Abstract
CTP synthase is encoded by the pyrG gene and catalyzes the conversion of UTP to CTP. A Lactococcus lactis pyrG mutant with a cytidine requirement was constructed, in which β-galactosidase activity in a pyrG-lacLM transcriptional fusion was used to monitor gene expression of pyrG. A 10-fold decrease in the CTP pool induced by cytidine limitation was found to immediately increase expression of the L. lactis pyrG gene. The final level of expression of pyrG is 37-fold higher than the uninduced level. CTP limitation has pronounced effects on central cellular metabolism, and both RNA and protein syntheses are inhibited. Expression of pyrG responds only to the cellular level of CTP, since expression of pyrG has no correlation to alterations in UTP, GTP, and ATP pool sizes. In the untranslated pyrG leader sequence a potential terminator structure can be identified, and this structure is required for regulation of the pyrG gene. It is possible to fold the pyrG leader in an alternative structure that would prevent the formation of the terminator. We suggest a model for pyrG regulation in L. lactis, and probably in other gram-positive bacteria as well, in which pyrG expression is directly dependent on the CTP concentration through an attenuator mechanism. At normal CTP concentrations a terminator is preferentially formed in the pyrG leader, thereby reducing expression of CTP synthase. At low CTP concentrations the RNA polymerase pauses at a stretch of C residues in the pyrG leader, thereby allowing an antiterminator to form and transcription to proceed. This model therefore does not include any trans-acting protein for sensing the CTP concentration as previously proposed for Bacillus subtilis.
All living organisms need purine and pyrimidine nucleotides as building blocks for synthesis of DNA, RNA, and several coenzymes. The de novo synthesis of the pyrimidine ribonucleotide CTP occurs in Lactococcus lactis, as in all other organisms investigated so far, through a linear pathway starting with synthesis of carbamoyl phosphate and ending with an amination of UTP to form CTP (Fig. 1). The pyrG gene is not linked to any other gene involved in pyrimidine metabolism (35). It encodes the CTP synthase, the terminal enzyme in this pathway, which is an amidotransferase that converts UTP, glutamine, and ATP to CTP, glutamate, and ADP. A mutation in the pyrG gene makes an otherwise pyrimidine prototrophic organism dependent on a supply of cytidine from the medium. The availability of pyrimidines controls expression of pyrimidine biosynthetic genes in gram-positive bacteria such as Bacillus subtilis, Enterococcus faecalis, and L. lactis, since growth in the presence of a pyrimidine source reduces expression of the pyr genes (9, 22, 29). Regulation of the pyrimidine biosynthetic genes has been investigated in great detail in B. subtilis. It has been shown that PyrR, an RNA binding protein that senses the concentration of UMP in the cell, regulates pyr gene expression through an attenuator mechanism (33). When the UMP pool is high, PyrR and UMP form a complex that binds to a specific site in the mRNA leader of the pyr genes (17, 18). The leader sequences can form both terminator and antiterminator structures, and binding of the PyrR-UMP complex destabilizes the antiterminator, thus resulting in termination of pyr transcription (34). In the presence of low UMP concentrations, PyrR does not bind the mRNA, giving transcriptional readthrough. PyrR is also found in a number of other gram-positive organisms, including L. lactis (22).
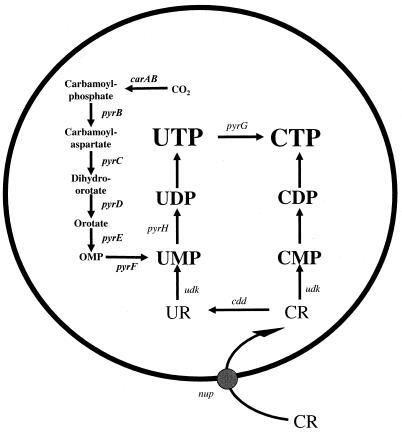
FIG. 1.
Pathways for the synthesis of CTP and the salvage of cytidine in L. lactis. UR, uridine; CR, cytidine; carAB, carbamoylphosphate synthase gene; pyrB, aspartate transcarbamylase gene; pyrC, dihydroorotase gene; pyrD, dihydroorotate dehydrogenase gene; pyrE, orotate phosphoribosyltransferase gene; pyrF, OMP decarboxylase gene; pyrH, UMP kinase gene; pyrG, CTP synthase gene; udk, uridine kinase gene; cdd, cytidine deaminase gene; nup, cytidine/purine nucleoside transporter gene.
Little is known about the regulation of pyrG in gram-negative bacteria. For Salmonella enterica serovar Typhimurium, however, it has been shown that CTP synthase activity is decreased 11-fold when cytidine is included in the growth medium (37). Similar observations with cytidine repression have been made for the gram-positive bacterium B. subtilis (2, 25). By monitoring gene expression from a pyrG promoter fragment in different genetic backgrounds, it has been shown that expression of pyrG is increased during limitation of cytidine nucleotides (25). However, no direct determinations of nucleotide pool sizes were performed. A terminator was identified in the pyrG mRNA leader, and deletion of this terminator abolished pyrG regulation, resulting in high expression both during pyrimidine starvation and during growth in the presence of pyrimidines. Further mutational analysis of the pyrG leader has identified two segments that are required for normal regulation of pyrG, and these nucleotides are conserved in the mRNA pyrG leaders from gram-positive bacteria (26). The conserved nucleotides are the first four nucleotides in the transcript (GGGC) and six nucleotides in the terminator stem (GCUCCC). No antiterminator structure was identified in the B. subtilis pyrG leader, which led to the proposal that an as-yet-unidentified protein senses the CTP concentration in the cell and during CTP starvation the protein binds to the pyrG leader, thus preventing termination and allowing transcription of pyrG.
In this study, regulation of the pyrG gene from L. lactis, including a correlation between pyrG expression and the CTP pool, was investigated. Expression from a pyrG::lacLM fusion during CTP limitation was determined, and it was found that a decreased CTP pool size results in increased pyrG expression. To gain insight into the mechanism behind pyrG regulation, the rate of synthesis from the pyrG gene after a sudden drop in the CTP pool was studied, and it was found that pyrG expression is not linear during the first almost 2.5 h of CTP limitation. Profound alterations in macromolecular biosynthesis affecting both RNA and protein synthesis were observed during CTP limitation. A model for the regulation of pyrG is presented, in which a terminator-antiterminator mechanism independent of additional protein factors is responsible for the regulation. A similar mechanism can be proposed for pyrG regulation in a number of gram-positive organisms.
MATERIALS AND METHODS
Growth conditions.
Escherichia coli cells were grown in Luria-Bertani medium at 37°C with vigorous shaking. L. lactis cultures were grown in SA defined medium with 1% glucose (GSA medium) (12), 1% galactose, or 1% maltose at 30°C. Erythromycin was added to a concentration of 2 or 150 μg/ml for L. lactis and E. coli, respectively. Unless noted otherwise, cytidine was added to 20 μg/ml. 5-Bromo-4-chloro-3-indolyl-β-d-galactoside was added to a final concentration of 90 μg/ml.
