Abstract
Adipokinetic hormone (AKH) is the main hormone involved in the acute regulation of hemolymph lipid levels in several insects. In adult Manduca sexta AKH promotes a rapid phosphorylation of “Lipid storage protein-1”, Lsd1, and a concomitant activation of the rate of hydrolysis of triglycerides by the main fat body lipase. In contrast, in the larval stage AKH modulates hemolymph trehalose levels. The present study describes the sequence of a full length Lsd1 cDNA obtained from M. sexta fat body and investigates a possible link between Lsd1 expression and the distinct effects of AKH in larva and adult insects. The deduced protein sequence showed a high degree of conservation compared to other insect Lsd1s, particularly in the central region of the protein (amino acids 211–276) in which the predicted lipid binding helices are found. Lsd1 was absent in feeding larva and its abundance progressively increased as the insect develops from the non-feeding larva to adult. Contrasting with the levels of protein, Lsd1 transcripts were maximal during the feeding larval stages. The subcellular distribution of Lsd1 showed that the protein exclusively localizes in the lipid droplets. Lsd1 was found in the fat body but it was undetectable in lipid droplets isolated from oocytes or embryos. The present study suggests a link between AKH-stimulated lipolysis in the fat body and the expression of Lsd1.
Keywords: Lipid storage droplet protein, Lsd, fat body, Manduca sexta, lipolysis, lipid mobilization, adipokinetic hormone, AKH
INTRODUCTION
The insect fat body is the main storage site for both lipids, in the form of triacylglycerols (TG), and carbohydrates, as glycogen. These energy reserves are particularly important to meet the energy requirements imposed by the developing embryo, flight and non-feeding periods. The tobacco hornworm, Manduca sexta, feeds constantly during the larval stages of development resulting in the accumulation of large amounts of TG in the fat body. Lipid reserves are mostly used to support the energy requirements of the adult insect (moth) that feeds occasionally (Ziegler 1991; Arrese et al. 2001). Mobilization of the energy reserves is controlled to a great extent by the neuropeptide adipokinetic hormone (AKH) (Gäde and Auerswald 2003). AKHs are small neuropeptides (8–10 amino acids) present in all life stages of insects, and they are stored and secreted from a distinct region of the corpora cardiaca (CC) into the hemolymph (Veelaert et al., 1998). In M. sexta the same AKH peptide is found in the corpora cardiaca from larval and adult insects. However, the function of the hormone is different in the two different life stages. Thus, during the larval stages AKH induces the hydrolysis of glycogen through the activation of glycogen-phosphorylase, whereas in the adult stage it promotes a lipolytic response (TG hydrolysis) (Ziegler et al. 1990). The nature of the cellular changes that accompany the larval-adult stages and modify the AKH response of adipocytes is not known. The present study investigates a possible role of the expression of the fat body protein lipid storage droplet protein 1 (Lsd1) in the distinct metabolic responses elicited by AKH on larval and adult insects. The study of Lsd1 expression is important for several reasons. First, TG accumulates as lipid droplets within the cytoplasm of fat body adipocytes (Willott et al. 1988) and Lsd1 is one of the few lipid droplet specific proteins found in insects (Miura et al. 2002; Teixeira et al. 2003). The lipid droplets are spherical bodies of several microns of diameter composed of a core of TG surrounded by a surface layer of phospholipid and proteins (Murphy 2001; Liu et al. 2004; Beller et al. 2006). The surface layer of the lipid droplet restricts the access of the lipases to the TG core and, thus, it represents a barrier for the hydrolysis and subsequent mobilization of TG. Recent studies have identified Lsd1 as a major player in the lipolytic response promoted by AKH in adult M. sexta (Patel et al. 2005). AKH stimulation of fat body cells produces a rapid activation of cAMP-dependent protein kinase A (PKA), through cAMP levels, and a sustained increase in calcium influx (Arrese et al. 1999). In vivo and in vitro studies have shown that PKA-dependent phosphorylation of Lsd1 is strongly associated with the increase in lipase activity induced by AKH (Patel et al. 2005; Patel et al. 2006; Arrese et al. 2008). Up to 70% of the increase in the lipolytic activity induced by AKH appear to be due to changes in the lipid droplets (Patel et al. 2006) rather than in cytosolic proteins. Most of the lipolytic activity of the fat body is associated to a single lipase, called TG-lipase (TGL), recently identified as the homolog of CG8552 from Drosophila melanogaster (Arrese et al. 2006). TGL is a cytosolic protein that is in vitro phosphorylated, but not activated, by PKA (Patel et al. 2004; Arrese et al. 2006; Patel et al. 2006). Because Lsd1 is the main substrate of PKA action induced by AKH, and Lsd1 phosphorylation activates TG-lipase, it is inferred that Lsd1 is a major regulator of the mechanism of activation of lipolysis in M. sexta. To gain further insights into the possible roles of Lsd1 in the metabolic responses of the fat body we have cloned and sequenced M. sexta Lsd1 and studied its subcellular localization. In this study we also show the developmental changes in the expression of Lsd1 at both mRNA and protein levels. The results are discussed in the context of a possible role of Lsd1 expression as a key factor in defining the metabolic state of the fat body and the function of AKH.
MATERIALS AND METHODS
Materials
[32P]-orthophosphate was purchased from MP Biochemicals (Irvine, CA). Protease and phosphatase inhibitors were purchased from Sigma-Aldrich (St. Louis, MO). M. sexta adipokinetic hormone (AKH) was obtained from Peninsula Laboratories (Belmont, CA). Electrophoresis items were from Invitrogen (Carlsbad,CA). Trypsin sequencing grade was purchased from Promega (Madison, WI). All other chemicals were of analytical grade.
Insects
M. sexta eggs were purchased from Carolina Biological supplies (NC) and larvae were reared on artificial diet (Bell and Joachim 1976). Adult insects were maintained at room temperature without food.
