Abstract
The serine/threonine protein phosphatases are targeted to specific subcellular locations and substrates in part via interactions with a wide variety of regulatory proteins. Understanding these interactions is thus critical to understanding phosphatase function. Using an iterative affinity purification/mass spectrometry approach, we generated a high density interaction map surrounding the protein phosphatase 2A catalytic subunit. This approach recapitulated the assembly of the PP2A catalytic subunit into many different trimeric complexes but also revealed several new protein-protein interactions. Here we define a novel large multiprotein assembly, referred to as the striatin-interacting phosphatase and kinase (STRIPAK) complex. STRIPAK contains the PP2A catalytic (PP2Ac) and scaffolding (PP2A A) subunits, the striatins (PP2A regulatory B‴ subunits), the striatin-associated protein Mob3, the novel proteins STRIP1 and STRIP2 (formerly FAM40A and FAM40B), the cerebral cavernous malformation 3 (CCM3) protein, and members of the germinal center kinase III family of Ste20 kinases. Although the function of the CCM3 protein is unknown, the CCM3 gene is mutated in familial cerebral cavernous malformations, a condition associated with seizures and strokes. Our proteomics survey indicates that a large portion of the CCM3 protein resides within the STRIPAK complex, opening the way for further studies of CCM3 biology. The STRIPAK assembly establishes mutually exclusive interactions with either the CTTNBP2 proteins (which interact with the cytoskeletal protein cortactin) or a second subcomplex consisting of the sarcolemmal membrane-associated protein (SLMAP) and the related coiled-coil proteins suppressor of IKKɛ (SIKE) and FGFR1OP2. We have thus identified several novel PP2A-containing protein complexes, including a large assembly linking kinases and phosphatases to a gene mutated in human disease.
Protein phosphatase 2A (PP2A)1 is a major eukaryotic serine/threonine phosphatase that has been implicated in the control of cell growth, proliferation, and differentiation (1–4). The catalytic subunit of PP2A is represented by two genes in humans (gene names are in the supplemental materials; gene products here are referred to as PP2Acα and PP2Acβ) sharing 97% identity at the protein level (5). Many mutually exclusive protein complexes containing the PP2A family catalytic subunits have been characterized biochemically (3, 6). The PP2A catalytic (PP2Ac) subunit binds directly to the PP2A A scaffolding subunit (two 85% identical proteins, PP2A Aα and PP2A Aβ, are present in human cells), to form the so-called PP2A dimeric core (7–9). The core serves as a platform for the association of a regulatory or B subunit to generate a trimeric complex important for substrate recruitment and subcellular targeting. Four families of B subunits exist in human cells (B, B`, B", and B‴; the B‴ members are commonly known as striatins), altogether coded for by at least 15 genes (for reviews, see Refs. 2, 3, and 6; for trimer structure, see Refs. 10 and 11). Several splice variants and post-translational modifications have been described for components of the PP2A holoenzyme, adding another level of complexity to the regulation and specificity of the phosphatases. Non-trimeric PP2A family-containing complexes have also been reported (in addition to the dimeric core of PP2Ac·PP2A A (12)). For example, the antiapoptotic protein alpha4 can interact directly with the PP2A catalytic subunit in the absence of the scaffolding subunit (13, 14). In addition, Mob3, a small molecular weight protein of the Mob family (also known as phocein or preimplantation antigen 3), stably assembles with striatin (B‴) molecules, PP2Ac, and PP2A A in a complex containing at least four proteins (15, 16). Recent studies have highlighted the role of the regulatory subunits as key determinants of specificity and biological activity. For example, PP2A B`δ targets the Cdc25 protein for dephosphorylation during mitosis (17, 18). In addition, the PP2A B`γ subunit was demonstrated to be targeted by the small T antigen of SV40 in human cell transformation (19). Intracellular localization is also important, and a splice variant of PP2A Bβ2, containing an N-terminal mitochondrial localization signal, assembles a holoenzyme involved in neuronal survival signaling (20).
Given the relevance of the composition of protein complexes in the biological functions of PP2A, it is important to devise approaches to characterize the many PP2A-containing molecular assemblies. Affinity purification coupled to mass spectrometry (AP-MS) is a powerful method to identify and characterize interaction partners (21–26). However, although a single AP-MS can successfully identify multiple interactors, this method is uninformative regarding the relationships between components of multiple multiprotein complexes containing a protein of interest. For example, AP-MS of PP2Ac or PP2A A would yield identification of both B and B` proteins in the same purification, yet B and B` proteins are mutually exclusive binding partners of the PP2Ac·PP2A A core and are never found in the same complex (2). Generating a high density interaction map in which most, and ideally all, interacting partners are purified in parallel experiments consisting of reciprocal purifications can help to resolve the components of individual complexes. The strategy utilized here is depicted in Fig. 1A. Briefly the protein of interest is epitope-tagged and stably expressed in human cells. The protein and its interacting partners are purified, subjected to proteolytic digestion, and identified using mass spectrometry (gel-free method). High confidence interactors are in turn cloned, expressed, and purified, and their interacting partners are identified. The process may be repeated in an iterative fashion to generate high density local interactomes centered around proteins of interest. The high density of the interactions allows for a more precise understanding of the multitude of alternative complexes containing a particular protein of interest, a problem not always addressed in lower density global interaction studies (27–30).
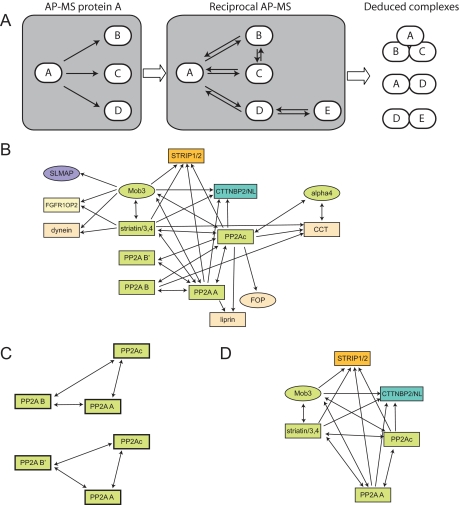
Fig. 1.
TAP interaction network for PP2A phosphatases. A, iterative TAP tagging approach to identify protein complexes. Protein A is stably expressed in HEK293 cells, and interaction partners B, C, and D are detected by LC-MS/MS. Proteins B, C, and D are in turn cloned and stably expressed, and their interactors are identified. Here proteins B and C only interacted with each other and with protein A. Protein D interacted with the new protein E, which was in turn tested by TAP tagging. The mass spectrometric data for this high density network suggest the association of A in two mutually exclusive complexes: (a) a trimer with B and C or (b) a dimer with D. The association of protein D in a dimer with A is likely to be mutually exclusive with its association with protein E in a separate complex. B, phosphatase interaction network for PP2A detected by TAP tagging. The proteins used as baits are shown in green. For display purposes (and because we have not detected evidence for paralog-specific interactions with TAP tagging), proteins belonging to the same paralogy group (defined in supplemental Table S3) are collapsed into a single node (in this case, they are represented by rectangles, whereas proteins without paralogs are shown as ovals). The directionality of the arrows represents the bait to prey relationship. C, classical PP2A trimers detected by reciprocal TAP tagging. D, PP2Ac, PP2A A, striatin4, and Mob3 interact with two novel protein families, STRIP1/STRIP2 (orange) and CTTNBP2/CTTNBP2NL (blue).
Here we utilized tandem affinity purification (TAP) and FLAG affinity purification (31, 32) to produce a high density interaction map surrounding the PP2A catalytic subunit (we previously reported on the same approach used to study PP4c (33)). Our map recapitulated previously characterized assemblies for PP2A but also allowed the discovery of a large complex surrounding the previously characterized PP2Ac·PP2A A·striatin·Mob3 assembly. This complex, which we refer to as striatin-interacting phosphatase and kinase (STRIPAK), contains the uncharacterized proteins FAM40A/FAM40B, which we have renamed striatin-interacting proteins 1 and 2 (STRIP1 and STRIP2). In addition, STRIPAK contains all members of the GCK-III subfamily of Ste20 protein kinases (STK24, STK25, and MST4) as well as cerebral cavernous malformation 3 (CCM3; gene name PDCD10), a protein encoded by a gene mutated in familial cases of cerebral cavernous malformations (34, 35). In addition, we present evidence that STRIPAK can establish mutually exclusive interactions with the cortactin-binding proteins cortactin-binding protein 2 (CTTNBP2)/cortactin-binding protein 2, N-terminal-like (CTTNBP2NL) or with an assembly of sarcolemmal membrane-associated protein (SLMAP) and the related proteins SIKE/FGFR1OP2.
EXPERIMENTAL PROCEDURES
cDNA and Stable Cell Lines—
Constructs used in this study are described in supplemental Table S2; all inserts were sequenced in their entirety. Briefly, coding sequences for proteins of interest were amplified by PCR using Pfu Ultra (or other high fidelity polymerases) from a HeLa cell cDNA library (Stratagene) or cDNA clones from the mammalian gene collection or IMAGE collection. Whenever human cDNAs were available they were used; in some cases, the mouse ORF was utilized instead. Our original clone for mouse Fam40A/STRIP1 lacked the initiator methionine and a glutamic acid codon as compared with the longest mouse cDNA (GenBank™ accession number NM_153563). A full-length construct of mFam40A/STRIP1 was therefore generated by adding coding sequences for methionine and glutamic acid in the PCR primer. The resulting sequence corresponds to GenBank accession number NM_153653 (coding sequence). Inserts were cloned in-frame into pcDNA3-FLAG, pcDNA3–3HA, or pcDNA3-NTAP vectors. Constructs were sequenced and found to match the sequence of the original cDNAs with the exception of MST4, which carried a single amino acid substitution (Ile-356 to Thr; no wild type clones were obtained). These constructs were stably expressed in low passage HEK293 cells as described previously (33), and pools of selected cell lines were generated. Expression was monitored by Western blotting using normal rabbit serum (for TAP-tagged constructs) or an antiserum directed against the FLAG epitope, respectively. To determine the approximate percentage of the FLAG-tagged proteins solubilized in our extraction procedure, we separately analyzed the pellet and supernatant following lysis; in each case, at least half of the protein was extracted, consistent with the previous annotation of many of the complex components as distributed in both membrane-associated and soluble compartments.
