Abstract
Bacterial toxin-antitoxin protein pairs (TA pairs) encode a toxin protein, which poisons cells by binding and inhibiting an essential enzyme, and an antitoxin protein, which binds the toxin and restores viability. We took an approach that did not rely on sequence homology to search for unidentified TA pairs in the genome of Escherichia coli K-12. Of 32 candidate genes tested, ectopic expression of 6 caused growth inhibition. In this report, we focus on the initial characterization of yeeV, ykfI, and ypjF, a novel family of toxin proteins. Coexpression of the gene upstream of each toxin restored the growth rate to that of the uninduced strain. Unexpectedly, we could not detect in vivo protein-protein interactions between the new toxin and antitoxin pairs. Instead, the antitoxins appeared to function by causing a large reduction in the level of cellular toxin protein.
Bacterial toxin-antitoxin pairs (TA pairs) consist of a stable toxin protein that can cause cell death by disrupting an essential cellular process, coupled with a labile antitoxin protein that can bind to and block activity of the toxin (16, 18). Also known as addiction modules, TA pairs were first identified on plasmids and characterized for their role in postsegregational killing (39). After cell division, daughter cells that do not inherit a copy of a plasmid expressing an addiction module can no longer produce antitoxin protein. Hence, following rapid degradation of residual antitoxin protein, the remaining excess toxin protein is free to bind and inhibit the cellular target, providing a selection mechanism for plasmid maintenance in the population.
More recently, several chromosomal genes similar to plasmid-borne addiction modules have been identified (19, 21, 26, 29). The cellular function of these chromosomally encoded TA pairs has not been clearly defined. Two models have been suggested by studies of proteins involved in the Escherichia coli stringent response (11). One hypothesis, stemming from investigation of mazE/mazF (chpAI/chpAK), a pair of genes located in the relA operon, is that genomic TA pairs also function as addiction modules (2). Global gene expression is down regulated in response to amino acid starvation as part of the stringent response (11). In a manner analogous to postsegregational killing, this transcriptional attenuation could result in the inability to replace the rapidly degraded antitoxin MazE, leading to MazF-mediated cell death (2, 16). It has been proposed that, under some circumstances, it may be evolutionarily advantageous for a fraction of cells to undergo programmed cell death in order to provide nutrients for the remainder of the population (25). Alternatively, examination of the toxin-antitoxin gene pair relB/relE (12, 17, 19, 31, 32) supports the model that, unlike plasmid-based toxins, the function of the chromosomal TA pairs is not bacterial “apoptosis” but to modulate the rate of metabolic processes in response to environmental stress (18). Mutant alleles of relB confer a defect in the stringent response termed a delayed relaxed phenotype, in which cells are unable to efficiently resume protein synthesis after readdition of amino acids (4, 15, 24). Excess RelE protein has been shown to result in a decrease in the rate of protein synthesis both in vitro and in vivo, and this inhibition is neutralized by the addition of the antitoxin RelB (12, 31). Furthermore, recent evidence suggested that the mechanism of protein synthesis inhibition by RelE is via sequence-specific cleavage of mRNA in the ribosomal A site, with preference for the stop codon UAG (32). The ability of RelB and RelE to reversibly inhibit translation may provide a mechanism to slow the rate of translation in response to nutrient deprivation. Although the precise mechanism of toxicity has not been reported for MazF, overexpression of MazF disrupts both translation and replication in a manner that is reversible by MazE, suggesting this TA pair could also play a similar role in stress adaptation (18, 31).
The cellular target and mechanism of toxicity for the majority of toxin proteins are unknown. However, where a target has been identified, the toxin has been shown to function by interacting with essential proteins known or suspected to be good targets for antimicrobial agents. For example, CcdB inhibits DNA gyrase activity (6, 28), and PemK is thought to poison cells via the replicative helicase DnaB (33). Therefore, identification and study of additional members of this class of proteins may help validate novel targets for antimicrobial therapy or provide insight into novel mechanisms of bacterial cell death.
We used an approach that did not rely upon sequence homology to investigate whether additional TA protein pairs are present in the E. coli genome. All pairs of genes in the annotated genome of E. coli K-12 (7) that fit the highly conserved size and genetic organization of known TA pairs were identified. Of 32 genes tested, ectopic expression of 6 genes resulted in growth inhibition and/or a reduction in the number of CFU. Paired antitoxin function was demonstrated for several of these, as defined by the ability to restore normal growth upon coexpression. In this report, we describe the initial characterization of a novel family of three homologous TA pairs.