M .sexta Lsd1 cDNA sequencing
Total RNA was isolated from the fat bodies of adult of M. sexta using Trizol reagent (Invitrogen). From total RNA, poly(A)+ RNA was subsequently isolated using Poly(A) Purist MAG (Ambion). mRNA was reversed transcribed using oligod(T)18-primer. The resulting cDNA was used as template in PCR reactions using the following the forward and reverse primers: 1f, 5’-AACCGAACATGCCGAGGCT-3’ and 540r, 5’-TGGTCAGGGGGTAGATACTTG-3’. These primers were designed based on two short sequences (239 and 216 bp, respectively) obtained from a search of 95,458 M. sexta expressed sequence tags (EST) using conserved regions of insect Lsd1s. M. sexta EST were obtained, and kindly provided to us, by Dr Haobo Jiang using parallel pyrosequencing (Zou et al. 2008). A cDNA of approximately 560 bp was amplified, cloned into pCR-II TOPO vector (Invitrogen), and sequenced using the Core Facility ABI Model 3730 DNA Analyzer. To clone the cDNA encoding the Lsd1 protein, we designed gene specific primers to determine the 5’-and 3’-ends of the transcript using RACE PCR. The SMART RACE cDNA Amplification kit (BD Biosciences) was used according to manufacturer’s instructions. 3’-RACE was performed using the specific forward primer 540f, 5’-CGACAAGTATCTACCCCCTGACCAC-3’ with reverse primers provided with the kit. For 5’-RACE the reverse specific primer (135r, 5’-ACCTCCAGCCTCGGCATGTTCGGTTTTTG-3’) and nested primer (125r, 5’-GCATGTTCGGTTTTTGGCTTCGAGTCA-3’) were used with the forward primer and nested forward primer supplied with the SMART RACE cDNA Amplification kit. The 5’-and 3’-RACE PCR products were cloned into the pGEM Easy Vector (Promega) and sequenced in both directions. The continuity of the 5’and 3’-RACE products was determined with gene specific primers performing PCR on ds cDNA prepared as indicated above. The coding region of M. sexta Lsd1 was amplified from cDNA by PCR using the forward and reverse primers 5’-GTGACTCGAAGCCAAAAACCGAACATGCCG-3’ and 5’-GTTCAG CCCGTTGATAGCCGCTAATGCGTC-3’, respectively. The 1119bp PCR product was cloned into the pGEM Easy Vector and the sequence was confirmed in both directions.
RT-PCR expression analysis
For expression analysis of Lsd1 transcripts during development, total RNA was extracted from a pool of dissected fat bodies (n=3) from various stages using the Trizol reagent (Invitrogen). Both the reverse transcription and PCR amplification were performed as “one-step” reaction using OneStep RT-PCR Kit (Qiagen) in a final volume of 50 μl. Two μg of total RNA and Lsd1 specific primers (forward, 5’-CACGCAGTGGAAAAGAGTCA-3’ and reverse, 5’-TGGTCAGGGGGTAGATACTTG- 3’) that were used as a final concentration of 0.6 μM were added to the reaction mixture prepared as manufacturer’s instructions. The simultaneous detection of the transcript of ribosomal protein S3 (MsrpS3) was performed by adding primers specific to rpS3 (forward, 5’-GAGTTTTCAAGGCGGAACTCAATG-3’, and reverse 5’-ACGAACTTCATGGACTTGGCTCTC-3’) that were used as a final concentration of 0.4 μM. Reverse transcription step was done at 50 °C for 30 min, followed by incubation at 95°C for 15 min. Amplification step of the duplex PCR included 22 cycles of 30 s denaturing at 94 °C, 30 s annealing at 56°C, and 1 min extension at 72 °C. The primer concentrations and cycling conditions were optimized in preliminary experiments to avoid saturation. RT-PCR products were separated on 2% agarose gels containing 0.5 μg/ml ethidium bromide and photographed over UV light. The intensity of the bands was determined by densitometry using Un-Scan-It software. The relative level of Lsd1 mRNA was normalized with the transcript of MsrpS3. Two independent sets of total RNA were independently analyzed at least by duplicate.
In vivo protein phosphorylation and subcellular fractionation
Experimental insects were injected with 250μCi of [32P]-orthophosphate, and 90 min later with 100 pmol of AKH. Fat body tissue was dissected 10 min after the hormonal injection. Tissue from two insects was pooled and homogenized with a Potter-Elvehjem glass homogenizer fitted with Teflon pestle, using 6ml of homogenization buffer (HB) (20mM Tris, pH 7.4, 0.25M sucrose, 1mM EDTA, 0.1mM benzamidine, 10mg/l leupeptine, 1mg/l aprotonin, 0.1% 2-mercaptoethanol, 2mM imidazole, 2mM sodium fluoride, 1.5mM sodium molybdate, 1mM sodium orthovanadate, and 4mM sodium potassium tartrate). The homogenate was overlaid with 2ml of buffer without sucrose, and centrifuged (100,000 x g for 1 hr). Three fractions were collected: fat cake, infranatant and pellet. The fat cake was resuspended in HB and sucrose concentration was adjusted to 15% (w/v). A layer of 2 ml buffer without sucrose was laid on top and samples were centrifuged in SW40 rotor at 100,000 x g for 1hr. Purified lipid droplets were collected from the top and resuspended in HB. Typically lipid droplets of two insect fat bodies were resuspended in 0.5 ml of buffer. The pellet was resuspended in 15 ml of HB and re-centrifuged at 100,000 x g for 1hr. The resulting pellet was dissolved in 1ml of HB and centrifuged at 500 x g for 15min and the supernatant was used as the membrane fraction. Subcellular fractions were separated by SDS-PAGE on 10% gels. Proteins were stained with Coomassie Blue. Gel was dried and phosphorylation was visualized by autoradiography.
Peptide mass fingerprinting
Lipid droplet-associated proteins were separated on 10 % SDS-PAGE and the Lsd1 doublet bands visualized by Coomassie staining, excised, minced and destained using 100 % acetonitrile, followed by four washes with water. The gel pieces corresponding to the 42.8kDa and 44.2kDa protein bands were incubated for 20 min in 500 μL of 100 mM ammonium bicarbonate followed by 20 min incubation with 500 μL of 50% acetonitrile in 50 mM ammonium bicarbonate. Gel pieces were dried under vacuum, rehydrated and digested with 50 ng/μL trypsin in 25 mM ammonium bicarbonate at 4°C overnight. Peptides were extracted and analyzed by Maldi-tof mass spectrometry in the Core Facility using the department ABI Voyager De PRO Maldi-MS and α-cyano-4-hydroxycinnamic acid as matrix.