Antibodies—
Rabbit anti-GST-alpha4 (rabbit 2972, a kind gift from Dr. N. Sonenberg) recognizes a single major band in HEK293 cell extract (not shown). Commercial antibodies were as follows (catalog numbers are in parentheses): anti-PP2Acα (610555), anti-striatin (610838), and anti-MST4 (612684) were from BD Transduction Laboratories; anti-PP2A A was from Upstate Biotechnology (07-250); anti-SG2NA (STRN3) was from Cell Signaling Technology (S68); anti-PP4R2 was from Bethyl Laboratories (A300-838A), anti-FLAG was from Sigma (F3165); and anti-HA was from Covance Research Products (MMS-101P). Secondary antibodies for immunoblotting were donkey anti-mouse IgG and donkey anti-rabbit IgG, both conjugated to horseradish peroxidase (GE Healthcare). For the IP/Western of MST4 in Fig. 3A, the anti-mouse IgG TrueBlot Ultra horseradish peroxidase conjugate from eBioscience (88-8817-31) was used as a secondary antibody.
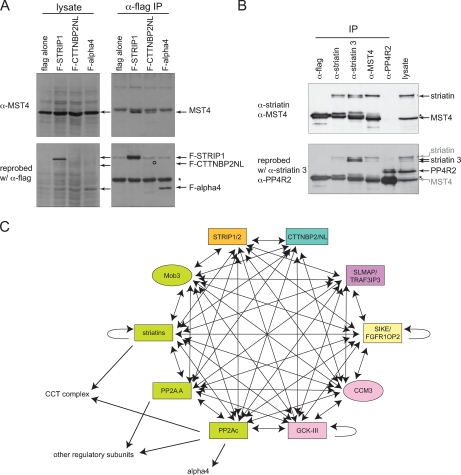
Fig. 3.
STRIPAK complex interactions. A, interaction of MST4 with STRIPAK. FLAG purification was performed as in Fig. 2, and detection of endogenous MST4 in immunoprecipitates was performed. B, validation of the interactions among endogenous proteins. Lysate from untransfected HEK293 cells (1 mg per IP) was used for immunoprecipitation using the indicated antibodies and protein G-Sepharose (whole cell lysate = 30 μg). Proteins were directly eluted in Laemmli sample, separated by SDS-PAGE, transferred to nitrocellulose, and detected by immunoblotting with specified antibodies. PP4R2 is a PP4c-interacting protein used here as a negative control. C, summary of the interactions detected by AP-MS with FLAG-tagged proteins. Green nodes represent the previously characterized association between PP2Ac, PP2A A, striatins, and Mob3; pink indicates previously characterized interactions between CCM3 and the Ste20 kinases of the CGK-III subgroup. The directionality of the arrows represents the bait to prey relationship. When evidence of self-interaction within a protein family exists, self-pointing arrows are depicted.
Tandem Affinity Purification and Mass Spectrometric Analysis—
Tandem affinity purification was performed essentially as described previously (33), and proteins were eluted by incubation in 25 mm EGTA in 50 mm ammonium bicarbonate, pH 8. Sequencing grade modified trypsin (0.5–1 μg; Promega) was added directly to the eluate. Digestion was performed overnight at 37 °C. Following digestion, the sample was lyophilized and then resuspended in reversed-phase HPLC buffer A1 (20 μl; 0.4% AcOH, 0.005% heptafluorobutyric anhydride in H2O). Prior to loading onto the reversed-phase column, the sample was centrifuged at 13,000 rpm for 10 min, and the supernatant was transferred to a fresh tube. Microcapillary reversed-phase columns (75-μm inner diameter, 363-μm outer diameter; Polymicro Technology) were cut to a final length of 15–20 cm, and spray tips were pulled in-house by hand. Columns were packed in-house (12 cm) with Magic C18 100-Å, 5-μm silica particles (Michrom) using a pressure bomb. Prior to loading the sample, columns were equilibrated in HPLC buffer A1. Half of the sample was applied to the column using a pressure bomb and then washed off line in buffer A1 + 5% acetonitrile for 30–60 min. The LC column was then placed in front of a Finnigan LCQ mass spectrometer programmed for data-dependent MS/MS acquisition (one survey scan, three MS/MS of the most abundant ions). After sequencing the same m/z species (±3 Da) three times, it was placed on an exclusion list for 3 min. Peptides were eluted from the reversed-phase column using a multiphasic elution gradient (5–14% acetonitrile over 5 min, 14–40% over 60 min, and 40–80% over 10 min). The remaining half of the sample was processed in the same manner. To prevent cross-contamination, each sample was analyzed on a freshly prepared reversed-phase column. The MS searches were performed as in Gingras et al. (33) such that the results are directly comparable: raw files generated by Xcalibur (Finnigan) were converted to the mzXML format (36), and combined runs (from the same sample) were searched using SEQUEST against the human International Protein Index (IPI) database, version 3.01 (55,140 entries were searched). SEQUEST searches were performed without constraining for the number of tryptic termini, with a mass tolerance on the precursor ion of ±2, and with methionine oxidation (+16) as a variable modification. SEQUEST html output was analyzed with PeptideProphet (37) and ProteinProphet (38) using the default parameters of each program. Supplemental Table S4 contains the experimental data; supplemental Tables S6 and S7 list the proteins identified on the basis of a single peptide together with additional evidence for the interaction. Supplemental Fig. S7 presents the annotated spectra of those proteins identified by a single unique peptide.
FLAG Affinity Purification, Immunoprecipitation Using Antibodies to Endogenous Proteins, and Mass Spectrometric Analysis—
FLAG affinity purification was performed essentially as described previously (32) with the following modifications. Detergent concentration in the lysis buffer was 0.5% Nonidet P-40, the lysis buffer was added at 4 ml/g of wet cell pellet, and cells were subjected to passive lysis (30 min) followed by one freeze-thaw cycle. For the AP-MS of endogenous proteins, 0.3 mg of cell pellet from untransfected HEK293 cells was resuspended in 2.7 ml of lysis buffer, and cell extract was prepared as above. For each IP, 7 mg of crude lysate was used. The lysate was subjected to two preclearing steps: one on 20 μl of packed protein G beads for 1 h and one on 20 μl of packed protein G beads preincubated with 4 μg of anti-HA antibody for 1 h. IP was performed by adding anti-MST4 (16 μl), anti-striatin (8 μl), or anti-FLAG (8 μl) antibodies and protein G-Sepharose (25 μl of packed beads) to the precleared lysate and incubating for 2 h. Beads were washed three times in lysis buffer and three times in 50 mm ammonium bicarbonate. Samples (from the FLAG or endogenous immunoprecipitations) were eluted with ammonium hydroxide, lyophilized in a SpeedVac, resuspended in 50 mm ammonium bicarbonate (pH 8–8.3), and incubated at 37 °C with trypsin overnight. The ammonium bicarbonate was evaporated, and the samples were resuspended in HPLC buffer A2 (2% acetonitrile, 0.1% formic acid) and then directly loaded onto capillary columns packed in-house with Magic 5 μm, 100 Å, C18AQ. MS/MS data were acquired in data-dependent mode (over a 2-h acetonitrile 2–40% gradient) on a ThermoFinnigan LTQ or a ThermoFinnigan LTQ-Orbitrap equipped with a Proxeon NanoSource and an Agilent 1100 capillary pump. mzXML files were generated from ThermoFinnigan *.RAW files using the ReAdW tool available in the TransProteomic Pipeline platform. The searched database contained the human IPI fasta sequence file (version 3.38; 70,856 entries plus the same number in reverse were searched) and common contaminants and was appended with a reversed version of the same IPI database. mzXML files were searched with X!Tandem (39) with k-score plug-in (40) using the following parameters: b- and y-ion series, partial trypsin digestion, allowing for one missed cleavage site, and methionine oxidation and N-terminal acetylation specified as variable modifications. The fragment mass tolerance was 0.8 Da (monoisotopic mass), and the mass window for the precursor was from −1 to 4 Da (average mass) in the case of LTQ data and from −100 to 100 ppm (monoisotopic) in the case of LTQ-Orbitrap data.
Search results were converted into pepXML format using Tandem2XML. PeptideProphet (37) was run on each result set, generating probability scores for each search result that are added to the pepXML documents. In the case of Orbitrap data, the high mass accuracy option was used. The resulting lists of peptides were assembled into proteins using ProteinProphet (38). ProteinProphet files were parsed and exported into a local Mysql database for further analysis and extraction of spectral count information.
Data were exported into Excel files and manually curated. Biological replicates were analyzed for FLAG-PP2Ac (n = 3) and FLAG alone (n = 3) to verify the reproducibility of the procedure (not shown). To generate lists of high confidence specific interactors, proteins detected in any of the FLAG alone samples were subtracted from the final list. Only those proteins that were detected with spectral counts >20 with at least two of the baits are reported. The error rate is 0% as measured by both ProteinProphet and using decoy scores. Supplemental Table S5 contains the experimental data; supplemental Tables S6 and S7 list the proteins identified on the basis of a single peptide together with additional evidence for the interaction. Supplemental Fig. S7 presents the annotated spectra of those proteins identified by a single unique peptide. Supplemental Table S8 contains the MS data for the endogenous IPs along with the annotated spectra for those proteins identified on the basis of a single unique peptide. High confidence interactions reported here were submitted to the BioGrid (41).
Immunoprecipitation/Western Blot—
Lysates from stable cell lines (prepared as above) were incubated with FLAG M2-agarose beads for 1.5 h, washed as above, and directly eluted by boiling in Laemmli or Criterion XT sample buffer. Alternatively lysates from untransfected HEK293 cells were incubated with protein G-Sepharose (GE Healthcare) and antibodies to endogenous striatin (5 μl), striatin3 (2.5 μl), MST4 (2.5 μl), PP4R2 (5 μl), or the FLAG epitope (2.5 μl) for 2 h and washed three times in lysis buffer and once in PBS prior to elution in 25 μl of 2× Laemmli sample buffer. Precipitated proteins were separated via SDS-PAGE, transferred to nitrocellulose, and detected with antibodies directed against the FLAG or HA epitopes or endogenous proteins.
RESULTS
A PP2A High Density Interaction Network—
The catalytic subunits of PP2A (PP2Acα and PP2Acβ) were cloned in-frame C-terminal to a TAP cassette and stably expressed in HEK293 cells. Cells were lysed, and proteins recovered following dual purification were analyzed by mass spectrometry as described previously (33). After background subtraction (as in Ref. 33), we identified high confidence interactors for these catalytic subunits (Table I). Several of the interactors (alpha4 and the multisubunit CCT chaperonin complex) were detected previously with PP4c (note that we previously detected PP2Ac, PP4c, and PP6c peptides in immunoprecipitates from another protein, hTip41, but that we have not detected peptides for hTip41 in purifications of the catalytic subunits) (33). All other interactors were specific for PP2Acα and PP2Acβ. The two PP2A catalytic subunits yielded essentially the same interactors as expected from previous studies (42, 43).