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this work are described in Table 1. Plasmid pCJ24 carries an internal fragment of pyrG and was constructed by digesting pSH105 (35) with HindIII and BamHI and subsequently transferring a 1.1-kb fragment to the integration vector pSMA500 (19) cleaved with HindIII and BamHI. The ligated plasmid was transformed to competent ABLE K cells (Stratagene, La Jolla, Calif.) and plated on Luria-Bertani plates containing 150 μg of erythromycin per ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli ABLE K | Stratagene | |
| L. lactis | ||
| MG1363 | Wild type | 8 |
| MB109 | MG1363 cdd | 21 |
| MB420 | MG1363 pyrB::ISS1 | 23 |
| CJ217 | MB109 pyrG::pCJ24 | This study |
| CJ233 | MB109/pAK80 | This study |
| CJ238 | MB109/pCJ29 | This study |
| CJ240A | MB109 pyrG::pCJ30 | This study |
| CJ295 | MB109 pyrG::ISS1 | This study |
| CJ298 | CJ295/pCJ29 | This study |
| CJ300 | MB420/pCJ29 | This study |
| Plasmids | ||
| pGh9:ISS1 | Used for ISS1 mutagenesis | 20 |
| pAK80 | Contains promoterless lacLM genes | 11 |
| pSMA500 | Contains promoterless lacLM genes and no origin for replication in L. lactis | 19 |
| pMOSBlue | Plasmid vector | Amersham Biosciences |
| pCR2.1-TOPO | Plasmid vector | Invitrogen |
| pSH105 | Contains 1.4-kb internal fragment of pyrG | 35 |
| pCJ24 | Internal pyrG fragment from pSH105 in pSMA500 | This study |
| pCJ29 | pyrG promoter fragment (−667 to +297) in pAK80 | This study |
| pCJ30 | pyrG promoter fragment (−667 to +297) in pSMA500 | This study |
| pCJ34 | pyrG promoter fragment (−667 to +82) in pAK80 | This study |
| pCJ35 | pyrG promoter fragment (−667 to +69) in pAK80 | This study |
| pCJ36 | pyrG promoter fragment (−667 to +45) in pAK80 | This study |
The promoter region upstream of pyrG, including the 5′ end of pyrG, was amplified by PCR with primers pyrGupF2 (5′-CCCAAGCTTGGAAAATAGACAAAGCCC-3′) and pyrG9b1-Bam (5′-CGGGATCCTCAGTAACAAAGACTTCACC-3′). The PCR product was cloned by using the pMOS Blue blunt-ended cloning kit from Amersham Biosciences. The cloned fragment was transferred as a HindIII-BamHI fragment to the promoter-probe vector pAK80 as well as pSMA500, resulting in pCJ29 and pCJ30, respectively.
Plasmids containing parts of the pyrG leader were cloned in the TOPO TA cloning kit from Invitrogen. The cloned fragments were made by PCR with primers pyrGupF2 and either pyrGdel_1 (5′-TCTCGGATCCAAAGAAATAACTGGGAAA-3′), pyrGdel_2 (5′-CCTCGGATCCAAAAACAAAAACAGCTCCCC-3′), or pyrGdel_3 (5′-TCTCGGATCCGGGGAGCCTACCGTTACTG-3′). HindIII-BamHI fragments were transferred to the vector pAK80, resulting in pCJ34, pCJ35, and pCJ36, respectively.
DNA isolation.
Chromosomal DNA from L. lactis was isolated as previously described (13). Plasmid DNA from E. coli cells was purified by using the Qiagen plasmid midi kit.
Transformation.
E. coli cells were made competent with CaCl2 and transformed as described previously (30). L. lactis cells were transformed by electroporation as described previously (10).
Isolation of pyrG mutants.
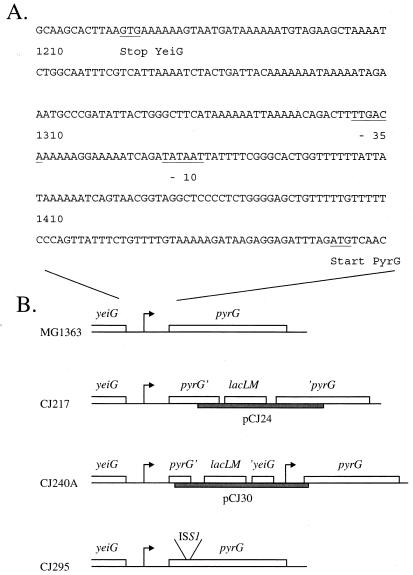
An L. lactis strain with a transposon inserted in the pyrG gene was isolated in a cdd derivative of MG1363 (MB109). The mutant was isolated from a transposon library by using pGh9:ISS1 as previously described (15). The library was isolated on GSA defined medium containing 20 μg of cytidine per ml. From this library, 5 ml of GSA medium containing 20 μg of uracil per ml was inoculated to an optical density at 436 nm (OD436) of 0.1 and grown for 5 h at 37°C. Ampicillin was added to 100 μg/ml, and the culture was incubated overnight at 37°C. Cells were harvested and washed with 0.9% NaCl, and 100 μl from this cell suspension as well as 100 μl from a 10× dilution was plated on GSA plates with 50 μg of cytidine per ml and incubated for 3 days at 37°C. Colonies were screened for growth on plates with and without cytidine or uracil. Fifteen colonies with a specific cytidine requirement were restreaked twice at 28°C to select for excision of the plasmid and twice at 37°C to cure the plasmid from the strain. An isolated strain with a cytidine requirement and an erythromycin-sensitive phenotype was kept as CJ295. The ISS1 element was inserted in the pyrG open reading frame as verified by PCR on chromosomal DNA from CJ295. To isolate mutants with the reporter genes lacLM under control of the pyrG promoter, competent L. lactis cells carrying a cdd mutation (MB109) were transformed with plasmid pCJ24 or pCJ30, resulting in CJ217 and CJ240A, respectively. After transformation, erythromycin-resistant colonies were isolated and purified on GSA plates with erythromycin and cytidine. The presence of the integrated plasmid in the pyrG region was verified by the cytidine requirement of strain CJ217 as well as by PCR analysis of chromosomal DNA from CJ217 and CJ240A. The genetic organizations of the pyrG region in CJ217, CJ240A, and CJ295 are shown in Fig. 2.
FIG. 2.
(A) Nucleotide sequence of the noncoding region in front of pyrG. The numbers refer to the sequence found in the EMBL data library (accession number AJ010153). The −10 and −35 sequences of the putative promoter are shown. (B) Genetic maps of the pyrG regions in the strains used in this study. Grey boxes indicate plasmids with the reporter genes lacLM integrated in the chromosome. Arrows indicate the position of the putative pyrG promoter. The maps are not drawn to scale.
Growth experiments and enzyme assay.
Starting with fresh colonies from GSA plates with erythromycin and cytidine, the isolated pyrG mutants were grown for 6 to 8 h at 30°C in 1 ml of GSA medium with erythromycin and cytidine. Different dilutions of the growing culture were added to 10 ml of medium and grown overnight. From an exponentially growing culture, new medium was inoculated to an OD436 of 0.05. The growth experiments were performed in glass or plastic flasks without stirring. For β-galactosidase activity determinations, 35 ml of culture was harvested, washed with 0.9% NaCl, and resuspended in 1 ml of Z buffer.
β-Galactosidase activity was determined at 30°C as previously described (30) except that the cell density was measured at 436 nm and the specific activity was determined as OD420/(OD436 per minute per milliliter of culture).
RNA extraction.
L. lactis RNA was harvested from strain MG1363 grown exponentially in SA glucose medium to an OD450 of approximately 0.8. Total RNA from 200 ml of culture was isolated by using the Fast Prep system (BIO101) with the protocols of the manufacturer.
RT-PCR.
L. lactis RNA was used as the template in the Titan one-tube reverse transcription-PCR (RT-PCR) system from Boehringer Mannheim (product number 1 888 382) in accordance with the protocols of the manufacturer. As a control, conventional PCR was conducted on both chromosomal DNA and the RNA. The following primer pairs were used: A, SLLH17 (5′-GGCAAATTGATATTGCACTTG-3′) and SLLH18 (5′-AAAAAGAATGTTGTCTACGGCTTGG-3′); B, SLLH19 (5′-CAACTAAGTATATTTTCGTCACTGG-3′) and pyrG9b (5′-TCAGTAACAAAAACTTCCCC-3′); C, SLLH17 and pyrGb9; and D, pyrG8a (5′-GGCAAAAAATTCTTCGTT-3′) and SLLH7 (5′-TACAAAAGATTTTGGGC-3′).
Determination of intracellular nucleoside triphosphate concentrations.
Nucleotides were extracted from [33P]orthophosphate-labeled cell cultures and separated by thin-layer chromatography as previously described (23).
Determination of RNA and protein synthesis.