Western blotting
Polyclonal antibodies against Lsd1 were generated in rabbit at Cocalico Biologicals (Reamstown, PA). The antibody was obtained using a mixture of two peptides (KVVHLVNYTHTDLPRC and TYLEHLAIFLAGNEEREKC) that were coupled to KLH. For Western Blotting, proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. The blots were developed with ECL chemiluminescence reagents (Amershan Pharmacia, Piscataway, NJ) and exposed to X-ray films.
Other methods
Protein concentrations were determined by the Bradford dye-binding assay using bovine serum albumin as standard. SDS-PAGE was performed according to Laemmli. The proteins were visualized by Coomassie Brilliant Blue R staining. The deduced amino acid sequence was obtained using the translate tool at ExPASy (http://ca.expasy.org/tools/dna.html). Open reading frame identification was done by using ORF Finder at NCBI (http://www.ncbi.nlm.nih.gov). Predictions of Lsd1 phosphorylation sites were obtained as described (Blom et al. 2004) using NetPhosk 1.0 at (http://www.cbs.dtu.dk/services/NetPhosK).
RESULTS
Cloning and analysis of M. sexta Lsd1 cDNA sequence
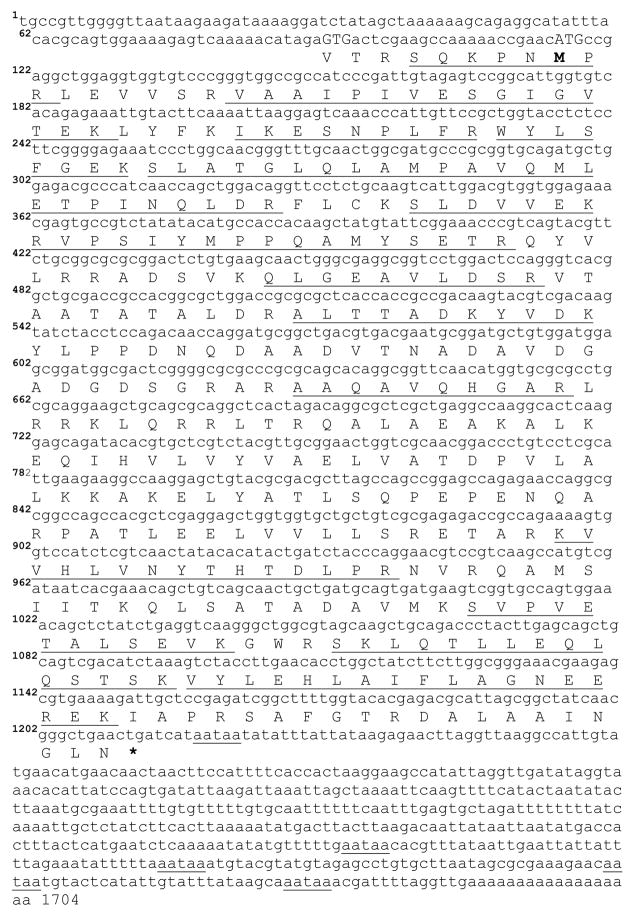
A previous study involving in vivo and in vitro experiments with lipid droplets led to the identification of Lsd1, a lipid droplet associated protein whose level of phosphorylation correlates with the activation of lipolysis (Patel et al. 2005). In this study we have used RT-PCR methods to clone M. sexta (Ms) Lsd1 cDNA using cDNA synthesized from fat body mRNA and primers that were designed as described in methods. The product from this reaction comprised a single band on an agarose gel stained with ethidium bromide and was cloned into pCR-II TOPO vector and sequenced. The sequence of this partial clone - 557 bp- was used to design gene-specific primers for 5’- and 3’- RACE (Frohman et al. 1988) and obtain the 5’-and 3’-ends of Lsd1 cDNA. The full-length coding sequence deduced from these studies is shown in Figure 1. The 5’-RACE reactions produced two products, ~150 and ~200 bp, that were purified and used for a second round of PCR performed with nested primers as indicated in methods. These PCR reactions resulted in single bands that were cloned into the pGEM vector and sequenced. The nucleotide sequence of the shortest product showed that this was a truncated form of the longest product (~ 200bp). On the other hand, 3’-RACE amplified a single product of ~900 bp, which was cloned and sequenced. These studies provided an MsLsd1 cDNA sequence of 1704 bases. The sequence has an open reading frame (ORF) of 1098bp (positions 116–1213) encoding a 365 amino acid protein (Lsd1) with a theoretical mass of 40.4kDa and isoelectric point of 9.18 (Fig 1). Two alternative initiation sites (GTG) located at positions 68 and 92 that use a non-ATG initiation codon (Fig 1) were also identified. In this case the encoded proteins would contain 381 and 373 amino acids in length with theoretical mass of 42.3kDa and 41.3 kDa, respectively. These alternative initiation codons are located upstream and in the same reading frame as the first ATG initiation codon (position 116).
Figure 1. Nucleotide and deduced amino acid sequence of Lsd1 from M. sexta.
cDNA nucleotide (1–1704) is shown above the deduced amino acid sequences (1–373). Amino acid residues are aligned with the first nucleotide of each codon. The stop codon TGA is marked by an asterisk. The amino acid sequence underlined represents the matched peptides obtained from the Maldi-tof analysis of the tryptic digest of the gel excised protein. The possible polyadenylation signals are shown underlined. MsLsd1 cDNA sequence has been deposited to GenBank (Accesion number EU809925).
The 3’non-coding region of MsLsd1 consists of 473 nucleotides and a poly(A) tail of 18 residues. Five copies of a predicted polyadenylation signaling site AATAA were found at positions 15, 45, 111, 155, 485 upstream of the poly(A) tail.