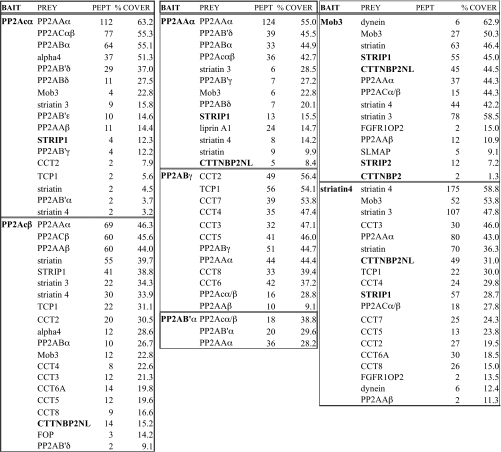
Table I.
Human PP2A-type phosphatase interaction network
Common names for the bait and prey proteins are listed alongside the spectral counts (PeptideProphet probability value >0.9; PEPT) and sequence coverage (% COVER) for the prey proteins. Results were sorted according to decreasing prey protein coverage. Preys in bold are STRIP1 and STRIP2 as well as CTTNBP2 and CTTNBP2NL, which are studied further in Fig. 2. For identifications based on a single unique peptide, a list of the experimental evidence is presented in supplemental Tables S6 and S7; annotated spectra are shown in supplemental Fig. S7.
To characterize phosphatase complex organization, we generated a high density interaction network surrounding these phosphatases. To this end, we cloned into the TAP vector and stably expressed in HEK293 cells several of our identified interactors (alpha4, PP2A Bα, PP2A B`α, the PP2A B‴ striatin4, and Mob3; gene names are in supplemental Table S1). TAP and mass spectrometric analysis of these interactors yielded a detailed map of the interactions surrounding the PP2A catalytic subunits (Table I and Fig. 1B). We identified several different prototypical trimeric complexes containing the PP2Ac catalytic subunit, the PP2A A scaffolding subunit, and regulatory B, B`, or B‴ (striatin4) subunits. Note that we did not detect association with the B" family in this set of experiments, but we did see these in subsequent purifications using FLAG affinity purification (supplemental Table S5). Each regulatory subunit associated specifically with the PP2Ac·PP2A A core and did not associate with other families of regulatory subunits (see below for discussion of striatin dimerization). Overall this experiment recapitulated the known organization of the PP2A trimers, which are composed of one catalytic protein, a single scaffolding protein, and a single regulatory molecule (for the B and B` families; Fig. 1C). In addition, we recapitulated alpha4 interaction with PP2Ac in the absence of PP2A A, representing an apparently mutually exclusive interaction between alpha4 and the PP2A A and B subunits. These results indicate that our approach can successfully identify complexes described previously by biochemical approaches and that this type of analysis can be extended to identify potentially novel interactions.
In addition to the classical PP2A family trimers, a number of additional proteins were also detected in our network. For example, several peptides for liprin α (PPFIA1) were identified in PP2A Aα purifications (and a few in PP2Ac purifications). Liprins are multidomain scaffolding proteins first identified as interactors of the leukocyte antigen-related (protein tyrosine phosphatase, receptor type F) tyrosine phosphatase (40) linked to synaptic functions in several organisms (40, 44). A recent publication reported the interaction of liprin α with the B`γ PP2A regulatory subunit (45). Another interaction partner for PP2Acβ is FOP (also see supplemental Table S5), a protein associated with CAP350, required for microtubule anchoring to the centrosome (46). FOP was initially discovered as a fusion partner for fibroblast growth factor receptor 1 (FGFR1) in stem cell myeloproliferative disorders and confers dimerization (and constitutive activation) to FGFR1 (47–49). Other than the liprins and FOP, all other new PP2A interactors co-purified with striatin4 and Mob3; these interactors are described in more detail below. FOP and liprins did not associate with Mob3 and striatin4 and will be the topic of a separate follow-up study.
PP2A, Striatins, and Mob3 Stably Associate with Novel Components—
We were particularly intrigued by an apparent higher order complex containing the catalytic and scaffolding PP2A subunits, the regulatory B‴ molecule striatin4, and the Mob3 protein. Three striatin proteins exist in humans (gene names STRN, STRN3, and STRN4). Striatin expression levels are highest in brain where it has been implicated in synaptic functions (50–54). Earlier co-precipitation studies (16) identified the small Mob3 protein (referred to in the initial publications as phocein or hMob1, gene name MOBKL3 (15, 55)) as a striatin-interacting partner. Multiple Mob-like proteins exist in higher eukaryotes that appear to regulate the related nuclear DumbBell forming (Dbf)2-related and large tumor suppressor family of kinases through direct binding (56). Other striatin interactors detected in these earlier studies by co-immunoprecipitation and two-dimensional gel electrophoresis remained uncharacterized (16).
Our TAP tagging and AP-MS analysis of striatin4 and Mob3 (as well as PP2Ac and PP2A A) yielded many peptides corresponding to two different protein families: 1) FAM40A/FAM40B, which we have renamed STRIP1 and STRIP2 (also see below), and 2) CTTNBP2NL/CTTNBP2 (Fig. 1, B and D; lower amounts of peptides for additional unrelated proteins were also detected). STRIP1 and STRIP2 are highly related proteins (68% identity; supplemental Fig. S1) of unknown function with no significant homology to other proteins in the human genome and no recognizable sequence motifs. However, the STRIP1/2 proteins bear 13% identity with the yeast protein Far11, which has been linked to pheromone signaling, and 21% identity with the Neurospora crassa HAM-2 gene product, which is implicated in hyphal fusion (supplemental Fig. S1). CTTNBP2 and CTTNBP2NL are also related; the N terminus of CTTNBP2 is 38% identical and 53% similar to CTTNBP2NL over 649 aa (supplemental Fig. S2). This N-terminal portion is predicted to contain a ∼200-aa stretch of coiled-coil motifs. A shorter variant of CTTNBP2 (also known as CBP90), corresponding to the N-terminal 630 aa, was first identified as a protein interacting with the Src homology 3 domain of cortactin (cortical actin-binding protein) in an overlay assay and GST pulldown (57). This interaction is apparently mediated by a proline-rich region at the C terminus of CTTNBP2 that is conserved in CTTNBP2NL (supplemental Fig. S2). Note that we did not detect cortactin in any of our interaction studies.
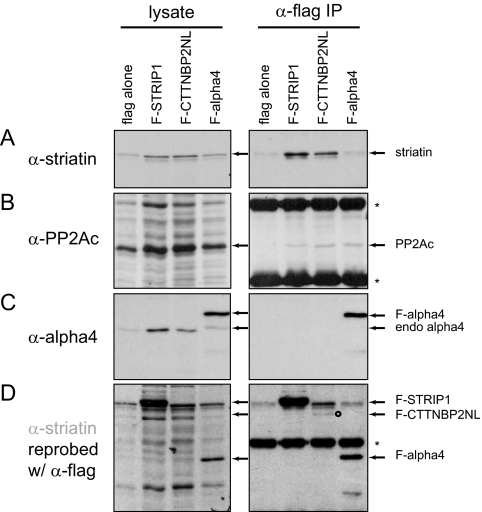
Because we obtained many more peptides for STRIP1 (FAM40A) and CTTNBP2NL than for their respective paralogs in the TAP experiments, we used these two proteins for subsequent IP/Western validation studies. STRIP1 and CTTNBP2NL were cloned into the pcDNA3-FLAG vector (see “Experimental Procedures”) and stably expressed in HEK293 cells. Lysates were subjected to immunopurification using FLAG M2-agarose beads followed by immunoblotting using antibodies directed against endogenous striatin and PP2Ac (the interaction of STRIP1 with PP2A Aα is shown in supplemental Fig. S3). As a control, cells were transfected with the FLAG vector alone or FLAG-alpha4 (according to our MS results, alpha4 interacts with PP2Ac but not with striatin). As shown in Fig. 2 (and despite the low expression of FLAG-CTTNBP2NL) STRIP1 and CTTNBP2NL co-precipitated striatin and PP2Ac, although the percentage of PP2Ac (and PP2A A; supplemental Fig. S3) co-precipitated was low. This is consistent with our MS results in which the number of STRIP1 and CTTNBP2NL peptides was highest in striatin4 and Mob3 purifications. The relatively low abundance of STRIP1 and CTTNBP2NL peptides in PP2Ac and PP2A A purifications (13 each with PP2Ac) may be accounted for by alternate complexes containing the same PP2Ac and PP2A A subunits (2–4).
Fig. 2.
Association of STRIP and CTTNBP2NL with PP2Ac and striatin. Extracts were prepared from pools of cells stably expressing FLAG alone, FLAG-STRIP1, FLAG-CTTNBP2NL, or FLAG-alpha4, and FLAG purification on M2-agarose beads was performed on 300 μg of lysate per condition (whole cell lysate was 25 μg). Proteins were eluted directly in Laemmli sample buffer, separated by SDS-PAGE, transferred to nitrocellulose, and detected by immunoblotting with antibodies to the endogenous striatin (A), PP2Ac (B), alpha4 (C), or FLAG tag (D). The position of the proteins is indicated by an open arrow, and asterisks denote the IgG heavy and light chains. FLAG-tagged CTTNBP2NL is expressed at very low levels, and its position is indicated by an open circle.
Interaction Partners for STRIP1 and CTTNBP2NL—
We next focused on the characterization of proteins associating with STRIP1 and CTTNBP2NL. To increase throughput and potentially enrich for weaker interactors, we utilized a purification strategy using a single N-terminal FLAG epitope. Although this approach yielded a significant increase in background contaminants (32, 58), it also allowed us to obtain significantly increased peptide coverage for each of the proteins in our complex. FLAG-tagged STRIP1 and CTTNBP2NL stably expressed in HEK293 cells (as in Fig. 2) were used for AP-MS, and contaminant proteins were subtracted as described under “Experimental Procedures” to generate an annotated list of interactors (Table II and supplemental Table S5).
