available, it may be possible to identify differences that could be responsible for this divergent dependence upon MSH.
M. smegmatis mutants devoid of MSH are much more sensitive to peroxide than the parent strain (10, 13), and we would therefore expect that MSH is of critical importance in protecting M. tuberculosis from oxidative compounds produced by mammalian host cells during infections. Current studies indicate that dormant (nonreplicating persistent) M. tuberculosis cells are metabolically active (16) and, therefore, must maintain a reducing intracellular redox environment. Since MSH and its disulfide reductase form the thiol redox buffer in mycobacteria (7, 12), we postulate that MSH biosynthesis drug targets may be particularly relevant to the treatment of dormant tuberculosis. The present experiments validate the MSH biosynthetic pathway as a source of potential drug targets in the treatment of tuberculosis and suggest that MshC is an especially attractive candidate.
Recently Sassetti et al. (15) reported an assessment of essential genes in M. tuberculosis by using transposon site hybridization. Although subject to some uncertainty, this method did identify Rv2130c (mshC/cysS2) as an essential gene, and the present results confirm that finding.
Acknowledgments
This research was funded by the National Institutes of Health (grant AI49174). R.C.F. was a consultant for eXegenics Inc. during the initial phase of these studies.
We thank J. S. Cox for the transducing mycobacteriophage strains, Teresa Koledin for technical assistance, and Yossef Av-Gay for helpful discussions.
REFERENCES
- 1.Bardarov, S., S. Bardarov, Jr., M. S. Pavelka, Jr., V. Sambandamurthy, M. Larsen, J. Tufariello, J. Chan, G. Hatfull, and W. R. Jacobs, Jr. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007-3017. [DOI] [PubMed] [Google Scholar]
- 2.Bardarov, S., J. Kriakov, C. Carriere, S. Yu, C. Vaamonde, R. A. McAdam, B. R. Bloom, G. F. Hatfull, and W. R. Jacobs, Jr. 1997. Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10961-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchmeier, N., A. Blanc-Potard, S. Ehrt, D. Piddington, L. Riley, and E. A. Groisman. 2000. A parallel intraphagosomal survival strategy shared by Mycobacterium tuberculosis and Salmonella enterica. Mol. Microbiol. 35:1375-1382. [DOI] [PubMed] [Google Scholar]
- 4.Buchmeier, N. A., G. L. Newton, T. Koledin, and R. C. Fahey. 2003. Association of mycothiol with protection of Mycobacterium tuberculosis from toxic oxidants and antibiotics. Mol. Microbiol. 47:1723-1732. [DOI] [PubMed] [Google Scholar]
- 5.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 6.Koledin, T., G. L. Newton, and R. C. Fahey. 2002. Identification of the mycothiol synthase gene (mshD) encoding the acetyltransferase producing mycothiol in actinomycetes. Arch. Microbiol. 178:331-337. [DOI] [PubMed] [Google Scholar]
- 7.Newton, G. L., K. Arnold, M. S. Price, C. Sherrill, S. B. delCardayré, Y. Aharonowitz, G. Cohen, J. Davies, R. C. Fahey, and C. Davis. 1996. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 178:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newton, G. L., and R. C. Fahey. 2002. Mycothiol biochemistry. Arch. Microbiol. 178:388-394. [DOI] [PubMed] [Google Scholar]
- 9.Newton, Gerald L., Teresa Koledin, Batia Gorovitz, Mamta Rawat, Robert C. Fahey, and Yossef Av-Gay. 2003. The glycosyltransferase gene encoding the enzyme catalyzing the first step of mycothiol biosynthesis (mshA). J. Bacteriol. 185:3476-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newton, Gerald L., Mia D. Unson, Sara J. Anderberg, Joseph A. Aguilera, Nancy N. Oh, Stephen B. delCardayre, Yossef Av-Gay, and Robert C. Fahey. 1999. Characterization of a Mycobacterium smegmatis mutant defective in 1-d-myo-inosityl-2-amino-2-deoxy-α-d-glucopyranoside and mycothiol biosynthesis. Biochem. Biophys. Res. Commun. 255:239-244. [DOI] [PubMed] [Google Scholar]
- 11.Parish, Tanya, and Neil G. Stoker. 2000. glnE is an essential gene in Mycobacterium tuberculosis. J. Bacteriol. 182:5715-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel, M. P., and J. S. Blanchard. 1999. Expression, purification, and characterization of Mycobacterium tuberculosis mycothione reductase. Biochemistry 38:11827-11833. [DOI] [PubMed] [Google Scholar]
- 13.Rawat, Mamta, Gerald L. Newton, Mary Ko, Gladys J. Martinez, Robert C. Fahey, and Yossef Av-Gay. 2002. Mycothiol-deficient Mycobacterium smegmatis mutants are hypersensitive to alkylating agents, free radicals, and antibiotics. Antimicrob. Agents Chemother. 46:3348-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sareen, D., M. Steffek, G. L. Newton, and R. C. Fahey. 2002. ATP-dependent L-cysteine:1d-myo-inosityl 2-amino-2-deoxy-α-d-glucopyranoside ligase, mycothiol biosynthesis enzyme MshC, is related to class I cysteinyl-tRNA synthetases. Biochemistry 41:6885-6890. [DOI] [PubMed] [Google Scholar]
- 15.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 16.Wayne, L. G., and C. D. Sohaskey. 2001. Nonreplicating persistence of Mycobacterium tuberculosis. Annu. Rev. Microbiol. 55:139-163. [DOI] [PubMed] [Google Scholar]