For pulse-labeling of RNA, 600 μl of culture was mixed with 1 μl of [14C]adenine (50 μCi/ml) and 3 μl of 10 mM adenine to a final adenine concentration of 55 μM. Pulse-labeling of proteins was done by adding 2.6 μl of [14C]leucine (50 μCi/ml) to 220 μl of culture (the concentration of leucine in SA medium is 0.8 mM [12]). After 10 min of labeling with either [14C]adenine or [14C]leucine, 200 μl of culture was transferred to a tube with 3 ml of cold 5% trichloroacetic acid (TCA) and put on ice for 0.5 to 1.5 h. The precipitated macromolecules were collected on a membrane filter (0.45-μm pore size; Schleicher & Schuell, Dassel/Reliehausen, Germany), washed twice with cold 5% TCA and once with boiling water, and left to air dry (5). The radioactivity on the filters was counted in an Instant Imager.
mRNA half-life determinations.
Determination of mRNA half-life was based on a method described previously (36). A 1.65-ml portion of culture was mixed with 11 μl of [14C]adenine (50 μCi/ml) and 8 μl of 10 mM adenine and pulse-labeled for 4 min. At time zero, actinomycin D was added to a final concentration of 4 μg/ml and nalidixic acid was added to a final concentration of 20 μg/ml in order to stop transcription. At different time intervals between 0 and 10 min and at 60 min, 200 μl of labeled culture was mixed with 3 ml of cold 5% TCA and left on ice for 30 to 60 min. Precipitated RNA was collected on a membrane and treated as described above. The mean of the values obtained at 8, 10, and 60 min after inhibition of transcription was taken to represent stable RNA, a value which was subtracted from the values obtained at 0, 1, 2, 4, and 6 min.
RESULTS
Identification of the pyrG transcript.
The region upstream of pyrG was sequenced and found to contain a potential promoter sequence with consensus −10 and −35 regions separated by 17 nucleotides. A 950-bp DNA fragment covering bp −667 to +297 with respect to +1, thus including the promoter region and the 5′ end of the pyrG open reading frame, was cloned in the pAK80 promoter-probe vector (pCJ29). The cloned fragment has promoter activity, as it results in expression of the lacLM reporter genes (compare CJ238 with CJ233 in Table 2).
TABLE 2.
Regulation of pyrG::lacLM fusion in L. lactis
| Strain | Genotype | Plasmid | Addition(s) to mediuma (μg/ml) | Td (min)b | β-Galactosidase activity (U/OD436 unit)c | Relative expression |
|---|---|---|---|---|---|---|
| CJ217 | cdd pyrG::lacLM | None | CR (20) | 56 ± 3 | 3.7 ± 1.4 | 1 |
| CR (20), UR (500) | 109 ± 7 | 55 ± 8 | 15 | |||
| CR (20), AR (100) | 110 ± 14 | 24 ± 5 | 6 | |||
| CR (20), CdR (500) | 151 ± 10 | 57 ± 1 | 15 | |||
| CJ298 | cdd pyrG::ISS1 | pCJ29 | CR (20) | 59 ± 4 | 71 ± 19 | 1 |
| CR (20), UR (500) | 119 ± 8 | 1,275 ± 98 | 18 | |||
| CR (20), AR (100) | 114 ± 10 | 380 ± 93 | 5 | |||
| CR (20), CdR (500) | 166 ± 8 | 1,150 ± 44 | 16 | |||
| CJ238 | cdd | pCJ29 | None | 60 ± 2 | 76 ± 8 | 1 |
| CR (20) | 58 ± 1 | 47 ± 6 | 0.62 | |||
| CR (50) | 59 ± 2 | 46 ± 5 | 0.60 | |||
| CR (500) | 59 ± 2 | 48 ± 5 | 0.63 | |||
| UR (50) | 59 ± 3 | 78 ± 9 | 1 | |||
| CJ233 | cdd | pAK80 | None | 57 ± 1 | <0.02 |
Abbreviations: CR, cytidine; UR, uridine; AR, adenosine; CdR, deoxycytidine. The nucleosides were added to exponentially growing cells at an OD436 of 0.2.
Td, doubling time GSA in defined medium. The data are averages and standard deviations from three to five independent experiments.
The cells were harvested at an OD436 of 0.8. The data are averages and standard deviations from three to five independent experiments.
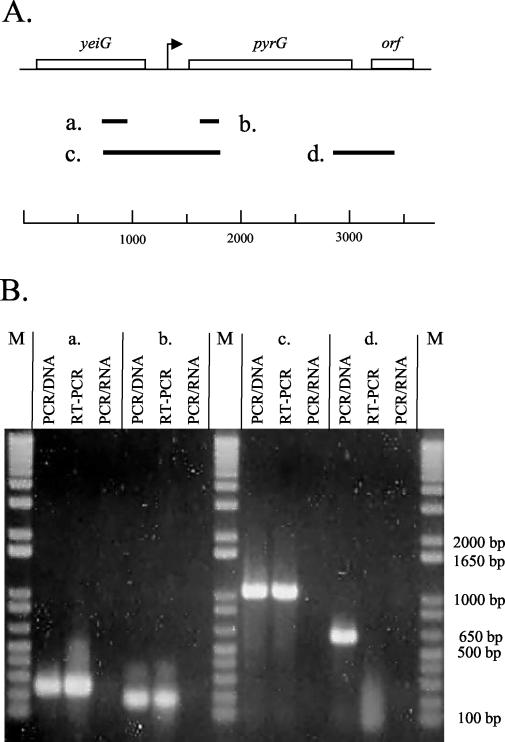
No putative terminator can be identified after the gene upstream of pyrG, and it cannot be ruled out that upstream transcription may contribute to the overall expression of pyrG. To test whether transcription initiated upstream of the pyrG promoter may contribute to the overall expression of pyrG, an RT-PCR experiment was conducted. Total RNA isolated from the wild-type strain L. lactis MG1363 was analyzed by RT-PCR as described in Materials and Methods, and the results are presented in Fig. 3. It can be seen that it is possible to obtain RT-PCR products spanning the intergenic region of pyrG and yeiG (Fig. 3, PCR product c.). It is not possible to obtain an RT-PCR product including both pyrG and the downstream open reading frame (Fig. 3, PCR product d.). This indicates that in vivo upstream-initiated transcription does contribute to the overall expression of pyrG and that all pyrG transcription is terminated immediately after the pyrG open reading frame.
FIG. 3.
RT-PCR analysis of transcription of yeiG, pyrG, and orf in L. lactis. (A) Physical map of the pyrG region from L. lactis. Numbering refers to the DNA sequence submitted to the EMBL data library and assigned accession number AJ010153. The theoretical PCR products are shown. a, PCR fragment obtainable with oligonucleotides SLLH17 and SLLH18; b, PCR fragment obtainable with oligonucleotides SLLH19 and pyrG9b; c, PCR fragment obtainable with oligonucleotides SLLH17 and pyrG9b; d, PCR fragment obtainable with oligonucleotides pyrG8a and SLLH7. (B) Agarose gel electrophoresis of the RT-PCR products. Lanes PCR/DNA, ordinary PCR with DNA as the template; lanes RT-PCR, RT-PCR with RNA as the template; PCR/RNA, PCR with RNA as the template. a, b, c, and d refer to the theoretical PCR fragments shown in panel A. Lanes M, 1-kb Plus DNA ladder from GibcoBRL.
The 950-bp fragment with the pyrG promoter was also transferred to a plasmid that is unable to replicate in L. lactis. Integration of this plasmid in the chromosome resulted in a strain with a functional pyrG open reading frame expressed only from the promoter present on the cloned fragment (CJ240A) (Fig. 2). This strain grows at a normal rate in the absence of exogenous cytidine, showing that the promoter immediately in front of pyrG has adequate strength to ensure sufficient production of CTP.
The expression of pyrG is repressed by cytidine.