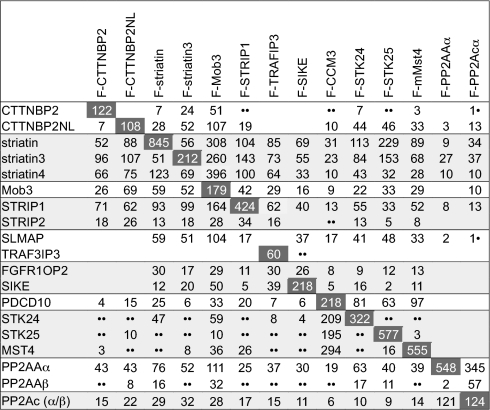
Table II.
Matrix distribution of spectral counts across FLAG pulldowns (columns) for one representative experiment
Hits are listed in rows; horizontal lines separate groups of paralogs. Dark gray shading indicates spectral counts for the bait. A complete table is presented in the supplemental materials. When multiple entry numbers matched the same gene, only the top entry is reported.
• For identifications based on a single unique peptide, a list of the experimental evidence is presented in supplemental Tables S6 and S7; annotated spectra are shown in supplemental Fig. S7.
•• ProteinProphet detected spectra corresponding to this protein, but no spectrum could uniquely be assigned to this entry.
As shown in Table II, purification of the FLAG-tagged mouse STRIP1 protein efficiently recovered PP2A catalytic and scaffolding subunits as well as all three striatins and Mob3, confirming these interactions (Fig. 1). CTTNBP2NL and CTTNBP2 were also detected in STRIP1 AP-MS, indicating that these proteins can be present within the same complex as STRIP1. Additional proteins were also confidently identified by several peptides (Table III and text below for a brief description of these proteins), including SLMAP; two small related proteins, SIKE and FGFR1OP2; members of the GCK-III subfamily of the Ste20 kinases (STK24, STK25, and MST4); and the CCM3 protein. Purification of CTTNBP2NL also recovered all of these proteins (including STRIP1/FAM40A and STRIP2/FAM40B) with the exception of SLMAP, FGFR1OP2, and SIKE (Figs. 3 and 4). A brief description of each of these interactors follows.
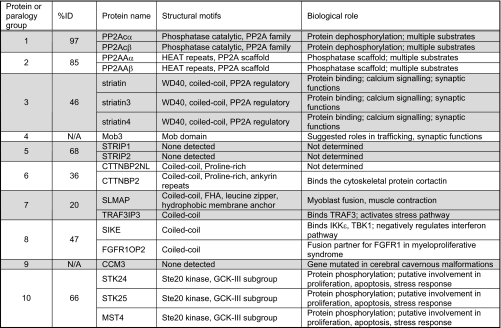
Table III.
STRIPAK components
STRIPAK and associating partners comprise 10 protein families (paralogy groups) and 20 proteins. The percentage of identity between the most distant family members (%ID) is indicated as well as structural motifs and biological functions assigned to each of the proteins. See text and the supplemental material for more details. N/A, not applicable.
Fig. 4.
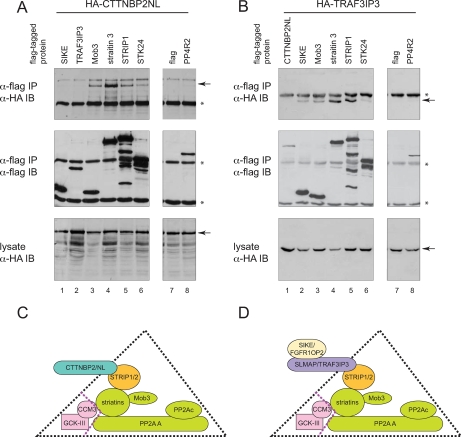
Mutually exclusive associations with STRIPAK. A and B, STRIPAK assembles with CTTNBP2NL or TRAF3IP3 in a mutually exclusive manner. Immunoprecipitation on anti-FLAG-Sepharose beads was performed on lysate from HEK293 cells transiently co-expressing the indicated FLAG- and HA-tagged constructs. To monitor specificity of the interactions, negative controls included FLAG alone as well as FLAG-PP4R2. Immune complexes were resolved by SDS-PAGE followed by transfer to nitrocellulose. Co-precipitation of HA-tagged proteins was detected by immunoblotting (IB) for the HA epitope (top panels; position of the tagged protein is indicated by an arrow. Asterisks indicate the heavy chains.). The precipitated FLAG-tagged protein was detected with anti-FLAG antibodies (middle panels). The expression of the HA-tagged protein in all samples is comparable (bottom panels). C and D, model for mutually exclusive interactions with STRIPAK. Our data suggest that STRIP proteins (orange) associate with the striatins, Mob3, and the phosphatases (green) in a stable complex. Associated with this complex are the CCM3 and GCK-III kinases (pink); these are likely to be more weakly associated as they were not recovered in TAPs. This core complex, STRIPAK, may associate with CTTNBP2 family proteins (C) or with a second complex containing SLMAP and SIKE family proteins (D).
SLMAP was initially identified as a component of the cardiac sarcolemma involved in myoblast fusion and muscle contraction (59–61). Four SLMAP splicing variants have been studied; all encode a putative transmembrane domain toward the C terminus as well as two putative leucine zippers but vary in their N termini: one of the longest isoforms encodes a putative FHA domain and localizes to the centrosome (62).
SIKE is a small coiled coil-containing protein first characterized as an interaction partner for the kinases TBK1 and IKKɛ and as a negative regulator of the interferon pathway (63). SIKE is 50% identical to another co-precipitating protein, FGFR1OP2. SIKE and FGFR1OP2 orthologs have been detected throughout metazoa, but no functions have been ascribed to these proteins in model organisms (supplemental Fig. S4). Interestingly FGFR1OP2 is implicated in the 8p11 myeloproliferative syndrome, a rare and aggressive hematological malignancy characterized by the fusion of unrelated genes to the FGFR1, resulting in the dimerization and activation of FGFR1 in a ligand-independent manner. The FGFR1OP2 fusion involves two coiled-coil domains from FGFR1OP2 that likely promote dimerization of the FGFR1OP2-FGFR1 protein (64).
The CCM3 protein is encoded by the gene PDCD10, mutations of which are found in families with cerebral cavernous malformations (34, 35, 65). Cerebral cavernous malformations are a type of angioma affecting 0.5–1% of the population (66–68). Although most individuals are asymptomatic, roughly 30% will develop symptoms ranging from headaches to seizures, strokes, and even death. The CCM3 protein was demonstrated previously to interact with the GCK-III subgroup of Ste20 family kinases both in yeast two-hybrid assays (where the interaction was first reported) and by co-immunoprecipitation followed by Western blotting (69, 70). Consistent with this, several peptides for these three Ste20 kinases (STK24, STK25, and MST4) were found in association with STRIP1 and CTTNBP2NL in our study. These three kinases define a subgroup of the germinal center kinases (GCK-III) characterized by an N-terminal kinase domain followed by a C-terminal tail of unknown function. The GCK-III kinases have been linked to proliferation, apoptosis, and stress response, but the substrates and molecular mechanism of action remain largely unknown (71–77).
The interaction of STRIP1 and CTTNBP2NL with endogenous MST4 was confirmed by immunoprecipitation/immunoblotting (Fig. 3A); FLAG-alpha4, which was used as a negative control, failed to co-precipitate MST4. We also monitored the interactions among endogenous proteins. Antisera directed against MST4, striatin, and striatin3 (but not anti-PP4R2 or anti-FLAG) all recovered striatin and striatin3 from untransfected HEK293 cells (Fig. 3B), indicating that the interaction detected between the MST4 kinase and striatins is not due to the presence of an epitope tag or mild overexpression. We also subjected the anti-MST4 and anti-striatin3 immunoprecipitates to mass spectrometry and were able to recover many of the binding partners also seen in FLAG AP-MS (supplemental Table S8).
In summary, AP-MS of STRIP1 and CTTNBP2NL confirmed the interactions with PP2Ac, PP2A A, striatins, and Mob3 but also revealed that these proteins are capable of interacting with each other. In addition, SLMAP, SIKE, FGFR1OP2, CCM3, and the GCK-III subfamily of kinases were able to interact with these proteins and are putative components of this complex (Figs. 3 and 4).
Reciprocal Interactions among Complex Components—
To further characterize the many newly detected protein-protein interactions, FLAG-PP2Acα, FLAG-PP2Acβ, FLAG-PP2A Aα, FLAG-striatin, FLAG-striatin3, FLAG-STK24, FLAG-STK25, FLAG-CTTNBP2, and FLAG-SIKE were stably expressed in HEK293 cells for a second round of interaction studies. We could not express SLMAP in HEK293 cells (supplemental Fig. S6 and data not shown) and therefore cloned and expressed the only related family member of SLMAP, namely TRAF3IP3 (supplemental Fig. S5). SLMAP and TRAF3IP3 share 22% identity at the amino acid level (a stretch of 343 amino acids exhibits 28% identity and 50% similarity), and both proteins contain predicted coiled-coil regions. TRAF3IP3, initially cloned as a TRAF3 interactor involved in signaling via the c-Jun N-terminal kinase (JNK) pathway, is normally restricted to cells of lymphoid origin and is not endogenously expressed in HEK293 cells. At the onset of these studies (and because the similarity between SLMAP and TRAF3IP3 is fairly low) we did not know whether TRAF3IP3 would interact with the same partners as SLMAP. With the inclusion of FLAG-TRAF3IP3, the list of baits represents at least one member for each of the paralogous protein families detected in our STRIP1 and CTTNBP2NL AP-MS analyses.
The interaction of each of these proteins with endogenous striatin was confirmed by IP/Western from each of the FLAG-tagged stable cell lines (supplemental Fig. S6 and data not shown). AP-MS for each of the FLAG-tagged proteins was also performed to detect their associations with each other (Table II; a complete list of the MS data is in supplemental Table S5; interactions of some of these proteins with STK24 were reported previously by Ewing et al. (78), but the low density of the data did not allow determination of complex composition). Striatins as well as the Mob3 protein, the PP2A scaffolding subunit, CCM3, and the Ste20 kinases of the GCK-III group co-precipitated every other protein in this large group, indicating that these proteins have the ability to form interactions with each member of the complex. In addition, as shown above and as reported previously, we observed that the striatins can associate with each other; striatin brought down unique peptides for striatin3 and striatin4 and vice versa. The PP2Ac·PP2A A·striatin·Mob3·STRIP·CCM3·GCK-III proteins thus form a large multiprotein complex in vivo that we refer to as the STRIPAK complex.