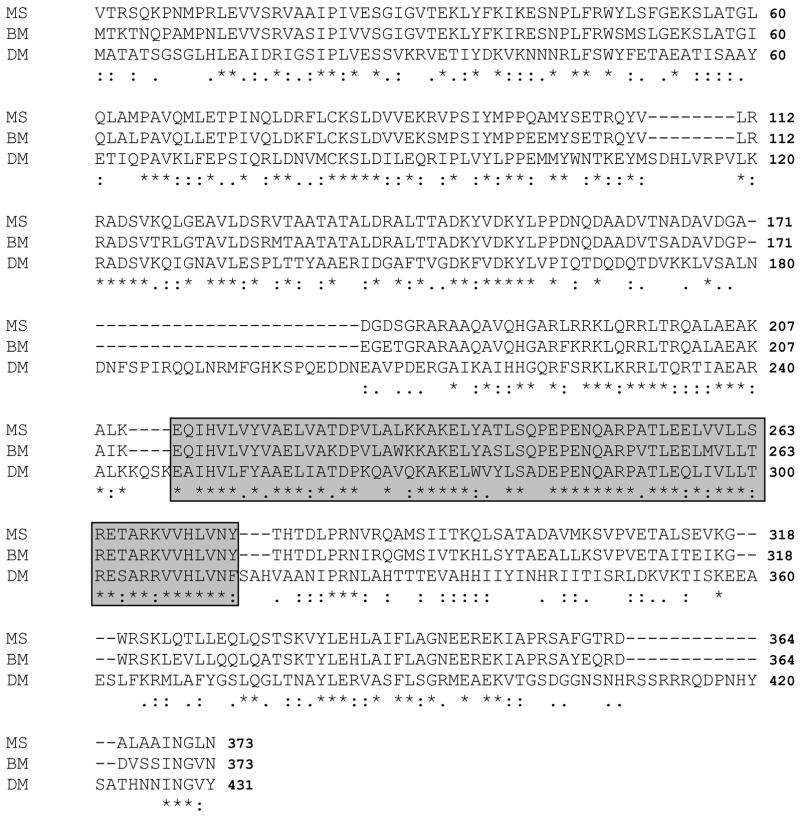
Alignment of MsLsd1 with other insect Lsd1s shows a high degree of conservation (Fig 2 and Supplementary Data), particularly in the central region of the protein (amino acids 211–276) in which the predicted lipid binding helices are found (Arrese et al. 2008).
Figure 2. Alignment of Lsd1 deduced amino acid sequences of M. sexta, B. mori and D. melanogaster.
MS (M. sexta, accession number EU809925), BM (B. mori, accession number NP_001040143) and DM (D. melanogaster, accession number NP_732904). Identical (*) and similar (.) residues are indicated underneath the sequences. The alignment was performed using the program CLUSTALW available at www.ch.emb.net.org. The highly conserved region that is predicted to be involved in lipid binding (Arrese et al., 2008) is framed.
Comparison between the peptide mass fingerprint and deduced amino acid sequence of M. sexta Lsd1
MsLsd1 migrates on a 10% acrylamide-SDS-PAGE as a doublet with apparent masses of 42.8kDa and 44.2kDa (Patel et al. 2005). Maldi-tof analysis of the individual bands of the doublet produced nearly identical spectra (data not shown). The experimentally determined peptide masses obtained from the Maldi-tof spectra of the Lsd1 bands was compared with the theoretical peptides obtained from the deduced amino acid sequence (Fig 1) using FindMod at ExPASy (http://ca.expasy.org/tools/dna.html). Several of the peptides found in the Ms Lsd1 translate were identified in both spectra. The spectra of 42.8 kDa Lsd1 resulted in sixteen matching peptides that are shown underlined in Fig 1. Fourteen of those matching peptides were also identified in the spectra of 44.2 kDa Lsd1. Interestingly a peak with a mass of 957.49, which matched the sequence SQKPNMPR, was observed in the spectra of both Lsd1 isoforms. The SQKPNMPR sequence is absent in the 365 amino acid translate that starts at the ATG initiation codon, position 116 (Fig 1). However, the alternative translates starting on the GTG codons located at positions 68 or 92 contain the SQKPNMPR sequence (Fig 1). Therefore, the comparison of the mass spectrometry data and the deduced amino acid sequence of the 1704 bp transcript suggests that translation of the Lsd1 mRNA could indeed start at the alternative GTG initiation codons.
Subcellular localization of Lsd1. Effect of AKH
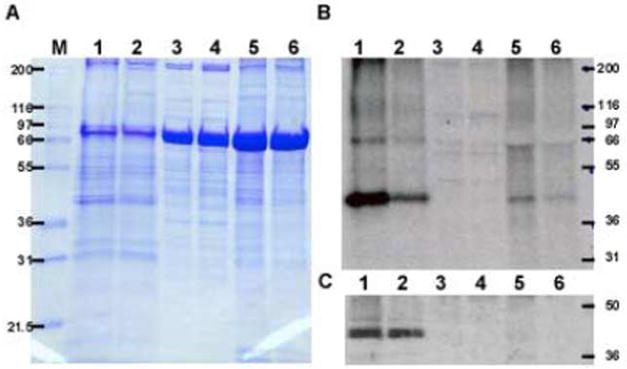
The distribution of Lsd1 among subcellular fractions of fat body cells was investigated. Fat body homogenates prepared with tissue dissected from adult insects under non-stimulated (control) and AKH stimulated conditions were fractionated by ultracentrifugation. The abundance of Lsd1 was determined by immunoblotting using an anti-M.sexta Lsd1 antibody. Possible AKH-dependent changes in the intracellular distribution of Lsd1 were monitored by determining the intensity of phosphorylation of Lsd1 in control and AKH treated insects, as previously described (Patel et al. 2005). Hormonal stimulation was done for 20 min, and non-stimulated tissue provided the level of protein phosphorylation corresponding to basal conditions. Fat body homogenates prepared with tissue from each condition were fractionated into subcellular fractions - cytosol, lipid droplets, and membranes - by centrifugation. Proteins from each subcellular fraction were separated by SDS-PAGE. Coomassie blue stained gels and the corresponding autoradioagram are shown in Fig 3A-B. Confirming previous observations, the autoradiograms of 32P-phosphoproteins of AKH lipid droplets showed the change in the phosphorylation state of Lsd1, the protein that migrates as a close doublet of 42.8/44.2 kDa. The immunoblot analysis shown in Fig 3C showed the occurrence of Lsd1 in the lipid droplets fraction. A comparison between control and stimulated conditions indicated that AKH did not alter the levels of Lsd1 associated to the lipid droplets (Fig 3C).