To investigate whether pyrG expression is affected by addition of cytidine, strain CJ238, containing the plasmid with the pyrG promoter controlling expression of lacLM, was grown in the presence of cytidine (Table 2). Expression from the reporter genes was reduced by 40% compared to growth in the absence of cytidine. Uridine does not repress pyrG expression. These results suggest that expression of pyrG is regulated at the level of transcription by the availability of cytidine compounds, as proposed for B. subtilis (25).
Limitation of cytidine availability results in increased pyrG expression.
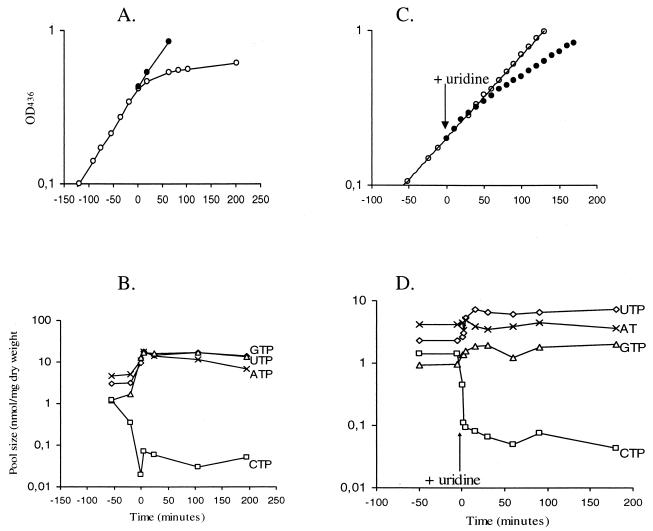
In order to test whether pyrG expression in L. lactis is subject to changes during CTP depletion, a chromosomal fusion which carries the lacLM reporter genes under control of the pyrG promoter was constructed. Strain CJ217 (Fig. 2) was constructed by a chromosomal integration of plasmid pCJ24, which carries an internal fragment of pyrG in front of the reporter genes. This strain contains a truncated pyrG gene and has a cytidine requirement for growth. Moreover, cytidine degradation is prevented, since the strain has no cytidine deaminase activity due to an inactive cdd gene. As shown previously, an L. lactis cdd pyrG mutant grown with cytidine has a growth rate and intracellular nucleotide concentrations similar to those of the wild-type strain (23). The pyrG::lacLM fusion strain was grown with a limited concentration of cytidine in the growth medium, resulting in a decline in growth rate when the supplied cytidine had been utilized (Fig. 4A). Determination of the intracellular concentrations of nucleoside triphosphates revealed that the cells were indeed starved for CTP, since the concentration of CTP was below the level of detection at the onset of the growth arrest (Fig. 4B). The β-galactosidase activity encoded by the lacLM reporter genes was assayed in order to monitor expression of the pyrG gene. No increase in expression was detected, even after 24 h of CTP starvation (specific activity, 1.3 U/OD unit), compared to cells grown without cytidine limitation (specific activity, 3.7 U/OD unit), showing that expression from the pyrG gene is not induced when the cell is completely starved for cytidine.
FIG. 4.
Growth curves (A and C) and nucleotide pool size changes (B and D) during starvation or limitation for cytidine. A cdd pyrG::lacLM mutant was grown in GSA defined medium. (A and B) Cytidine starvation. Cells were grown either with 20 μg of cytidine per ml (•) or with a limited amount of cytidine (○). Time zero indicates the point of reduced growth rate at an OD436 of 0.4. (C and D) Cytidine limitation. Cells were grown with cytidine (○) or with cytidine plus uridine (•). Uridine (500 μg/ml) was added at time zero at an OD436 of 0.2.
In a previous study, the pyrG gene of B. subtilis was shown to be induced in growing cells during pyrimidine limitation (25). Addition of high concentrations of nucleosides to a pyrG mutant grown with cytidine has been identified as a way to reduce the CTP concentration in L. lactis (23). The only nucleosides that do not inhibit growth of a cytidine-requiring strain are thymidine and deoxyuridine. The expression of the pyrG gene during growth with high concentrations of purine or pyrimidine nucleosides and consequently a low CTP pool size was therefore tested. On plates with cytidine and large amounts of (deoxy)pyrimidine or (deoxy)purine nucleosides, growth of the pyrG mutant CJ217 (pyrG-lacLM) is inhibited, as it takes several days for the mutant to show visible growth. Moreover, expression of the pyrG-lacLM fusion was clearly induced as judged by 5-bromo-4-chloro-3-indolyl-β-d-galactoside indicator plates, since the colonies turned blue when growth was inhibited by nucleosides compared to the same strain growing in the presence of only cytidine.
The effect on pyrG expression in a L. lactis pyrG mutant during CTP limitation was also investigated in liquid medium with cytidine. As shown in Table 2, the addition of uridine, deoxycytidine, or adenosine to strain CJ217 resulted in a decrease in growth rate and increased expression from the pyrG-lacLM chromosomal fusion when measured after two generations of growth with CTP limitation. Uridine and deoxycytidine resulted in a 15-fold induction, whereas adenosine increased expression only sixfold. In order to test whether all regulatory signals needed for pyrG regulation are present on the sequence immediately in front of the pyrG open reading frame, a pyrG mutant with an ISS1 element inserted in the gene (CJ295) (Fig. 2) was transformed with the promoter fusion clone pCJ29. The cloned promoter fragment is responsible for the observed regulation of pyrG, since CTP limitation by uridine, adenosine, or deoxycytidine addition resulted in induction ratios of 18-, 5-, and 16-fold, respectively. These ratios are identical to those observed for the chromosomal pyrG::lacLM fusion in CJ217. These results show that all required regulatory elements are present immediately upstream of pyrG and furthermore that the regulatory mechanism is fully functional even when multiple copies of the elements are present in the cell.
Determination of nucleotide pool sizes and pyrG transcriptional rate during CTP limitation.
Expression from the pyrG-lacLM chromosomal fusion during growth inhibition by uridine was analyzed in detail. Ribonucleoside triphosphate pool sizes during growth inhibition by uridine were determined, and the results clearly show that addition of uridine results in an instantaneous lowering of the CTP pool by more than one order of magnitude (Fig. 4C and D). Simultaneously, a threefold increase in UTP concentration and a twofold increase in GTP concentration were observed. No significant change in the ATP pool size was observed.
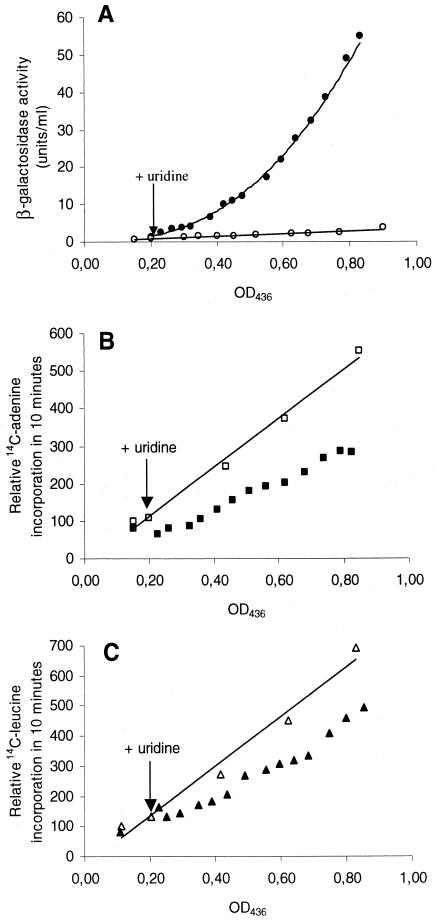
Figure 5A shows the rate of synthesis from the reporter genes lacLM under control of the pyrG promoter during CTP limitation. The slope of the curve for uninduced cells is 3.1, which, as expected, is similar to the specific activity found in Table 2 for CJ217 (3.7 ± 1.4 U/OD unit). After uridine was added and the CTP concentration was lowered, pyrG expression immediately increased ninefold to 28 U/OD unit. Expression from the pyrG-lacLM gene fusion did not, however, attain a new steady-state level but gradually increased until 140 min of CTP limitation, when a final induction of 37-fold was reached. Growth of the pyrG mutant for more than two generations after this point resulted in no further increase in expression; i.e., full induction of the pyrG-lacLM fusion is first observed after more than 1.5 generations of growth during CTP limitation.