Interestingly AP of CTTNBP2 (as with CTTNBP2NL above) did not contain detectable amounts of SLMAP, SIKE, or FGFR1OP2. Conversely immunoprecipitates of the SLMAP-related protein TRAF3IP3 did not yield detectable amounts of CTTNBP2 or CTTNBP2NL but did yield large quantities of SIKE and FGFR1OP2. SIKE immunoprecipitates contained a large number of SLMAP and FGFR1OP2 peptides but no detectable CTTNBP2 or CTTNBP2NL peptides. These data suggest that the association of CTTNBP2NL or CTTNBP2 with the remainder of the STRIPAK complex may preclude the association of SLMAP/TRAF3IP3 and SIKE/FGFR1OP2 and vice versa; i.e. these interactions appear to be mutually exclusive. To validate these findings, transient transfections of either an HA-tagged version of CTTNBP2NL or of TRAF3IP3 with a panel of FLAG-tagged proteins (SIKE, Mob3, striatin3, STRIP1, STK24, CTTNBP2NL, TRAF3IP3, PP4R2, or FLAG alone) was performed. Recombinant FLAG-tagged proteins were immunoprecipitated, and the co-precipitation of the HA-tagged proteins was detected by immunoblotting with an antibody against the HA epitope. As expected, both HA-CTTNBP2NL and HA-TRAF3IP3 were efficiently recovered in Mob3, striatin3, STRIP1, and STK24 pulldowns but not in the negative controls (FLAG alone and PP4R2; Fig. 4, A and B). In agreement with our mass spectrometry results, however, HA-CTTNBP2NL was not detectable in SIKE or TRAF3IP3 pulldowns (Fig. 4A, top panel, lanes 1 and 2). Conversely HA-TRAF3IP3 was not detectable in CTTNBP2NL pulldowns but was easily detected in SIKE pulldowns (Fig. 4B, lanes 1 and 2) consistent with the notion that SIKE/FGFR1OP2 and SLMAP/TRAF3IP3 can be present in the same complex but do not co-exist with CTTNBP2NL/CTTNBP2.
Taken together, our data highlight a stable, core complex containing PP2A catalytic and scaffolding subunits, striatin(s), Mob3, and STRIP (Fig. 4, C and D). Because we did not detect CCM3 and the Ste20 kinases in our TAP experiments, they are not likely to be necessary for the formation of this core complex and may be associated with it more loosely to form the STRIPAK complex. The STRIPAK assembly appears to associate either with a member of the CTTNBP2 family or with SLMAP/TRAF3IP3 in combination with SIKE/FGFR1OP2.
DISCUSSION
CCM3 and Its GCK-III Kinase Subfamily Partners Are Physically Associated with PP2A Complexes—
Here we report the identification of a large molecular assembly (STRIPAK) that links the PDCD10 gene product CCM3 and associated GCK-III kinases to a PP2A·striatin·Mob3·STRIP complex. Mutations in the CCM1, CCM2, and CCM3 genes account for nearly all cases of familial cerebral cavernous malformations (68). Previous proteomics studies detected an interaction between CCM3, CCM1, and CCM2, suggesting a common mode of action for all three proteins (70, 79). However, here we demonstrate that (at least in HEK293 cells) CCM3 preferentially associates with GCK-III kinases and our newly defined PP2A·striatin·Mob3·STRIP complex. No CCM1 or CCM2 peptides were detected in our analyses either with CCM3 or with other STRIPAK components. However, we did obtain evidence for an interaction between CCM3 and CCM2 when performing AP-MS using CCM2 (rather than CCM3) as a bait. In that case, FLAG-tagged CCM2 pulldowns yielded numerous peptides for CCM1; yet a small number of CCM3 peptides were also detected (not shown). These data suggest that CCM1 and CCM2 form an abundant complex that likely associates with CCM3 in substoichiometric amounts, whereas a large portion of CCM3 is associated with the STRIPAK complex. Hence we posit that members of the PP2A·striatin·Mob3·STRIP complex should be analyzed for their involvement in non-familial cases of CCM where the genetic basis for the disease is unknown (mutations in CCM1–3 only account for 60% of sporadic cases (68)).
Interactions between GCK-III Kinases and PP2A—
It is intriguing that the STRIPAK complex contains both phosphatase and kinase components. Associations between PP2A and several other kinases have been reported previously. Casein kinase II, the ribosomal S6 protein kinase (S6K1), the calcium-calmodulin-dependent kinase IV (CaMKIV), and the Raf protein were all described as interactors for the catalytic subunit of PP2A (80–83) but were not identified as interaction partners for PP2A in this study. This apparent discrepancy may be due to differences in the biological system used (for example the initial identification of the CaMKIV-PP2A interaction was performed in T cells (80)) or more likely due to the differential sensitivity of the methods of analysis used. Previous studies detected interactions by affinity purification of one component (usually the kinase) followed by immunoblotting for the other factor (PP2A). By contrast, we relied on immunoprecipitation of the PP2A phosphatase coupled to direct MS analysis. Because PP2A is a highly abundant protein and is capable of interacting with multiple binding partners, the fraction of PP2A complexed with a specific protein may be below the detection limits of our method. In this regard, even the interactions of PP2Ac with the Ste20 kinases were not detected in every analysis; co-precipitation of the GCK-III kinases was much more readily detected when starting from striatin or other complex components that reside predominantly in STRIPAK.
The presence of the GCK-III kinases and the PP2A phosphatase within a single complex raises the intriguing question of the enzyme/substrate relationships. Are the kinases regulating the phosphatases or vice versa? Are the substrates for these two enzymes the other STRIPAK components, or are the substrates yet to be identified? Interestingly many of the STRIPAK components are phosphoproteins (16, 70, 76, 84, 85); known phosphopeptides are displayed in supplemental Figs. S1, S2, and S4. In addition, the striatins and the Mob3 protein were previously suggested to be PP2A targets as their phosphorylation (as detected by mobility shift on SDS-PAGE and by 32P labeling) increased following treatment with the PP2A inhibitor okadaic acid (16). Phosphorylation sites were identified on each of the three striatins and are located at amino acids Ser-245, Ser-229, and Ser-206, respectively (86–90). Further studies will be aimed at defining the dependence of these phosphorylation events on the phosphatases and kinases associated to STRIPAK.
What Is the Function of STRIPAK?—
Although our study allowed us to define a macromolecular complex(es) in which a large fraction of the CCM3 protein resides, the function of this complex remains to be elucidated. Several of the complex components are localized to both the cytosol and intracellular membranes, and reports have linked them to intracellular trafficking in mammalian cells. For example, Mob3 (phocein) shares limited homology to clathrin adaptors and can interact with Eps15 (a protein involved in clathrin-mediated endocytosis) and the nucleoside-diphosphate kinase (whose product, GTP, is required for dynamin-dependent synaptic vesicle recycling) (91, 92). Similarly an IP/MS experiment with a peripheral endosome protein, GIPC, recently identified both STRN and STRN3 as binding partners (93). This is significant as GIPC regulates endocytosis and trafficking of the nerve growth factor receptor TrkA.
Interestingly STRIP1/2 orthologs are conserved from yeasts to humans; the budding yeast STRIP ortholog, Far11 (pheromone arrest 11) is a component of a protein complex (Far3–11) implicated in cell cycle arrest following pheromone signaling (94). Some sequence similarity can be detected between an additional Far protein, Far8, and the striatins (although the WD domain is not present in Far8). Limited sequence similarity is also detected between Far9/Vps64 and Far10 with SLMAP. Finally a link between the PP2A A ortholog (Tpd3) and Far11 was detected by both yeast two-hybrid assay (95) and TAP-MS,2 indicating that portions of the STRIPAK assembly are conserved throughout evolution.
Two of the STRIPAK components (STRIP and striatin) have orthologs that have been isolated independently by forward genetics screens in filamentous fungi. The only Sordaria macrospora striatin ortholog, PRO11 regulates fungal cell differentiation and is required for fertility and fruiting body formation (96). The human striatin cDNA can functionally replace the PRO11 gene product, indicating functional conservation (96). The only STRIP ortholog in Neurospora crassa, HAM-2 has also been cloned from a genetic screen: HAM-2 mutants are female sterile, are slow growing, have short hyphae, and have a pronounced defect in hyphal fusion (97). Importantly directed mutations in the striatin homolog HAM-3 and the SLMAP/VPS64 homolog HAM-4, which has an FHA domain, result in strains with a fusion phenotype identical to that of HAM-2 mutants.3 These results indicate that components of the STRIPAK complex share similar functions in mediating hyphal fusion. As hyphal fusion may be a model system to study membrane fusion events, it is tempting to speculate that the STRIPAK complex may affect cell fusion in other systems. Consistent with this possibility, overexpression of SLMAP prevents the myoblast to myotube transition in C2C12 cells without precluding the expression of differentiation markers (61).
Although this study highlighted the composition of STRIPAK, the topology of this large multiprotein complex remains to be defined. For example, the striatins were shown to dimerize (or perhaps multimerize) (Ref. 16 and Tables I and II), raising the possibility that the STRIPAK complex may be asymmetric. The presence of so many paralogous protein species may indicate that subcomplexes containing different paralogs perform slightly different functions. For example, the functions of the three Ste20 kinases found in STRIPAK are likely very different: STK25 and to a lesser extent MST4 but not STK24 have been found associated with the Golgi apparatus and are required for Golgi integrity (84) An in-depth analysis will be required to further elucidate the function of the various paralogs within this complex.
Supplementary Material
Acknowledgments
We are grateful to Dr. N Sonenberg for the kind gift of anti-alpha4 antibody; D. Fermin and G. Liu for programming assistance; and Drs. L. Hood, K. Colwill, and T. Pawson for access to the mammalian gene collection clones. We thank A. Simonin, C. G. Rasmussen, Dr. N. L. Glass, and A. B. Mak for sharing unpublished data. We are grateful to Dr. D. Ceccarelli for critical reading of the manuscript and to Dr. S. Angers for helpful discussions.