MATERIALS AND METHODS
Identification of candidate toxin genes.
A database containing several physical parameters for the proteins encoded by all predicted open reading frames (ORFs) in the E. coli genome (G. Schoenhals and K. J. Shaw, unpublished data) was used to identify all proteins 65 to 135 amino acids (aa) in length. The genes encoding these proteins were sorted by chromosomal position and scanned for pairs of ORFs that fell into the desired size range and were predicted to be in the same operon (intergenic space of ≤150 bp). The nomenclature and identification of all of the hypothetical E. coli genes identified via genomic sequencing and used in this study are found at the following websites: http://genolist.pasteur.fr/Colibri/ and http://bmb.med.miami.edu/EcoGene/EcoWeb/.
Subcloning of genes of interest.
The predicted ORF (start codon to stop codon) for each gene of interest was amplified by PCR from MG1655 genomic DNA. The 5′ end of upstream primers incorporated an EcoRI site (italics) as well as a consensus ribosome-binding site (bold), followed by 18 to 22 bp of gene sequence starting with ATG (e.g., GAATTC GGAGTGAAACG ATG…). The 5′ end of each downstream primer contained an XbaI site. After amplification, each PCR product was digested with EcoRI and XbaI (New England Biolabs, Inc., Beverly, Mass.) and ligated into one or both of the arabinose-inducible expression vectors pBAD18 (ampicillin resistance; high copy, ∼100 to 300 copies/cell) or pBAD33 (chloramphenicol resistance; low copy, ∼15 copies/cell) that had been digested with EcoRI and XbaI (20). Subcloning into pBAD33 required partial digestion with EcoRI due to the presence of a second cut site in the chloramphenicol cassette. Ligation mixtures were transformed into chemically competent E. coli TOP10 cells (Invitrogen Corporation, Carlsbad, Calif.), selecting for the appropriate plasmid-borne drug resistance markers. Positive clones were verified by DNA sequencing.
Toxicity assays.
Overnight cultures were diluted 100-fold in fresh medium and grown to log phase (optical density at 600 nm [OD600] = 0.4 to 0.5), then rediluted to OD600 = 0.01 in fresh medium with or without 0.2% l-(+)-arabinose (Sigma, St. Louis, Mo.). Optical density was monitored using a Spectronic 20D+; when the OD600 was ≥1.0, cultures were diluted 10-fold and reread to stay within the accurate range of the instrument. To quantitate CFU, cells were diluted in phosphate-buffered saline (pH 7.1; Invitrogen Corporation), plated on Luria-Bertani (LB) agar plus 100 μg of ampicillin/ml, and incubated overnight at 37°C. All cultures were grown at 37°C in LB plus 100 μg of ampicillin/ml or 30 μg of chloramphenicol/ml, as appropriate, with shaking (∼225 rpm).
Coexpression constructs.
Genes that are adjacent on the E. coli chromosome were amplified directly from MG1655 genomic DNA with the same primers as described above and ligated into pBAD18. Others were constructed using PCR-SOEing (22). As in the single-gene constructs, the upstream primer contained a consensus ribosome-binding site.
Cluster analysis.
Multiple sequence alignments were done using ClustalW version 1.8 (36). Formatting was done with BOXSHADE (http://www.ch.embnet.org/software/BOX_form.html).
Examination of toxin protein and mRNA levels.
Overnight cultures were diluted 100-fold into fresh LB plus ampicillin (100 μg/ml), grown to OD600 = 0.5, and then split into new tubes containing LB plus ampicillin (100 μg/ml) with or without 0.2% arabinose. After 30 min, the optical density was monitored and 0.5 ml of each culture was pelleted (5,000 × g for 5 min at 4°C). Supernatant was removed, and the pellet was dissolved in 100 μl of B-PER (Pierce Biotechnology Inc., Rockford, Ill.) plus 1× NuPAGE lithium dodecyl sulfate sample buffer (Invitrogen Corporation) per 1.0 OD600 unit. A 3.0-μl aliquot of each sample was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using a NuPage 12% bis-Tris gel with morpholineethanesulfonic acid running buffer (Invitrogen Corporation) and transferred to polyvinylidene difluoride membrane. Western blotting was performed following the manufacturer's instructions using purified murine monoclonal His6 antibody (Covance Inc., Princeton, N.J.) and anti-FLAG M2 monoclonal antibody (Sigma). Total RNA was isolated from equivalent samples using the Ambion NorthernMax kit according to the manufacturer's instructions (Ambion Inc., Austin, Tex.). Ten micrograms of RNA was used for each sample. Insert DNA isolated after digestion of each expression construct with EcoRI and XbaI was used as a template for synthesis of 32P-labeled DNA probes using the Radprime DNA labeling system (Invitrogen Corporation).