FIG. 5.
Synthesis of pyrG, RNA, and protein during CTP limitation. (A) Rate of synthesis from a pyrG::lacLM chromosomal fusion mutant (CJ217) grown in GSA defined medium with cytidine (○) or with cytidine and uridine (•). (B and C) Synthesis of RNA (squares) and protein (triangles), respectively, either during growth with cytidine (open symbols) or during growth with cytidine and uridine (CTP limitation) (closed symbols). Uridine (500 μg/ml) was added at an OD436 of 0.2. The amount of incorporated labeled adenine or leucine in a 10-min pulse-labeling is set to 100 for the first determination for cells grown only with cytidine in the medium and is plotted against the OD value after 5 min of labeling.
The expression of pyrG responds primarily to CTP and not UTP.
Transformation of the promoter fusion clone pCJ29 to a pyrimidine de novo mutant made it possible to determine pyrG expression during UTP limitation. A pyrB mutant grown with cytidine as a pyrimidine source will be subject to cytidine limitation in the presence of inosine, since cytidine and inosine transport is facilitated by the same transport system. As a result, the strain will have a decreased UTP pool and an increased CTP pool (23). As shown in Table 3, a fourfold increase in the CTP pool was followed by a sevenfold decrease in expression of the reporter genes, again suggesting that pyrG expression responds to the CTP concentration in the cell.
TABLE 3.
Effects of varying nucleotide pool sizes on expression from a pyrG::lacLM fusion.
| Strain | Genotype | Plasmid | Carbon sourcea | Cytidine (μg/ml) | Added inhibitorb | Td (min)c | β-Galactosidase activity (U/OD436 unit)d | Ribonucleotide concn (nmol/mg [dry wt])e
|
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| GTP | ATP | CTP | UTP | ||||||||
| CJ217 | cdd pyrG::lacLM | None | Glucose | 20 | None | 56 ± 3 | 3.7 ± 1.4 | 1.41 ± 0.30 | 5.91 ± 0.63 | 1.73 ± 0.48 | 3.56 ± 0.28 |
| Galactose | 20 | 157 ± 11 | 32 ± 2 | 0.64 ± 0.05 | 1.92 ± 0.12 | 0.65 ± 0.10 | 1.14 ± 0.19 | ||||
| Maltose | 20 | 111 ± 17 | 38 ± 6 | 0.74 ± 0.18 | 3.24 ± 0.60 | 0.51 ± 0.14 | 2.21 ± 0.29 | ||||
| CJ238 | cdd | pCJ29 | Glucose | 0 | None | 61 ± 2 | 76 ± 8 | 1.28 ± 0.11 | 8.02 ± 1.02 | 1.52 ± 0.32 | 4.22 ± 0.50 |
| Decoyinine | 181 ± 6 | 126 ± 1 | 0.95 ± 0.21 | 4.59 ± 0.16 | 0.88 ± 0.07 | 2.79 ± 0.43 | |||||
| 50 | None | 60 ± 2 | 46 ± 5 | 1.09 ± 0.10 | 6.50 ± 0.23 | 1.96 ± 0.26 | 3.37 ± 0.22 | ||||
| Decoyinine | 159 ± 7 | 44 ± 4 | 0.74 ± 0.12 | 6.47 ± 0.75 | 3.05 ± 0.17 | 2.86 ± 0.38 | |||||
| CJ300 | pyrB::ISS1 | pCJ29 | Glucose | 20 | None | 59 ± 3 | 96 ± 5 | 1.36 ± 0.46 | 5.78 ± 1.00 | 0.78 ± 0.30 | 1.27 ± 0.83 |
| Inosine | 215 ± 21 | 13 ± 1 | 2.43 ± 0.14 | 6.07 ± 2.47 | 4.91 ± 0.11 | 0.22 ± 0.06 | |||||
All strains were grown in defined medium with the indicated carbon source at 1%.
Decoyinine and inosine were added at an OD436 of 0.2. Decoyinine was added to a concentration of 5 μg/ml, and inosine was added at 100 μg/ml.
Td, doubling time. The data are averages and standard deviations from three to five independent experiments.
The cells were harvested at an OD436 of 0.8. The data are averages and standard deviations from three to five independent experiments.
The data are averages and standard deviations from three to five independent experiments.
Reduced nucleotide pool sizes, including CTP, during growth on galactose and maltose induce pyrG expression.
When grown on galactose or maltose as a carbon source, L. lactis shifts from homolactic fermentation to mixed acid fermentation with a reduction in growth rate. Growth of the pyrG::lacLM fusion strain in defined medium with cytidine and galactose or maltose as a carbon source increased the doubling time to 157 and 111 min, respectively, compared to 56 min when grown on glucose (Table 3). These doubling times are similar to previous observations of growth of MG1363 on defined medium (1, 24). The nucleoside triphosphate pools, and thus the CTP pool, are lower when the strain is grown on these carbon sources than when it is grown on glucose. The reduced pool sizes are followed by an increase in pyrG expression, as the lacLM reporter genes are induced almost 10-fold, suggesting that induction of pyrG expression is triggered by a lowered CTP pool size.
A correlation between CTP concentration and pyrG expression in L. lactis.
Distorted nucleotide pool sizes were also achieved by the use of decoyinine, an inhibitor of GMP synthase and hence of GTP synthesis. Addition of decoyinine to strain CJ238, which harbors plasmid pCJ29, reduced the growth rate threefold and not only reduced the GTP pool but also caused a general reduction in pool sizes (Table 3). These reduced pool sizes were followed by an increased pyrG expression from 76 to 126 U/OD unit. Next, the cells were inhibited with decoyinine in the presence of cytidine. Cells inhibited with decoyinine had a higher CTP pool when grown with cytidine and did not have increased pyrG expression, again suggesting a correlation between the CTP pool size and pyrG expression. The latter experiment also indicates that pyrG expression is not triggered by a reduced growth rate as such.
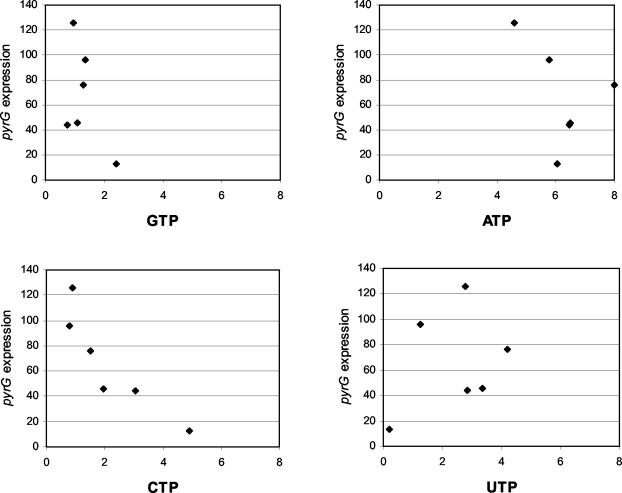
Figure 6 shows a plot of the activity from the pyrG promoter and the corresponding concentrations of the four nucleotides from the experiments with decoyinine and the pyrB mutant from Table 3. There is a clear correlation between the CTP pool size and expression of pyrG. The pyrG expression is elevated at low CTP concentrations and decreased at high CTP concentrations. No correlation between the ATP, GTP, and UTP pools and the expression of pyrG was found.
FIG. 6.
Correlation between the different nucleoside triphosphate concentrations and expression of pyrG in L. lactis. Plots of β-galactosidase activity as a measure for pyrG expression and nucleoside triphosphate concentrations for CJ238 and CJ300 under different growth conditions (Table 3) are shown. The nucleoside triphosphate concentration is given in nanomoles per milligram (dry weight), and pyrG expression is presented as units/OD unit.
Macromolecular biosynthesis is reduced during CTP limitation.