Footnotes
Published, MCP Papers in Press, September 8, 2008, DOI 10.1074/mcp.M800266-MCP200
The abbreviations used are: PP2A, protein phosphatase 2A; PP2Ac, PP2A catalytic; STRIPAK, striatin-interacting phosphatase and kinase; STRIP1, striatin-interacting protein 1 (formerly FAM40A); STRIP2, striatin-interacting protein 2 (formerly FAM40B); GCK-III, germinal center kinase, subgroup III; CCM, cerebral cavernous malformation; AP, affinity purification; TAP, tandem affinity purification; SIKE, suppressor of IKKɛ; IKKɛ, IκB kinase ɛ; SLMAP, sarcolemmal membrane-associated protein; IP, immunoprecipitation; IPI, International Protein Index; HA, hemagglutinin; CCT, chaperonin- containing TCP1; FOP, fibroblast growth factor receptor 1 oncogene partner; CTTNBP2, cortactin-binding protein 2; CTTNBP2NL, cortactin-binding protein 2, N-terminal-like; aa, amino acid(s); FHA, forkhead-associated; FGFR1, fibroblast growth factor receptor 1; CaMKIV, calcium-calmodulin-dependent kinase IV; STRN, striatin. See supplemental materials for gene names; LTQ, linear trapquadrupole; XML, eXtensive Markup Language.
A. B. Mak, unpublished data.
A. Simonin, C. G. Rasmussen, and N. L. Glass, personal communication.
This work was supported, in whole or in part, by National Institutes of Health Grant N01-HV-28179 from the NHLBI (to R. A.). This work was also supported by Canadian Institute for Health Research Grants MOP-84314 (to A.-C. G.) and MOP-81268 (to B. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Van Hoof, C., and Goris, J. ( 2004) PP2A fulfills its promises as tumor suppressor: which subunits are important? Cancer Cell 5, 105–106 [DOI] [PubMed] [Google Scholar]
- 2.Janssens, V., Goris, J., and Van Hoof, C. ( 2005) PP2A: the expected tumor suppressor. Curr. Opin. Genet. Dev. 15, 34–41 [DOI] [PubMed] [Google Scholar]
- 3.Virshup, D. M. ( 2000) Protein phosphatase 2A: a panoply of enzymes. Curr. Opin. Cell Biol. 12, 180–185 [DOI] [PubMed] [Google Scholar]
- 4.Mumby, M. ( 2007) PP2A: unveiling a reluctant tumor suppressor. Cell 130, 21–24 [DOI] [PubMed] [Google Scholar]
- 5.Stone, S. R., Hofsteenge, J., and Hemmings, B. A. ( 1987) Molecular cloning of cDNAs encoding two isoforms of the catalytic subunit of protein phosphatase 2A. Biochemistry 26, 7215–7220 [DOI] [PubMed] [Google Scholar]
- 6.Goldberg, Y. ( 1999) Protein phosphatase 2A: who shall regulate the regulator? Biochem. Pharmacol. 57, 321–328 [DOI] [PubMed] [Google Scholar]
- 7.Hemmings, B. A., Adams-Pearson, C., Maurer, F., Muller, P., Goris, J., Merlevede, W., Hofsteenge, J., and Stone, S. R. ( 1990) α- and β-forms of the 65-kDa subunit of protein phosphatase 2A have a similar 39 amino acid repeating structure. Biochemistry 29, 3166–3173 [DOI] [PubMed] [Google Scholar]
- 8.Mumby, M. C., and Walter, G. ( 1993) Protein serine/threonine phosphatases: structure, regulation, and functions in cell growth. Physiol. Rev. 73, 673–699 [DOI] [PubMed] [Google Scholar]
- 9.Xing, Y., Xu, Y., Chen, Y., Jeffrey, P. D., Chao, Y., Lin, Z., Li, Z., Strack, S., Stock, J. B., and Shi, Y. ( 2006) Structure of protein phosphatase 2A core enzyme bound to tumor-inducing toxins. Cell 127, 341–353 [DOI] [PubMed] [Google Scholar]
- 10.Xu, Y., Xing, Y., Chen, Y., Chao, Y., Lin, Z., Fan, E., Yu, J. W., Strack, S., Jeffrey, P. D., and Shi, Y. ( 2006) Structure of the protein phosphatase 2A holoenzyme. Cell 127, 1239–1251 [DOI] [PubMed] [Google Scholar]
- 11.Cho, U. S., and Xu, W. ( 2007) Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature 445, 53–57 [DOI] [PubMed] [Google Scholar]
- 12.Kremmer, E., Ohst, K., Kiefer, J., Brewis, N., and Walter, G. ( 1997) Separation of PP2A core enzyme and holoenzyme with monoclonal antibodies against the regulatory A subunit: abundant expression of both forms in cells. Mol. Cell. Biol. 17, 1692–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murata, K., Wu, J., and Brautigan, D. L. ( 1997) B cell receptor-associated protein alpha4 displays rapamycin-sensitive binding directly to the catalytic subunit of protein phosphatase 2A. Proc. Natl. Acad. Sci. U. S. A. 94, 10624–10629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, J., Peterson, R. T., and Schreiber, S. L. ( 1998) Alpha 4 associates with protein phosphatases 2A, 4, and 6. Biochem. Biophys. Res. Commun. 247, 827–832 [DOI] [PubMed] [Google Scholar]
- 15.Baillat, G., Moqrich, A., Castets, F., Baude, A., Bailly, Y., Benmerah, A., and Monneron, A. ( 2001) Molecular cloning and characterization of phocein, a protein found from the Golgi complex to dendritic spines. Mol. Biol. Cell 12, 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreno, C. S., Lane, W. S., and Pallas, D. C. ( 2001) A mammalian homolog of yeast MOB1 is both a member and a putative substrate of striatin family-protein phosphatase 2A complexes. J. Biol. Chem. 276, 24253–24260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margolis, S. S., Perry, J. A., Forester, C. M., Nutt, L. K., Guo, Y., Jardim, M. J., Thomenius, M. J., Freel, C. D., Darbandi, R., Ahn, J. H., Arroyo, J. D., Wang, X. F., Shenolikar, S., Nairn, A. C., Dunphy, W. G., Hahn, W. C., Virshup, D. M., and Kornbluth, S. ( 2006) Role for the PP2A/B56δ phosphatase in regulating 14-3-3 release from Cdc25 to control mitosis. Cell 127, 759–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forester, C. M., Maddox, J., Louis, J. V., Goris, J., and Virshup, D. M. ( 2007) Control of mitotic exit by PP2A regulation of Cdc25C and Cdk1. Proc. Natl. Acad. Sci. U. S. A. 104, 19867–19872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen, W., Possemato, R., Campbell, K. T., Plattner, C. A., Pallas, D. C., and Hahn, W. C. ( 2004) Identification of specific PP2A complexes involved in human cell transformation. Cancer Cell 5, 127–136 [DOI] [PubMed] [Google Scholar]
- 20.Dagda, R. K., Zaucha, J. A., Wadzinski, B. E., and Strack, S. ( 2003) A developmentally regulated, neuron-specific splice variant of the variable subunit Bβ targets protein phosphatase 2A to mitochondria and modulates apoptosis. J. Biol. Chem. 278, 24976–24985 [DOI] [PubMed] [Google Scholar]
- 21.Gingras, A. C., Gstaiger, M., Raught, B., and Aebersold, R. ( 2007) Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 8, 645–654 [DOI] [PubMed] [Google Scholar]
- 22.Gingras, A. C., Aebersold, R., and Raught, B. ( 2005) Advances in protein complex analysis using mass spectrometry. J. Physiol. 563, 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bauer, A., and Kuster, B. ( 2003) Affinity purification-mass spectrometry. Powerful tools for the characterization of protein complexes. Eur. J. Biochem. 270, 570–578 [DOI] [PubMed] [Google Scholar]
- 24.Dziembowski, A., and Seraphin, B. ( 2004) Recent developments in the analysis of protein complexes. FEBS Lett. 556, 1–6 [DOI] [PubMed] [Google Scholar]
- 25.Chang, I. F. ( 2006) Mass spectrometry-based proteomic analysis of the epitope-tag affinity purified protein complexes in eukaryotes. Proteomics 6, 6158–6166 [DOI] [PubMed] [Google Scholar]
- 26.Vasilescu, J., and Figeys, D. ( 2006) Mapping protein-protein interactions by mass spectrometry. Curr. Opin. Biotechnol. 17, 394–399 [DOI] [PubMed] [Google Scholar]
- 27.Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., Yang, L., Wolting, C., Donaldson, I., Schandorff, S., Shewnarane, J., Vo, M., Taggart, J., Goudreault, M., Muskat, B., Alfarano, C., Dewar, D., Lin, Z., Michalickova, K., Willems, A. R., Sassi, H., Nielsen, P. A., Rasmussen, K. J., Andersen, J. R., Johansen, L. E., Hansen, L. H., Jespersen, H., Podtelejnikov, A., Nielsen, E., Crawford, J., Poulsen, V., Sorensen, B. D., Matthiesen, J., Hendrickson, R. C., Gleeson, F., Pawson, T., Moran, M. F., Durocher, D., Mann, M., Hogue, C. W., Figeys, D., and Tyers, M. ( 2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 [DOI] [PubMed] [Google Scholar]
- 28.Gavin, A. C., Aloy, P., Grandi, P., Krause, R., Boesche, M., Marzioch, M., Rau, C., Jensen, L. J., Bastuck, S., Dumpelfeld, B., Edelmann, A., Heurtier, M. A., Hoffman, V., Hoefert, C., Klein, K., Hudak, M., Michon, A. M., Schelder, M., Schirle, M., Remor, M., Rudi, T., Hooper, S., Bauer, A., Bouwmeester, T., Casari, G., Drewes, G., Neubauer, G., Rick, J. M., Kuster, B., Bork, P., Russell, R. B., and Superti-Furga, G. ( 2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 29.Gavin, A. C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J. M., Michon, A. M., Cruciat, C. M., Remor, M., Hofert, C., Schelder, M., Brajenovic, M., Ruffner, H., Merino, A., Klein, K., Hudak, M., Dickson, D., Rudi, T., Gnau, V., Bauch, A., Bastuck, S., Huhse, B., Leutwein, C., Heurtier, M. A., Copley, R. R., Edelmann, A., Querfurth, E., Rybin, V., Drewes, G., Raida, M., Bouwmeester, T., Bork, P., Seraphin, B., Kuster, B., Neubauer, G., and Superti-Furga, G. ( 2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 30.Krogan, N. J., Cagney, G., Yu, H., Zhong, G., Guo, X., Ignatchenko, A., Li, J., Pu, S., Datta, N., Tikuisis, A. P., Punna, T., Peregrin-Alvarez, J. M., Shales, M., Zhang, X., Davey, M., Robinson, M. D., Paccanaro, A., Bray, J. E., Sheung, A., Beattie, B., Richards, D. P., Canadien, V., Lalev, A., Mena, F., Wong, P., Starostine, A., Canete, M. M., Vlasblom, J., Wu, S., Orsi, C., Collins, S. R., Chandran, S., Haw, R., Rilstone, J. J., Gandi, K., Thompson, N. J., Musso, G., St Onge, P., Ghanny, S., Lam, M. H., Butland, G., Altaf-Ul, A. M., Kanaya, S., Shilatifard, A., O'Shea, E., Weissman, J. S., Ingles, C. J., Hughes, T. R., Parkinson, J., Gerstein, M., Wodak, S. J., Emili, A., and Greenblatt, J. F. ( 2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 31.Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. ( 1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 32.Chen, G. I., and Gingras, A. C. ( 2007) Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods 42, 298–305 [DOI] [PubMed] [Google Scholar]