Figure 3. Subcellular localization of Lsd1 in adult M. sexta in basal and stimulated (AKH) conditions.
[32P]-Fat body homogenates were collected at 0 and 10 min after AKH treatment. Lipid droplets (lanes 1-2), cytosol (lanes 3–4), and membranes (lanes 5–6) were isolated from control and AKH treated fat bodies, and subjected to SDS-PAGE in 10% acrylamide gel. A) Coomassie Blue- stained gel. B) Autoradiogram of the gel shown in A. C) Western blot analysis using anti-Lsd1 polyclonal antibody preparation. Control samples are represented in lanes 2, 4, and 6, whereas AKH samples are represented in lanes 1, 3, and 5, respectively. Approximately 30–35 μg of total protein was loaded into each lane.
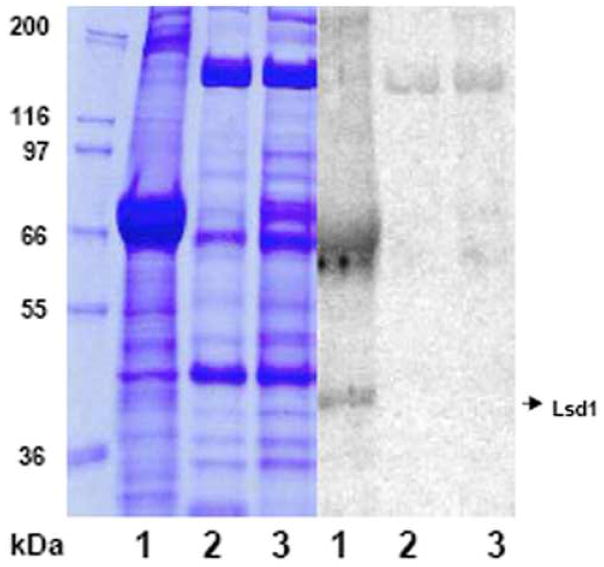
Moreover, Lsd1 was found associated to lipid droplets isolated from the fat bodies regardless of the sex of the insects. Lsd1 occurred in the lipid droplets of both adult males and female insects (Fig 3 and 5) whereas it was undetected in lipid droplets isolated from oocytes and embryos (Fig 5, lane 2 and 3).
Figure 5. SDS-PAGE and immunoblot analysis of Lsd1 level in the lipid droplets purified from adult female.
M. sexta. SDS-PAGE gel stained with Coomassie Blue (A) and immunoblot (B) of M. sexta lipid droplets collected from fat body (lane 1), oocytes (lane 2) and embryos (lane 3). Approximately 50 μg total protein was loaded in two identical gels. The second gel was used for Western blot.
Expression of Lsd1 during development
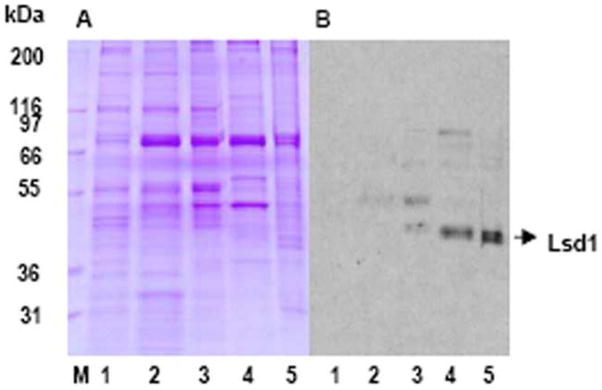
The expression of Lsd1 protein in different developmental stages of M. sexta was investigated by Western blotting. Figure 5 shows a Coomassie blue stained gel and a corresponding Western blot obtained with lipid droplets isolated from fat body of feeding larvae (early and late 5th instar larvae), wandering-5th instar larvae (2nd day), pupae (17th day) and adults (2nd day). As seen in Fig 4, the abundance of Lsd1, relative to total proteins of the lipid droplets, increased during development. Lsd1 was absent in the lipid droplets of the feeding larvae (Fig 4B, lane 1 and 2), whereas reached detectable levels in the non-feeding wandering-5th instar larvae (Fig 4B, lane 3) and attained a maximum level at the adult stage (Fig 4B, lane 5).
Figure 4. SDS-PAGE and immunoblot analysis of Lsd1 protein levels in the lipid droplets purified from fat body of M. sexta during development.
SDS-PAGE gel stained with Coomassie Blue (A) and immunoblot (B) of M. sexta lipid droplets collected from different developmental stages. Lane M) Molecular weight marker; 1) 5th instar (2nd day); 2) 5th instar (5th day); 3) Wanderer (2nd day); 4) Pupa (17th day) and 5) Adult (2nd day). Approximately 30 μg total protein was loaded in two identical gels. The second gel was used for Western blot (B).
Expression of Lsd1 mRNA levels during development
The fat body levels of Lsd1 mRNA to that of rpS3 (control) were determined by RT-PCR. rpS3 is a highly conserved protein component of the small ribosomal subunit (Jiang et al, 1996; Lyamouri et al. 2002). Manduca rpS3 has been previously used as control in studies dealing with fat body expression of proteins (Yu and Kanost, 1999, Jiang et al. 1999). RT-PCR was carried out in one step as described in Material and Methods. PCR reactions were carried out in the linear range of amplification which was determined in preliminary experiments. The relative levels of Lsd1 transcripts were estimated by quantification of gene-specific amplification products separated on agarose gels. RT-PCR analysis throughout development revealed that the transcript is present at all developmental stages, larvae, pupae, and adults (Fig 6). Feeding larvae (Fig 6, lanes 1 and 2) exhibited the highest level of Lsd1 mRNA. These levels decreased significantly in the non-feeding stages (Fig 6, lane 3, 4, and 5) when compared to controls. The difference observed between the relative levels of transcript in feeding larvae was not significant. However, significant differences were found between the non-feeding larvae and either pupae or adult. MsLsd1 transcript was also detected in the fat body of female moths as well as in oocytes (data not shown).