In experiments using nucleosides as inhibitors of cytidine metabolism, we saw no decrease in the growth rate for more than half an hour after the inhibiting nucleoside was added, even though the CTP concentration in the cell was below our detection limit (Fig. 4C and D). Intuitively, one would expect that during deprivation of CTP, central cellular processes such as RNA and protein synthesis would be affected, immediately resulting in a decrease in the rate by which the OD is increased. In L. lactis the amount of ribosomes, representing the vast majority of RNA in a cell, may not contribute significantly to the OD, as the amount of stable RNA in L. lactis has been determined to be only ca. 11% of the total dry weight (3, 6). Proteins comprise more than 50% of the total dry weight (3, 27), and therefore the rate of synthesis of proteins has a greater influence on OD measurements than synthesis of rRNA. A change in cell size may also contribute significantly to the OD, but the cells do not change in size during CTP limitation as observed in the microscope (data not shown). To explain the observed phenotypes of the pyrG mutant during CTP limitation, we therefore decided to determine the rates of synthesis of RNA and protein by pulse-labeling for 10 min with [14C]adenine or [14C]leucine, respectively (Fig. 5B and C). Limitation for CTP had an immediate effect on RNA synthesis, as the amount of RNA synthesized decreased to about 50% of the wild-type steady-state level. Since CTP is one of the substrates for RNA synthesis, this is not surprising. Protein synthesis was also affected during CTP limitation and was reduced to about 65% of the values obtained for cells not limited for CTP. A gradual increase in protein synthesis was seen, as after almost two generations of CTP limitation, the protein synthesis rate had increased from 65 to almost 75% of the reference level. However, protein synthesis in the time interval from 5 to 15 min after CTP limitation was at the level for the uninhibited cells, although RNA synthesis decreased to half its normal value. The delayed growth inhibition seen during uridine inhibition may therefore partly be due to unaffected protein synthesis during the first 15 min of CTP limitation. As unaffected protein synthesis could be explained by an increase in mRNA stability, we determined the mRNA half-life during CTP limitation. An mRNA half-life of 70 s was obtained both for the pyrG mutant after 30 and 100 min of CTP limitation and for the pyrG mutant with a normal CTP pool, suggesting that CTP limitation has no effect on mRNA half-life.
Identification of a terminator structure in the pyrG promoter region.
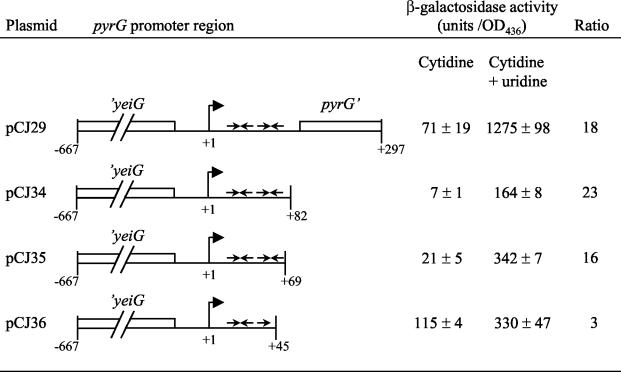
As in B. subtilis (25), a terminator structure can be predicted to form in the L. lactis pyrG leader sequence. The existence of this terminator in vivo was supported by deletion analysis of the pyrG leader and subsequent starvation for CTP in a pyrG background. The plasmids pCJ34 and pCJ35 contain the predicted terminator, and transcription is low in these promoter constructs when cytidine is present in the medium; CTP limitation increased expression about 20-fold (Fig. 7). The fragment in pCJ36 lacks half of the terminator stem-loop structure and has high expression from the reporter genes even when cells are grown in the presence of cytidine, suggesting that this structure is required for repression during high CTP concentrations. A threefold regulation, however, was still observed for this construct. This effect has also been reported for B. subtilis and may be a nonspecific effect of the pyrimidine starvation condition (25). Expression from pCJ29 is severalfold higher than expression from the three plasmids in Fig. 7. This is probably due to the presence of the pyrG open reading frame in pCJ29. There are very few base pairs between the stop codons and the ribosome binding site for the lacL gene in the pAK80 vector, which may cause an increase in expression from the reporter genes due to translational coupling to the upstream open reading frame. This is a common phenomenon with this vector (14).
FIG. 7.
Deletion analysis of the pyrG promoter region. The indicated pyrG promoter regions were cloned in the promoter-probe vector pAK80. The plasmids were transformed into a pyrG::ISS1 mutant and grown in defined medium either with cytidine or under CTP-limiting conditions (cytidine plus uridine) by the addition of uridine at a 25-fold excess at an OD436 of 0.2. Cells were harvested at an OD436 of 0.8, and the β-galactosidase activity was determined. Arrows indicate the two stem-loop structures predicted to form in the leader sequence. The activity of the negative control harboring only the vector pAK80 was determined to be less than 0.02 U/OD unit.
DISCUSSION
The work presented here shows that a promoter immediately upstream of the pyrG open reading frame has sufficient strength to ensure full expression of pyrG. On the other hand, an RT-PCR experiment indicated that transcription arising from upstream promoters could contribute to the overall expression of pyrG. From Table 2 it is evident that the expression of pyrG is increased approximately 20-fold when the fusion is present on a plasmid compared to a chromosomal localization, as judged by the β-galactosidase expression in CJ217 and CJ298 grown under identical conditions. Since the copy number of the plasmid is reported to be lower than 30, it can be concluded that all significant expression of pyrG arises from a promoter present on the cloned fragment, provided that no regulatory factor is titrated by the plasmid. This is not the fact, since the fold of regulation is the same on both chromosomal and plasmid fusions. Consequently, only the promoter immediately in front of pyrG has physiological relevance.
The results presented here clearly show that expression of the pyrG gene from L. lactis is regulated by the concentration of cytidine nucleotides within the cell, as illustrated in Fig. 6. The conclusion is based on a valid determination of intracellular nucleotide pools. The method is based on labeling of cultures with radioactive phosphate for two generations, immediate quenching and extraction of the nucleotides with 2 M formic acid, and separation of the nucleotides by thin-layer chromatography. The description of the method and the validation of the different steps in the method were presented in a recent paper (23). Reducing the CTP pool either by growth of a pyrG mutant with uridine at a concentration 25 times higher than that of cytidine or by growth with galactose or maltose as a carbon source results in increased pyrG expression. Under all of these growth conditions, the strains have lower growth rates than the uninduced control strain. However, reduction in growth rate is not the mechanism triggering pyrG induction, since decoyinine addition in the presence of cytidine inhibits growth but does not induce pyrG expression (Table 3). Growth inhibition is also observed when pyrimidine uptake in a pyrB mutant (CJ300) is inhibited by addition of inosine; however, this treatment does not reduce the CTP concentration in the cell and gives no pyrG induction (Table 3). The latter experiment also shows that pyrG expression is not induced by a decrease in the UTP pool; the regulation of pyrG is thus specific to cytidine nucleotides. Expression of pyrG in L. lactis is repressed by cytidine, as found in other bacteria (2, 25, 37). The CTP dependence of pyrG expression is similar to what has been proposed for pyrG from B. subtilis (25), but a correlation with CTP pool size was not demonstrated. In this work, nucleotide pool sizes during pyrimidine limitation have been determined and it has been shown directly that the concentration of CTP and pyrG expression are correlated in L. lactis. The fact that pyrG is not induced upon complete CTP starvation is most likely due to a general stop in transcription.
The rate of synthesis from pyrG during uridine-inhibited cytidine phosphorylation in a pyrG mutant was found not to be linear (Fig. 5A). Initially, pyrG is induced 9-fold, but the synthesis rate increases for 1.5 generations to a final induction ratio of 37-fold. This pattern of induction cannot be explained from altered macromolecular synthesis, since neither RNA nor protein synthesis changes with the same kinetics. The low CTP concentration during cytidine limitation is at the limit of detection, and the CTP concentration may therefore decrease further during the growth experiment after uridine addition, not reaching its final lower value until 1.5 generations of growth, where pyrG expression reaches a new steady-state level. It is important to emphasize that the concentration of CTP does not reflect the flow through the CTP pool. The small amount of cytidine phosphorylated to CTP during uridine inhibition is rapidly used for, e.g., RNA production, causing a high turnover of CTP.