- 33.Gingras, A. C., Caballero, M., Zarske, M., Sanchez, A., Hazbun, T. R., Fields, S., Sonenberg, N., Hafen, E., Raught, B., and Aebersold, R. ( 2005) A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Mol. Cell. Proteomics 4, 1725–1740 [DOI] [PubMed] [Google Scholar]
- 34.Guclu, B., Ozturk, A. K., Pricola, K. L., Bilguvar, K., Shin, D., O’Roak, B. J., and Gunel, M. ( 2005) Mutations in apoptosis-related gene, PDCD10, cause cerebral cavernous malformation 3. Neurosurgery 57, 1008–1013 [DOI] [PubMed] [Google Scholar]
- 35.Bergametti, F., Denier, C., Labauge, P., Arnoult, M., Boetto, S., Clanet, M., Coubes, P., Echenne, B., Ibrahim, R., Irthum, B., Jacquet, G., Lonjon, M., Moreau, J. J., Neau, J. P., Parker, F., Tremoulet, M., and Tournier-Lasserve, E. ( 2005) Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations. Am. J. Hum. Genet. 76, 42–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedrioli, P. G., Eng, J. K., Hubley, R., Vogelzang, M., Deutsch, E. W., Raught, B., Pratt, B., Nilsson, E., Angeletti, R. H., Apweiler, R., Cheung, K., Costello, C. E., Hermjakob, H., Huang, S., Julian, R. K., Kapp, E., McComb, M. E., Oliver, S. G., Omenn, G., Paton, N. W., Simpson, R., Smith, R., Taylor, C. F., Zhu, W., and Aebersold, R. ( 2004) A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 37.Keller, A., Nesvizhskii, A. I., Kolker, E., and Aebersold, R. ( 2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 38.Nesvizhskii, A. I., Keller, A., Kolker, E., and Aebersold, R. ( 2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 39.Craig, R., and Beavis, R. C. ( 2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 40.MacLean, B., Eng, J. K., Beavis, R. C., and McIntosh, M. ( 2006) General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics 22, 2830–2832 [DOI] [PubMed] [Google Scholar]
- 41.Breitkreutz, B. J., Stark, C., Reguly, T., Boucher, L., Breitkreutz, A., Livstone, M., Oughtred, R., Lackner, D. H., Bahler, J., Wood, V., Dolinski, K., and Tyers, M. ( 2008) The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 36, D 637–D640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou, J., Pham, H. T., and Walter, G. ( 2003) The formation and activity of PP2A holoenzymes do not depend on the isoform of the catalytic subunit. J. Biol. Chem. 278, 8617–8622 [DOI] [PubMed] [Google Scholar]
- 43.Ikehara, T., Shinjo, F., Ikehara, S., Imamura, S., and Yasumoto, T. ( 2006) Baculovirus expression, purification, and characterization of human protein phosphatase 2A catalytic subunits α and β. Protein Expr. Purif. 45, 150–156 [DOI] [PubMed] [Google Scholar]
- 44.Spangler, S. A., and Hoogenraad, C. C. ( 2007) Liprin-α proteins: scaffold molecules for synapse maturation. Biochem. Soc Trans. 35, 1278–1282 [DOI] [PubMed] [Google Scholar]
- 45.Arroyo, J. D., Lee, G. M., and Hahn, W. C. ( 2008) Liprin α1 interacts with PP2A B56γ. Cell Cycle 7, 525–532 [DOI] [PubMed] [Google Scholar]
- 46.Yan, X., Habedanck, R., and Nigg, E. A. ( 2006) A complex of two centrosomal proteins, CAP350 and FOP, cooperates with EB1 in microtubule anchoring. Mol. Biol. Cell. 17, 634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mikolajka, A., Yan, X., Popowicz, G. M., Smialowski, P., Nigg, E. A., and Holak, T. A. ( 2006) Structure of the N-terminal domain of the FOP (FGFR1OP) protein and implications for its dimerization and centrosomal localization. J. Mol. Biol. 359, 863–875 [DOI] [PubMed] [Google Scholar]
- 48.Popovici, C., Zhang, B., Gregoire, M. J., Jonveaux, P., Lafage-Pochitaloff, M., Birnbaum, D., and Pebusque, M. J. ( 1999) The t(6;8)(q27;p11) translocation in a stem cell myeloproliferative disorder fuses a novel gene, FOP, to fibroblast growth factor receptor 1. Blood 93, 1381–1389 [PubMed] [Google Scholar]
- 49.Guasch, G., Delaval, B., Arnoulet, C., Xie, M. J., Xerri, L., Sainty, D., Birnbaum, D., and Pebusque, M. J. ( 2004) FOP-FGFR1 tyrosine kinase, the product of a t(6;8) translocation, induces a fatal myeloproliferative disease in mice. Blood 103, 309–312 [DOI] [PubMed] [Google Scholar]
- 50.Moreno, C. S., Park, S., Nelson, K., Ashby, D., Hubalek, F., Lane, W. S., and Pallas, D. C. ( 2000) WD40 repeat proteins striatin and S/G(2) nuclear autoantigen are members of a novel family of calmodulin-binding proteins that associate with protein phosphatase 2A. J. Biol. Chem. 275, 5257–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Castets, F., Rakitina, T., Gaillard, S., Moqrich, A., Mattei, M. G., and Monneron, A. ( 2000) Zinedin, SG2NA, and striatin are calmodulin-binding, WD repeat proteins principally expressed in the brain. J. Biol. Chem. 275, 19970–19977 [DOI] [PubMed] [Google Scholar]
- 52.Castets, F., Bartoli, M., Barnier, J. V., Baillat, G., Salin, P., Moqrich, A., Bourgeois, J. P., Denizot, F., Rougon, G., Calothy, G., and Monneron, A. ( 1996) A novel calmodulin-binding protein, belonging to the WD-repeat family, is localized in dendrites of a subset of CNS neurons. J. Cell Biol. 134, 1051–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bartoli, M., Ternaux, J. P., Forni, C., Portalier, P., Salin, P., Amalric, M., and Monneron, A. ( 1999) Down-regulation of striatin, a neuronal calmodulin-binding protein, impairs rat locomotor activity. J. Neurobiol. 40, 234–243 [PubMed] [Google Scholar]
- 54.Benoist, M., Gaillard, S., and Castets, F. ( 2006) The striatin family: a new signaling platform in dendritic spines. J. Physiol. (Paris) 99, 146–153 [DOI] [PubMed] [Google Scholar]
- 55.Bailly, Y. J., and Castets, F. ( 2007) Phocein: a potential actor in vesicular trafficking at Purkinje cell dendritic spines. Cerebellum 23, 1–9 [DOI] [PubMed] [Google Scholar]
- 56.Hergovich, A., Cornils, H., and Hemmings, B. A. ( 2008) Mammalian NDR protein kinases: from regulation to a role in centrosome duplication. Biochim. Biophys. Acta 1784, 3–15 [DOI] [PubMed] [Google Scholar]
- 57.Ohoka, Y., and Takai, Y. ( 1998) Isolation and characterization of cortactin isoforms and a novel cortactin-binding protein, CBP90. Genes Cells 3, 603–612 [DOI] [PubMed] [Google Scholar]
- 58.Chen, G. I., Tisayakorn, S., Jorgensen, C., D’Ambrosio, L. M., Goudreault, M., and Gingras, A.-C. ( 2008) PP4R4/KIAA1622 forms a novel stable cytosolic complex with phosphoprotein phosphatase 4. J. Biol. Chem. 283, 29273–29284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wigle, J. T., Demchyshyn, L., Pratt, M. A., Staines, W. A., Salih, M., and Tuana, B. S. ( 1997) Molecular cloning, expression, and chromosomal assignment of sarcolemmal-associated proteins. A family of acidic amphipathic alpha-helical proteins associated with the membrane. J. Biol. Chem. 272, 32384–32394 [DOI] [PubMed] [Google Scholar]
- 60.Wielowieyski, P. A., Sevinc, S., Guzzo, R., Salih, M., Wigle, J. T., and Tuana, B. S. ( 2000) Alternative splicing, expression, and genomic structure of the 3` region of the gene encoding the sarcolemmal-associated proteins (SLAPs) defines a novel class of coiled-coil tail-anchored membrane proteins. J. Biol. Chem. 275, 38474–38481 [DOI] [PubMed] [Google Scholar]
- 61.Guzzo, R. M., Wigle, J., Salih, M., Moore, E. D., and Tuana, B. S. ( 2004) Regulated expression and temporal induction of the tail-anchored sarcolemmal-membrane-associated protein is critical for myoblast fusion. Biochem. J. 381, 599–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guzzo, R. M., Sevinc, S., Salih, M., and Tuana, B. S. ( 2004) A novel isoform of sarcolemmal membrane-associated protein (SLMAP) is a component of the microtubule organizing centre. J. Cell Sci. 117, 2271–2281 [DOI] [PubMed] [Google Scholar]
- 63.Huang, J., Liu, T., Xu, L. G., Chen, D., Zhai, Z., and Shu, H. B. ( 2005) SIKE is an IKK epsilon/TBK1-associated suppressor of TLR3- and virus-triggered IRF-3 activation pathways. EMBO J. 24, 4018–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grand, E. K., Grand, F. H., Chase, A. J., Ross, F. M., Corcoran, M. M., Oscier, D. G., and Cross, N. C. ( 2004) Identification of a novel gene, FGFR1OP2, fused to FGFR1 in 8p11 myeloproliferative syndrome. Genes Chromosomes Cancer 40, 78–83 [DOI] [PubMed] [Google Scholar]