Figure 6. MsLsd1 transcript levels during development.
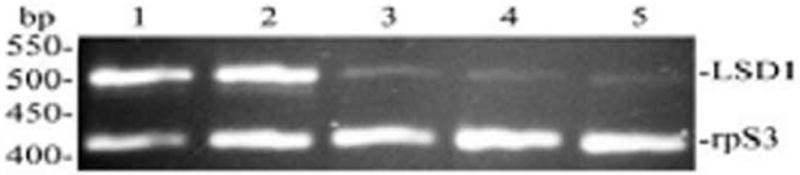
The relative level of the Lsd1 transcript was determined by RT–PCR in a duplex PCR that was performed using primers specific for MsrpS3 (M. sexta ribosomal protein S3) and MsLsd1. Amplification step of the duplex PCR included 22 cycles of 30 s denaturing at 94 °C, 30 s annealing at 56°C, and 1 min extension at 72 °C. The primer concentrations and cycling conditions were optimized in preliminary experiments to avoid saturation. RT-PCR products were separated on 2% agarose gels containing 0.5 μg/ml ethidium bromide and photographed over UV light. A representative gel image is shown. The expected sizes for Lsd1 and rpS3 PCR products are 497 and 415 bp, respectively. Two independent sets of total RNA were independently analyzed at least by duplicate.
DISCUSSION
Structure of MsLsd1
The complete M. sexta Lsd1 cDNA sequence contains a 115-nucleotide 5′ noncoding region, an open reading frame of 1098 nucleotides, and a 3′ untranslated sequence of 491 nucleotides. A comparison of the deduced amino acid sequence with the peptide mass fingerprint of Lsd1 isolated from the lipid droplets produced a significant number of matching peptides masses confirming that the cDNA obtained and sequenced corresponds to the Lsd1 transcript.
Multiple sequence analysis using the deduced protein sequence of MsLsd1 and seven Lsd1 sequences available from other insects showed highly significant similarity among the sequences. As expected, MsLsd1 showed the greatest degree of homology (84.1% identity) with the sequence of B. mori Lsd1, which is a 373-amino acid polypeptide (Fig 2). The high amino acid sequence similarity observed among Lsd1 proteins suggests that this protein has a highly conserved role in insects. Likewise, the protein sequence also contains the PAT domain, which is a partially conserved N-terminal sequence of approximately 100 residues characteristic of proteins found in the lipid droplets from vertebrates (pfam 03036) (Lu et al. 2001). Using predictive methods to estimate the secondary structure (Garnier et al. 1996), it was inferred that MsLsd1 would be a predominately (~60%) α-helical protein. This predicted structure is consistent with the secondary structure of Drosophila melanogaster (Dm) Lsd1 that was recently determined by circular dichroism (Arrese et al. 2008). Highly amphipathic and hydrophobic helical regions located between the residues 248 and 313 of DmLsd1 were proposed as the putative lipid binding motifs of Lsd1. These helical domains are also found in MsLsd1 (amino acids 211–276 in Fig 2) and belong to a highly conserved region (~70% identity, ~90 similarity).
As indicated in Results, MsLsd1 transcript also exhibits non-ATG initiation codons located upstream and in the same reading frame as the first ATG triplet where the translation of the 365-residues Lsd1 would begin. The initiation of translation at the alternative codons would produce proteins of 373 or 381 amino acid residues. In the vast majority of mRNA, translation is initiated at the first ATG codon. However, several cases in which triplets that differ in one nucleotide from ATG, such as ATT, CTG, and GTG, function as translation initiation sites (Kozak 1989a; 1989b; Hann 1994). Non-ATG triplets are generally used in addition to a downstream ATG initiation codon located in the same frame as the non-ATG codons and therefore the production of several isoforms is observed (Takahashi et al. 2005). If this were the case of MsLsd1, slightly different isoforms with calculated molecular masses of 40.4, 41.3 and 42.3kDa could be produced and would explain the origin of the doublet observed by SDS-PAGE. Comparison of the sequences flanking these putative translational start sites with the Drosophila consensus sequence, AAAAAT(C/A)AA(A/C)ATGACC (Cavener 1987) shows that GTG at position 92 and the first ATG located at 116 are in better context to initiate translation (eight out of twelve nucleotides are conserved) than GTG at position 68 in which only three of those nucleotides are conserved. Altogether, the doublet observed in SDS-PAGE, the analysis by mass spectrometry and the features of the DNA sequence suggest that translation of Lsd1 could indeed start at an alternative initiation codon.
The deduced amino acid sequence of MsLsd1 contains five serine residues (Ser41, 55, 94, 116, 321) that are predicted as likely PKA-phosphorylation sites. The alignment of Lsd1 sequences from eight insect species showed that three of them are highly conserved. Ser116 is 100% conserved, whereas Ser41 and Ser321 are conserved in seven out of eight sequences. Ser55 and Ser94 are conserved in half of the sequences available. On the other hand, Ser 20 of Dm Lsd1 is a conserved residue present in most Lsd1 sequences available, including B. mori, but it is absent in M. sexta Lsd1. In a recent study with recombinant Lsd1 from D. melanogaster we showed that PKA mediated phosphorylation of Ser20 triggered TGL activation (Arrese et al. 2008) and, given its conservation, we suggested that it could be involved in activation of the TGL. The sequence of M. sexta Lsd1 suggests that other PKA sites could be involved in activation of the lipolysis. Additional studies to identify the actual MsLsd1 phosphorylation sites involved in the lipolytic response will provide relevant information to the mechanism of activation of lipolysis.