When the kinetics of the effects on macromolecular synthesis of cytidine limitation by addition of uridine was monitored (Fig. 5B and C), it was found that the rate of RNA synthesis was immediately changed to 50% of that found in uninhibited cells. In contrast, the rate of protein synthesis was unchanged during the first 15 min of cytidine limitation. The unchanged protein synthesis was not due to an increased mRNA half-life under these growth conditions (data not shown), and therefore we must conclude that the amount of mRNA synthesized during the first 15 min is unaltered compared to that in the control cells without uridine. The 50% inhibition of RNA synthesis observed is thus proposed to be primarily inhibition of stable RNA synthesis. In the final steady state obtained after 1.5 generations of growth, the growth rate is half of the uninhibited growth rate and the number of ribosomes is expected to be reduced. The immediate stop in synthesis of rRNA after uridine addition would help the cells to a fast adjustment to the new growth conditions. The rate of total RNA synthesis during the period after uridine addition was 50% of the uninhibited rate. This is likely to reflect different proportions of stable RNA and mRNA rates during the experiment, as discussed above. In the final steady state, the protein synthesis rate is 75% of the uninhibited rate. These numbers also indicate that a major fraction of RNA made is mRNA and hence that cells with a lower growth rate contain fewer ribosomes and more protein per cell mass. The relationship between the growth rate of a bacterial cell and the content of stable RNA was identified for E. coli and S. enterica serovar Typhimurium by Maaløe and coworkers, and they found that synthesis of rRNA increased with the square of the growth rate (16, 31). Later this was shown to apply for many other bacteria, including L. lactis (4). On the other hand, there has been controversy in the literature as to whether nucleotide pools vary with the growth rate in the same way (7, 28). The different results obtained have been attributed to variations in the methods used for nucleotide pool determinations (32). The results obtained in this study for L. lactis suggest a correlation between growth rate and nucleoside triphosphate pools, as growth on galactose or maltose as a carbon source results in decreased pool sizes. The variance of nucleoside triphosphate pools in L. lactis, however, may not be due to the method used but rather may be a consequence of the relatively simple metabolism of this bacterium, in which sugar is fermented to lactate (homolactic growth, as seen with growth on glucose) or to other acids such as acetate and ethanol (mixed acid fermentation, as seen with growth on galactose or maltose). In E. coli, oxidative phosphorylation, a pathway not active in L. lactis, may have a large influence on the turnover rate of nucleoside triphosphate.
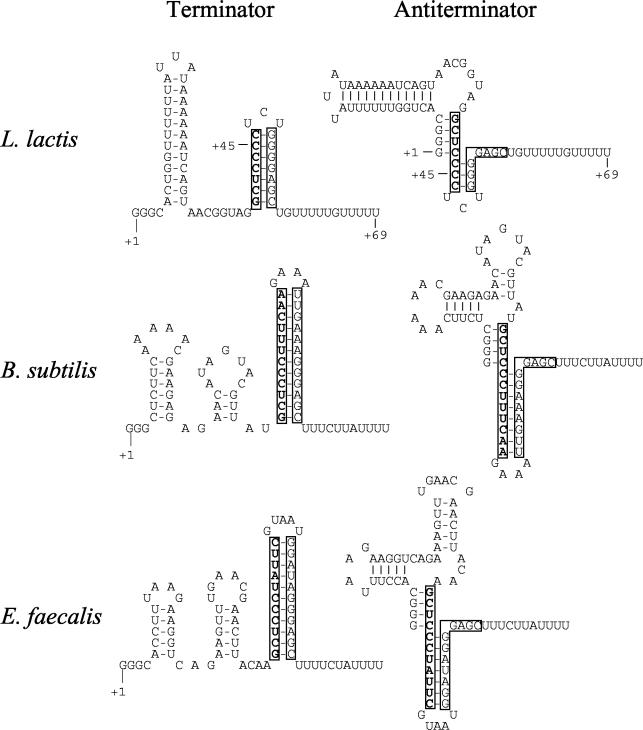
With respect to the mechanism for the CTP-regulated expression of pyrG, it has been shown to be due to termination and antitermination in B. subtilis, and a terminator structure has been identified in the untranslated leader sequence of pyrG (25). In L. lactis, deletion analysis of the pyrG leader also demonstrates that the terminator structure is required for regulation (Fig. 7). pyrG leaders from gram-positive bacteria contain several conserved segments and all have the potential to fold into transcription terminator structures, suggesting similar regulatory mechanisms for pyrG in these bacteria (25). Mutational analysis of the pyrG leader in B. subtilis has identified several interesting points regarding the mechanism of pyrG regulation. The first four nucleotides (GGGC) in the pyrG leader of B. subtilis are required for normal regulation of pyrG, as mutagenesis of these nucleotides results in increased termination under all growth conditions (26). Additionally, it was shown that antitermination during CTP starvation requires the nucleotides GCUCCC in the stem of the terminator structure. Deletion of the nucleotides between these two conserved segments has no effect on regulation of pyrG. It was proposed that a regulatory protein senses the CTP availability and binds to the conserved segments in the pyrG leader during CTP starvation, thereby preventing termination (25). However, the work presented here has revealed that the cloning of the pyrG leader on a plasmid does not lead to titration of an effector, since the fold regulation is conserved despite the copy number. Consequently, another explanation is possible, which does not include a regulatory protein but rather involves the formation of an antiterminator structure. Such a structure is presented in Fig. 8, and the structure includes base pairing between the two identified conserved segments. The predicted terminator-antiterminator structure can be identified in several gram-positive bacteria, including L. lactis, B. subtilis, E. faecalis, Streptococcus pyogenes, Lactobacillus plantarum, and Listeria monocytogenes, suggesting the presence of the structures in these organisms. In Fig. 8 the putative terminator and antiterminator structures of L. lactis, B. subtilis, and E. faecalis are shown. It is interesting that a mutation in the B. subtilis pyrG leader removing base pairs between the two conserved segments (e.g., changing one of the first three Gs to an A) abolishes pyrG regulation whereas a mutation that strengthens the antiterminator (e.g., changing the U at position +5 to a G) increases pyrG expression (26). The region between the two conserved segments is not needed for formation of the antiterminator, explaining the lack of homology in this part of the leader between gram-positive bacteria and the fact that this part can be deleted without loss of regulation (26).
FIG. 8.
Predicted structures of terminators and antiterminators in the 5′ pyrG leader sequences from L. lactis (accession number AJ010153) B. subtilis (accession number Z49782), and E. faecalis (accession number AE016950). The first nucleotide in the mRNA is marked +1. The +1 positions in E. faecalis and L. lactis are not supported by experimental evidence, whereas the +1 position of B. subtilis was determined by Meng and Switzer (25). The translational start codons are found further downstream. The stems in the terminator structure are marked by boxes in both the terminator and the antiterminator. +45 and +69 in the structure for L. lactis indicate the deletion points in plasmids pCJ35 and pCJ36 (Fig. 7).
We suggest the following model for pyrG regulation in L. lactis and probably in other gram-positive bacteria as well. At normal CTP concentrations the terminator in the pyrG leader is preferentially formed, thereby reducing expression of CTP synthase (Fig. 8). When the CTP concentration is reduced, RNA polymerase pauses at the stretch of C residues in the pyrG leader, thus allowing the antiterminator to form. Since the terminator and antiterminator are mutually exclusive structures (Fig. 8), transcription will proceed, thus allowing expression of the pyrG gene. We will pursue the verification of the model by in vivo and in vitro experiments.
Acknowledgments
This work was supported through the Center for Advanced Food Studies by the FØTEK program and the DFFE.