- 65.Craig, H. D., Gunel, M., Cepeda, O., Johnson, E. W., Ptacek, L., Steinberg, G. K., Ogilvy, C. S., Berg, M. J., Crawford, S. C., Scott, R. M., Steichen-Gersdorf, E., Sabroe, R., Kennedy, C. T., Mettler, G., Beis, M. J., Fryer, A., Awad, I. A., and Lifton, R. P. ( 1998) Multilocus linkage identifies two new loci for a mendelian form of stroke, cerebral cavernous malformation, at 7p15–13 and 3q25.2–27. Hum. Mol. Genet. 7, 1851–1858 [DOI] [PubMed] [Google Scholar]
- 66.Plummer, N. W., Zawistowski, J. S., and Marchuk, D. A. ( 2005) Genetics of cerebral cavernous malformations. Curr. Neurol. Neurosci. Rep. 5, 391–396 [DOI] [PubMed] [Google Scholar]
- 67.Revencu, N., and Vikkula, M. ( 2006) Cerebral cavernous malformation: new molecular and clinical insights. J. Med. Genet. 43, 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Labauge, P., Denier, C., Bergametti, F., and Tournier-Lasserve, E. ( 2007) Genetics of cavernous angiomas. Lancet Neurol. 6, 237–244 [DOI] [PubMed] [Google Scholar]
- 69.Ma, X., Zhao, H., Shan, J., Long, F., Chen, Y., Chen, Y., Zhang, Y., Han, X., and Ma, D. ( 2007) PDCD10 interacts with Ste20-related kinase MST4 to promote cell growth and transformation via modulation of the ERK pathway. Mol. Biol. Cell. 18, 1965–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Voss, K., Stahl, S., Schleider, E., Ullrich, S., Nickel, J., Mueller, T. D., and Felbor, U. ( 2007) CCM3 interacts with CCM2 indicating common pathogenesis for cerebral cavernous malformations. Neurogenetics 8, 249–256 [DOI] [PubMed] [Google Scholar]
- 71.Lin, J. L., Chen, H. C., Fang, H. I., Robinson, D., Kung, H. J., and Shih, H. M. ( 2001) MST4, a new Ste20-related kinase that mediates cell growth and transformation via modulating ERK pathway. Oncogene 20, 6559–6569 [DOI] [PubMed] [Google Scholar]
- 72.Osada, S., Izawa, M., Saito, R., Mizuno, K., Suzuki, A., Hirai, S., and Ohno, S. ( 1997) YSK1, a novel mammalian protein kinase structurally related to Ste20 and SPS1, but is not involved in the known MAPK pathways. Oncogene 14, 2047–2057 [DOI] [PubMed] [Google Scholar]
- 73.Dan, I., Ong, S. E., Watanabe, N. M., Blagoev, B., Nielsen, M. M., Kajikawa, E., Kristiansen, T. Z., Mann, M., and Pandey, A. ( 2002) Cloning of MASK, a novel member of the mammalian germinal center kinase III subfamily, with apoptosis-inducing properties. J. Biol. Chem. 277, 5929–5939 [DOI] [PubMed] [Google Scholar]
- 74.Qian, Z., Lin, C., Espinosa, R., LeBeau, M., and Rosner, M. R. ( 2001) Cloning and characterization of MST4, a novel Ste20-like kinase. J. Biol. Chem. 276, 22439–22445 [DOI] [PubMed] [Google Scholar]
- 75.Zhou, T. H., Ling, K., Guo, J., Zhou, H., Wu, Y. L., Jing, Q., Ma, L., and Pei, G. ( 2000) Identification of a human brain-specific isoform of mammalian STE20-like kinase 3 that is regulated by cAMP-dependent protein kinase. J. Biol. Chem. 275, 2513–2519 [DOI] [PubMed] [Google Scholar]
- 76.Pombo, C. M., Bonventre, J. V., Molnar, A., Kyriakis, J., and Force, T. ( 1996) Activation of a human Ste20-like kinase by oxidant stress defines a novel stress response pathway. EMBO J. 15, 4537–4546 [PMC free article] [PubMed] [Google Scholar]
- 77.Pombo, C. M., Force, T., Kyriakis, J., Nogueira, E., Fidalgo, M., and Zalvide, J. ( 2007) The GCK II and III subfamilies of the STE20 group kinases. Front. Biosci. 12, 850–859 [DOI] [PubMed] [Google Scholar]
- 78.Ewing, R. M., Chu, P., Elisma, F., Li, H., Taylor, P., Climie, S., McBroom-Cerajewski, L., Robinson, M. D., O’Connor, L., Li, M., Taylor, R., Dharsee, M., Ho, Y., Heilbut, A., Moore, L., Zhang, S., Ornatsky, O., Bukhman, Y. V., Ethier, M., Sheng, Y., Vasilescu, J., Abu-Farha, M., Lambert, J. P., Duewel, H. S., Stewart, II, Kuehl, B., Hogue, K., Colwill, K., Gladwish, K., Muskat, B., Kinach, R., Adams, S. L., Moran, M. F., Morin, G. B., Topaloglou, T., and Figeys, D. ( 2007) Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 3, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hilder, T. L., Malone, M. H., Bencharit, S., Colicelli, J., Haystead, T. A., Johnson, G. L., and Wu, C. C. ( 2007) Proteomic identification of the cerebral cavernous malformation signaling complex. J. Proteome Res. 6, 4343–4355 [DOI] [PubMed] [Google Scholar]
- 80.Westphal, R. S., Anderson, K. A., Means, A. R., and Wadzinski, B. E. ( 1998) A signaling complex of Ca2+-calmodulin-dependent protein kinase IV and protein phosphatase 2A. Science 280, 1258–1261 [DOI] [PubMed] [Google Scholar]
- 81.Westphal, R. S., Coffee, R. L., Jr., Marotta, A., Pelech, S. L., and Wadzinski, B. E. ( 1999) Identification of kinase-phosphatase signaling modules composed of p70 S6 kinase-protein phosphatase 2A (PP2A) and p21-activated kinase-PP2A. J. Biol. Chem. 274, 687–692 [DOI] [PubMed] [Google Scholar]
- 82.Heriche, J. K., Lebrin, F., Rabilloud, T., Leroy, D., Chambaz, E. M., and Goldberg, Y. ( 1997) Regulation of protein phosphatase 2A by direct interaction with casein kinase 2α. Science 276, 952–955 [DOI] [PubMed] [Google Scholar]
- 83.Abraham, D., Podar, K., Pacher, M., Kubicek, M., Welzel, N., Hemmings, B. A., Dilworth, S. M., Mischak, H., Kolch, W., and Baccarini, M. ( 2000) Raf-1-associated protein phosphatase 2A as a positive regulator of kinase activation. J. Biol. Chem. 275, 22300–22304 [DOI] [PubMed] [Google Scholar]
- 84.Preisinger, C., Short, B., De Corte, V., Bruyneel, E., Haas, A., Kopajtich, R., Gettemans, J., and Barr, F. A. ( 2004) YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3ζ. J. Cell Biol. 164, 1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schinkmann, K., and Blenis, J. ( 1997) Cloning and characterization of a human STE20-like protein kinase with unusual cofactor requirements. J. Biol. Chem. 272, 28695–28703 [DOI] [PubMed] [Google Scholar]
- 86.Munton, R. P., Tweedie-Cullen, R., Livingstone-Zatchej, M., Weinandy, F., Waidelich, M., Longo, D., Gehrig, P., Potthast, F., Rutishauser, D., Gerrits, B., Panse, C., Schlapbach, R., and Mansuy, I. M. ( 2007) Qualitative and quantitative analyses of protein phosphorylation in naive and stimulated mouse synaptosomal preparations. Mol. Cell. Proteomics 6, 283–293 [DOI] [PubMed] [Google Scholar]
- 87.Trinidad, J. C., Thalhammer, A., Specht, C. G., Lynn, A. J., Baker, P. R., Schoepfer, R., and Burlingame, A. L. ( 2008) Quantitative analysis of synaptic phosphorylation and protein expression. Mol. Cell. Proteomics 7, 684–696 [DOI] [PubMed] [Google Scholar]
- 88.Ballif, B. A., Villen, J., Beausoleil, S. A., Schwartz, D., and Gygi, S. P. ( 2004) Phosphoproteomic analysis of the developing mouse brain. Mol. Cell. Proteomics 3, 1093–1101 [DOI] [PubMed] [Google Scholar]
- 89.Villen, J., Beausoleil, S. A., Gerber, S. A., and Gygi, S. P. ( 2007) Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U. S. A. 104, 1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beausoleil, S. A., Villen, J., Gerber, S. A., Rush, J., and Gygi, S. P. ( 2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 91.Baillat, G., Gaillard, S., Castets, F., and Monneron, A. ( 2002) Interactions of phocein with nucleoside-diphosphate kinase, Eps15, and Dynamin I. J. Biol. Chem. 277, 18961–18966 [DOI] [PubMed] [Google Scholar]
- 92.Krishnan, K. S., Rikhy, R., Rao, S., Shivalkar, M., Mosko, M., Narayanan, R., Etter, P., Estes, P. S., and Ramaswami, M. ( 2001) Nucleoside diphosphate kinase, a source of GTP, is required for dynamin-dependent synaptic vesicle recycling. Neuron 30, 197–210 [DOI] [PubMed] [Google Scholar]
- 93.Varsano, T., Dong, M. Q., Niesman, I., Gacula, H., Lou, X., Ma, T., Testa, J. R., Yates, J. R., III, and Farquhar, M. G. ( 2006) GIPC is recruited by APPL to peripheral TrkA endosomes and regulates TrkA trafficking and signaling. Mol. Cell. Biol. 26, 8942–8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kemp, H. A., and Sprague, G. F., Jr. ( 2003) ar3 and five interacting proteins prevent premature recovery from pheromone arrest in the budding yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 23, 1750–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Uetz, P., Giot, L., Cagney, G., Mansfield, T. A., Judson, R. S., Knight, J. R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., Qureshi-Emili, A., Li, Y., Godwin, B., Conover, D., Kalbfleisch, T., Vijayadamodar, G., Yang, M., Johnston, M., Fields, S., and Rothberg, J. M. ( 2000) A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 [DOI] [PubMed] [Google Scholar]
- 96.Poggeler, S., and Kuck, U. ( 2004) A WD40 repeat protein regulates fungal cell differentiation and can be replaced functionally by the mammalian homologue striatin. Eukaryot. Cell 3, 232–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xiang, Q., Rasmussen, C., and Glass, N. L. ( 2002) The ham-2 locus, encoding a putative transmembrane protein, is required for hyphal fusion in Neurospora crassa. Genetics 160, 169–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.