Functional implications of Lsd1 expression and localization
Major behavioral and metabolic differences distinguish the larval and adult stages of M. sexta. Among those differences is the fact that larvae eat constantly and accumulate lipid reserves in the fat body, whereas adult insects consume the lipid stores to support the energy demands imposed by reproduction and flight (Ziegler 1985). A second major difference resides in the effect of AKH, which regulates the utilization of energy reserves in both larvae and adult insects. In larvae AKH activates glycogen-phosphorylase promoting the consumption of fat body glycogen, but it does not affect the fat body lipid content (Siegert and Ziegler 1983; Siegert 1987; 1995). In contrast, in adult M. sexta AKH promotes lipolysis and a two-fold increase in the hemolymph lipid levels (Arrese and Wells 1997). Since both larvae and moths have large amounts of TG stored in the fat body and in both stages of development the fat bodies are responsive to AKH the question is how AKH promotes lipid mobilization only in adult insects. Because TGL, the main lipase of M.sexta fat body (Arrese 2006), is found in both larval and adult fat body (unpublished results) it seems unlikely that TGL alone would have a major role in defining AKH as a lipolytic hormone. However, recent studies have suggested that Lsd1 could be a major player regulating lipid metabolism and AKH action in M. sexta. After the discovery of Lsd1 as lipid droplet protein involved in the AKH-dependent lipolytic response of M. sexta fat body (Patel et al. 2005), it has been shown that activation of the lipid droplets through AKH-induced phosphorylation accounts for ~70% of the activation of the AKH-lipolytic response (Patel et al. 2006) and also that Lsd1 phosphorylation enhances the activity of TGL (Arrese et al. 2008). The present study provides new evidence supporting a major role of Lsd1 in lipid metabolism in M. sexta, including a possible role in defining the action of AKH. We showed that Lsd1 is a protein specific to the lipid droplets whose appearance is detected when the insect is mobilizing TG stores (Fig 3 and 4). The fact that Lsd1 is absent in the feeding larvae (Fig 4) could explain why AKH does not induce lipid mobilization in larvae. In other words, the functional properties of Lsd1 and the fact that it is expressed in adult insects are consistent with the idea that Lsd1 expression could be necessary to allow an AKH-mediated lipolytic response. Further support for this hypothesis comes from the fact that oocytes and embryos, which also accumulate TG as lipid droplets, do not express Lsd1 (Fig 5). A different mechanism of TG hydrolysis seemingly independent of AKH could be regulating the utilization of TG in these compartments.
It is interesting to note that the highest levels of Lsd1 mRNA were found in the feeding larvae. Since the protein is not expressed in feeding larvae, this observation indicates that Lsd1 mRNA is synthesized early on in the development and stored in a translationally silent manner until the transition to the non-feeding wanderer stage. In fact, there seems to be an inverse correlation between protein (Fig 4) and mRNA (Fig 6) levels. The biological significance of this observation is unknown.
Lsd1 shares functional similarities with Perilipin A (PeriA), a member of the PAT family of proteins found in the lipid droplets of adipocytes in vertebrates. PeriA has a dual function on TG metabolism. In the unphosphorylated state PeriA acts as a barrier to TG lipases preventing hydrolysis of TG, whereas in the phosphorylated state it promotes TG hydrolysis (Brasaemle 2007). Lsd1 and Perilipin A have some structural features in common, such as the PAT domain, and also their involvement in activation of the lipolysis. However, the lack of Lsd1 in the lipid droplets of larvae is an indication that Lsd1 is not essential to shield TG from cytosolic lipases and therefore it would not have the dual role described for PeriA.
The present study suggests a link between AKH-stimulated lipolysis in fat body and the expression of Lsd1. Other gene products could be involved in determining the distinct metabolic responses elicited by AKH in larval and adult M. sexta fat bodies. However, the location, expression pattern and biochemical information gathered so far constitute compelling evidence pointing towards Lsd1 as a protein that could be defining the metabolic effect of AKH in M. sexta.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant GM 64677 and Oklahoma Agricultural Experiment Station. The authors are grateful to Elizabeth O’Connell for rearing the insects and providing technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arrese EL, Canavoso LE, Jouni ZE, Pennington JE, Tsuchida K, Wells MA. Lipid storage and mobilization in insects: current status and future directions. Insect Biochem Mol Biol. 2001;31:7–17. doi: 10.1016/s0965-1748(00)00102-8. [DOI] [PubMed] [Google Scholar]
- Arrese EL, Flowers MT, Gazard JL, Wells MA. Calcium and cAMP are second messengers in the adipokinetic hormone-induced lipolysis of triacylglycerols in Manduca sexta fat body. J Lipid Res. 1999;40:556–564. [PubMed] [Google Scholar]
- Arrese EL, Patel RT, Soulages JL. The main triglyceride-lipase from the insect fat body is an active phospholipase A(1): identification and characterization. J Lipid Res. 2006;47:2656–2667. doi: 10.1194/jlr.M600161-JLR200. [DOI] [PubMed] [Google Scholar]
- Arrese EL, Rivera L, Hamada M, Mirza S, Hartson SD, Weintraub S, Soulages JL. Function and structure of lipid storage droplet protein 1 studied in lipoprotein complexes. Arch Biochem Biophys. 2008;473:42–47. doi: 10.1016/j.abb.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese EL, Wells MA. Adipokinetic hormone-induced lipolysis in the fat body of an insect, Manduca sexta: synthesis of sn-1,2-diacylglycerols. J Lipid Res. 1997;38:68–76. [PubMed] [Google Scholar]
- Bell RA, Joachim FG. Techniques for rearing laboratory colonies of tobacco horn worms and pink bolloworms. Ann Entomol Soc Am. 1976;69:365–373. [Google Scholar]