We appreciate the technical assistance of Jeannette Lundin, the valuable help of Lise Schack (University of Copenhagen) with preparing polyethyleneimine plates, and the help of Ufuk Sumer with construction of plasmids with pyrG leader deletions. We acknowledge Steen Wadskov-Hansen for performing the RT-PCR experiment. We thank Martin Willemoës for reading the manuscript.
REFERENCES
- 1.Andersen, H. W., C. Solem, K. Hammer, and P. R. Jensen. 2001. Twofold reduction of phosphofructokinase activity in Lactococcus lactis results in strong decreases in growth rate and in glycolytic flux. J. Bacteriol. 183:3458-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asahi, S., M. Doi, Y. Tsunemi, and S. Akiyama. 1989. Regulation of pyrimidine nucleotide biosynthesis in cytidine deaminase-negative mutants of Bacillus subtilis. Agric. Biol. Chem. 53:97-102. [Google Scholar]
- 3.Benthin, S., U. Schulze, J. Nielsen, and J. Villadsen. 1994. Growth energetics of Lactococcus cremoris FD1 during energy-, carbon-, and nitrogen-limitation in steady state and transient cultures. Chem. Eng. Science 49:589-609. [Google Scholar]
- 4.Beresford, T., and S. Condon. 1993. Physiological and genetic regulation of rRNA synthesis in Lactococcus. J. Gen. Microbiol. 139:2009-2017. [DOI] [PubMed] [Google Scholar]
- 5.Edlin, G., and G. S. Stent. 1969. Nucleoside triphosphate pools and the regulation of RNA synthesis in E. coli. Proc. Natl. Acad. Sci. USA 62:475-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Even, S., N. D. Lindley, and M. Cocaign-Bousquet. 2001. Molecular physiology of sugar catabolism in Lactococcus lactis IL1403. J. Bacteriol. 183:3817-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaal, T., M. S. Bartlett, W. Ross, C. L. Turnbough, Jr., and R. L. Gourse. 1997. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278:2092-2097. [DOI] [PubMed] [Google Scholar]
- 8.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghim, S. Y., C. C. Kim, E. R. Bonner, J. N. D'Elia, G. K. Grabner, and R. L. Switzer. 1999. The Enterococcus faecalis pyr operon is regulated by autogenous transcriptional attenuation at a single site in the 5′ leader. J. Bacteriol. 181:1324-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Israelsen, H., S. M. Madsen, A. Vrang, E. B. Hansen, and E. Johansen. 1995. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl. Environ. Microbiol. 61:2540-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jensen, P. R., and K. Hammer. 1993. Minimal requirements for exponential growth of Lactococcus lactis. Appl. Environ. Microbiol. 59:4363-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johansen, E., and A. Kibenich. 1992. Characterization of Leuconostoc isolates from commercial mixed strain mesophilic starter cultures. J. Dairy Sci. 75:1186-1191. [Google Scholar]
- 14.Kilstrup, M., S. G. Jessing, S. B. Wichmand-Jørgensen, M. Madsen, and D. Nilsson. 1998. Activation control of pur gene expression in Lactococcus lactis: proposal for a consensus activator binding sequence based on deletion analysis and site-directed mutagenesis of purC and purD promoter regions. J. Bacteriol. 180:3900-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kilstrup, M., and J. Martinussen. 1998. A transcriptional activator, homologous to the Bacillus subtilis PurR repressor, is required for expression of purine biosynthetic genes in Lactococcus lactis. J. Bacteriol. 180:3907-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kjeldgaard, N. O., and C. G. Kurland. 1963. The distribution of soluble and ribosomal RNA as a function of growth rate. J. Mol. Biol. 6:341-348. [Google Scholar]
- 17.Lu, Y., and R. L. Switzer. 1996. Evidence that the Bacillus subtilis pyrimidine regulatory protein PyrR acts by binding to pyr mRNA at three sites in vivo. J. Bacteriol. 178:5806-5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu, Y., and R. L. Switzer. 1996. Transcriptional attenuation of the Bacillus subtilis pyr operon by the PyrR regulatory protein and uridine nucleotides in vitro. J. Bacteriol. 178:7206-7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madsen, S. M., B. Albrechtsen, E. B. Hansen, and H. Israelsen. 1996. Cloning and transcriptional analysis of two threonine biosynthetic genes from Lactococcus lactis MG1614. J. Bacteriol. 178:3689-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinussen, J., and K. Hammer. 1995. Powerful methods to establish chromosomal markers in Lactococcus lactis: an analysis of pyrimidine salvage pathway mutants obtained by positive selections. Microbiology 141:1883-1890. [DOI] [PubMed] [Google Scholar]
- 22.Martinussen, J., J. Schallert, B. Andersen, and K. Hammer. 2001. The pyrimidine operon pyrRPB-carA from Lactococcus lactis. J. Bacteriol. 183:2785-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinussen, J., S. L. Wadskov-Hansen, and K. Hammer. 2003. Two nucleoside uptake systems in Lactococcus lactis: competition between purine nucleosides and cytidine allows for modulation of intracellular nucleotide pools. J. Bacteriol. 185:1503-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melchiorsen, C. R., K. V. Jokumsen, J. Villadsen, H. Israelsen, and J. Arnau. 2002. The level of pyruvate-formate lyase controls the shift from homolactic to mixed-acid product formation in Lactococcus lactis. Appl. Microbiol. Biotechnol. 58:338-344. [DOI] [PubMed] [Google Scholar]
- 25.Meng, Q., and R. L. Switzer. 2001. Regulation of transcription of the Bacillus subtilis pyrG gene, encoding cytidine triphosphate synthetase. J. Bacteriol. 183:5513-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng, Q., and R. L. Switzer. 2002. cis-acting sequences of Bacillus subtilis pyrG mRNA essential for regulation by antitermination. J. Bacteriol. 184:6734-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neidhardt, F. C., and H. E. Umbarger. 1996. Chemical composition of Escherichia coli, p. 13-16. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington D.C.
- 28.Petersen, C., and L. B. Møller. 2000. Invariance of the nucleoside triphosphate pools of Escherichia coli with growth rate. J. Biol. Chem. 275:3931-3935. [DOI] [PubMed] [Google Scholar]
- 29.Potvin, B. W., R. J. Kelleher, Jr., and H. Gooder. 1975. Pyrimidine biosynthetic pathway of Bacillus subtilis. J. Bacteriol. 123:604-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schaechter, M., O. Maaløe, and N. O. Kjeldgaard. 1958. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J. Gen. Microbiol. 19:592-606. [DOI] [PubMed] [Google Scholar]
- 32.Schneider, D. A., T. Gaal, and R. L. Gourse. 2002. NTP-sensing by rRNA promoters in Escherichia coli is direct. Proc. Natl. Acad. Sci. USA 99:8602-8607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Switzer, R. L., R. J. Turner, and Lu Y. 1999. Regulation of the Bacillus subtilis pyrimidine biosynthetic operon by transcriptional attenuation: control of gene expression by an mRNA-binding protein. Prog. Nucleic Acid Res. Mol. Biol. 62:329-367. [DOI] [PubMed] [Google Scholar]
- 34.Turner, R. J., Y. Lu, and R. L. Switzer. 1994. Regulation of the Bacillus subtilis pyrimidine biosynthetic (pyr) gene cluster by an autogenous transcriptional attenuation mechanism. J. Bacteriol. 176:3708-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wadskov-Hansen, S. L., M. Willemoës, J. Martinussen, K. Hammer, J. Neuhard, and S. Larsen. 2001. Cloning and verification of the Lactococcus lactis pyrG gene and characterization of the gene product, CTP synthase. J. Biol. Chem. 276:38002-38009. [DOI] [PubMed] [Google Scholar]
- 36.Wang, W., and D. H. Bechhofer. 1996. Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain. J. Bacteriol. 178:2375-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.West, T. P., and G. A. O'Donovan. 1982. Repression of cytidine triphosphate synthetase in Salmonella typhimurium by pyrimidines during uridine nucleotide depletion. J. Gen. Microbiol. 128:895-899. [DOI] [PubMed] [Google Scholar]