- Beller M, Riedel D, Jansch L, Dieterich G, Wehland J, Jackle H, Kuhnlein RP. Characterization of the Drosophila lipid droplet subproteome. Mol Cell Proteomics. 2006;5:1082–1094. doi: 10.1074/mcp.M600011-MCP200. [DOI] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- Brasaemle DL. The perilipin family of structural lipid droplet proteins: Stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007:R700014–JLR700200. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- Cavener DR. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987;15:1353–1361. doi: 10.1093/nar/15.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gade G, Auerswald L. Mode of action of neuropeptides from the adipokinetic hormone family. Gen Comp Endocrinol. 2003;132:10–20. doi: 10.1016/s0016-6480(03)00159-x. [DOI] [PubMed] [Google Scholar]
- Garnier J, Gibrat JF, Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- Hann SR. Regulation and function of non-AUG-initiated proto-oncogenes. Biochimie. 1994;76:880–886. doi: 10.1016/0300-9084(94)90190-2. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Kanost MR. Primary structure of ribosomal proteins S3 and S7 from Manduca sexta. Insect Mol Biol. 1996;5:31–36. doi: 10.1111/j.1365-2583.1996.tb00038.x. [DOI] [PubMed] [Google Scholar]
- Jiang H, Wang Y, Kanost MR. Four serine proteinases expressed in Manduca sexta haemocytes. Insect Mol Biol. 1999;8:39–53. doi: 10.1046/j.1365-2583.1999.810039.x. [DOI] [PubMed] [Google Scholar]
- Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol Cell Biol. 1989a;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol. 1989b;9:5134–5142. doi: 10.1128/mcb.9.11.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamouri M, Enerly E, Lambertsson A. Organization, sequence, and phylogenetic analysis of the ribosomal protein S3 gene from Drosophila virilis. Gene. 2002;294:147–156. doi: 10.1016/s0378-1119(02)00763-1. [DOI] [PubMed] [Google Scholar]
- Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RG. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem. 2004;279:3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- Lu X, Gruia-Gray J, Copeland NG, Gilbert DJ, Jenkins NA, Londos C, Kimmel AR. The murine perilipin gene: the lipid droplet-associated perilipins derive from tissue-specific, mRNA splice variants and define a gene family of ancient origin. Mamm Genome. 2001;12:741–749. doi: 10.1007/s00335-01-2055-5. [DOI] [PubMed] [Google Scholar]
- Miura S, Gan JW, Brzostowski J, Parisi MJ, Schultz CJ, Londos C, Oliver B, Kimmel AR. Functional conservation for lipid storage droplet association among Perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J Biol Chem. 2002;277:32253–32257. doi: 10.1074/jbc.M204410200. [DOI] [PubMed] [Google Scholar]
- Murphy DJ. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res. 2001;40:325–438. doi: 10.1016/s0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- Patel R, Soulages JL, Wells MA, Arrese EL. cAMP-dependent protein kinase of Manduca sexta phosphorylates but does not activate the fat body triglyceride lipase. Insect Biochem Mol Biol. 2004;34:1269–1279. doi: 10.1016/j.ibmb.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Patel RT, Soulages JL, Arrese EL. Adipokinetic hormone-induced mobilization of fat body triglyceride stores in Manduca sexta: role of TG-lipase and lipid droplets. Arch Insect Biochem Physiol. 2006;63:73–81. doi: 10.1002/arch.20143. [DOI] [PubMed] [Google Scholar]
- Patel RT, Soulages JL, Hariharasundaram B, Arrese EL. Activation of the lipid droplet controls the rate of lipolysis of triglycerides in the insect fat body. J Biol Chem. 2005;280:22624–22631. doi: 10.1074/jbc.M413128200. [DOI] [PubMed] [Google Scholar]
- Siegert K, Ziegler R. A Hormone From The Corpora Cardiaca Controls Fat-Body Glycogen-Phosphorylase During Starvation In Tobacco Hornworm Larvae. Nature. 1983;301:526–527. [Google Scholar]
- Siegert KJ. Carbohydrate-Metabolism In Starved 5th Instar Larvae Of Manduca-Sexta. Archives Of Insect Biochemistry And Physiology. 1987;4:151–160. [Google Scholar]
- Siegert KJ. Carbohydrate-Metabolism During The Pupal Molt Of The Tobacco Hornworm, Manduca-Sexta. Archives Of Insect Biochemistry And Physiology. 1995;28:63–78. [Google Scholar]
- Takahashi K, Maruyama M, Tokuzawa Y, Murakami M, Oda Y, Yoshikane N, Makabe KW, Ichisaka T, Yamanaka S. Evolutionarily conserved non-AUG translation initiation in NAT1/p97/DAP5 (EIF4G2) Genomics. 2005;85:360–371. doi: 10.1016/j.ygeno.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Teixeira L, Rabouille C, Rorth P, Ephrussi A, Vanzo NF. Drosophila Perilipin/ADRP homologue Lsd2 regulates lipid metabolism. Mech Dev. 2003;120:1071–1081. doi: 10.1016/s0925-4773(03)00158-8. [DOI] [PubMed] [Google Scholar]
- Veelaert D, Schoofs L, De Loof A. Peptidergic control of the corpus cardiacum–corpora allata complex of locusts. Int Rev Cytol. 1998;182:249–302. doi: 10.1016/s0074-7696(08)62171-3. [DOI] [PubMed] [Google Scholar]
- Willott E, Bew LK, Nagle RB, Wells MA. Sequential structural changes in the fat body of the tobacco hornworm, Manduca sexta, during the fifth larval stadium. Tissue Cell. 1988;20:635–643. doi: 10.1016/0040-8166(88)90065-1. [DOI] [PubMed] [Google Scholar]
- Yu X, Kanost MR. Development expression of Manduca sexta hemolin. Arch Insect Biochem Physiol. 1999;42:198–212. doi: 10.1002/(SICI)1520-6327(199911)42:3<198::AID-ARCH4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Ziegler R. Metabolic energy expenditure and its regulation. Springer; Berlin: 1985. pp. 95–118. [Google Scholar]
- Ziegler R. Changes in lipid and carbohydrate metabolism during starvation in adult Manduca sexta. J Comp Physiol [B] 1991;161:125–131. doi: 10.1007/BF00262874. [DOI] [PubMed] [Google Scholar]
- Ziegler R, Eckart K, Law JH. Adipokinetic hormone controls lipid metabolism in adults and carbohydrate metabolism in larvae of Manduca sexta. Peptides. 1990;11:1037–1040. doi: 10.1016/0196-9781(90)90030-9. [DOI] [PubMed] [Google Scholar]
- Zou Z, Najar F, Wang Y, Roe B, Jiang H. Pyrosequence analysis of expressed sequence tags for Manduca sexta hemolymph proteins involved in immune responses. Insect Biochem Mol Biol. 2008;38:677–682. doi: 10.1016/j.ibmb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.