RESULTS
Identification of toxin genes.
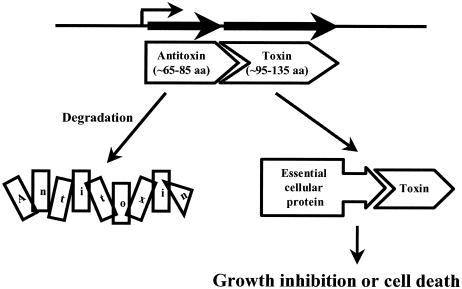
We used a method to identify genes encoding putative toxins that did not rely on sequence homology to known proteins. Most previously characterized TA pairs are cotranscribed and consist of a gene encoding a small (∼65- to 85-aa) antitoxin protein adjacent to a gene encoding a slightly larger (∼95- to 135-aa) toxin protein (Fig. 1). We identified 18 pairs of genes in the annotated genomic DNA sequence of E. coli K-12 (MG1655) that fit these size criteria and were linked closely enough to potentially be within the same operon (≤150 bp apart) (pspB-pspC, pspD-pspE, yafN-yafO, yeeT-yeeU, yeeU-yeeV, ybdJ-ybdF, ykfH-yafW, yafW-ykfI, ybgE-ybgC, ynaK-ydaY, yebG-yebF, yffM-yffN, yfhN-yfhF, ypjJ-yfjZ, yfjZ-ypjF, ygfY-ygfX, yheL-yheM, and yheM-yheN).
FIG. 1.
Characteristics and genetic organization of bacterial TA pairs.
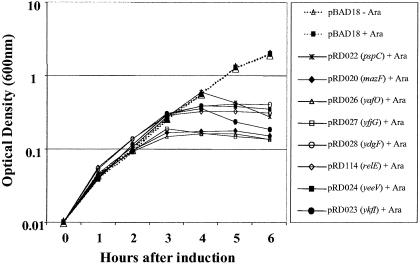
To determine which, if any, of the candidate genes encode toxin proteins, the predicted ORF for each of the genes listed above was amplified by PCR from E. coli MG1655 genomic DNA and subcloned into the arabinose-inducible expression vector pBAD18 (20) (Table 1). To assay for toxicity, log-phase cultures of E. coli TOP10 cells transformed with each expression construct were diluted to OD600 = 0.01 in fresh LB plus ampicillin (100 μg/ml) with or without arabinose, and the optical density was monitored over the course of 6 h. Six genes were identified whose overexpression resulted in significant inhibition of growth compared to the strain carrying the parent vector (Fig. 2). For five of the overexpressing strains, the growth rate diverged from that of the control strain after 3 h of induction, similar to that seen for strains overproducing RelE or MazF. Growth of the PspC overexpression strain was similar to that of the parent vector control for 4 h before slowing significantly.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Insert (description) | Source or reference |
|---|---|---|
| E. coli strains | ||
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacZ74 deoR recAI araD139 Δ(ara-leu)7697 galU galK rpsL endAI rupG | Invitrogen |
| MG1655 | F− λ−ilvG rfb-50 rph-1 | 7 |
| Plasmids and constructs | ||
| pBAD18 | None | 20 |
| pRD020 | mazF | This study |
| pRD022 | pspC | This study |
| pRD023 | ykfI | This study |
| pRD024 | yeeV | This study |
| pRD025 | ydjM | This study |
| pRD026 | yafO | This study |
| pRD027 | yfjG | This study |
| pRD028 | ydgF | This study |
| pRD029 | yafW | This study |
| pRD030 | yeeU | This study |
| pRD041 | ykfI-His6 | This study |
| pRD042 | yeeV-His6 | This study |
| pRD049 | yafW-ykfI | This study |
| pRD050 | yeeU-yeeV | This study |
| pRD052 | ypjF | This study |
| pRD061 | yafW-ykfI-His6 | This study |
| pRD062 | yeeU-yeeV-His6 | This study |
| pRD068 | ykfH | This study |
| pRD069 | yeeT | This study |
| pRD086 | yfjZ | This study |
| pRD095 | yeeU-BamHI-yeeV | This study |
| pRD103 | ypjJ | This study |
| pRD104 | yeeT-yeeV | This study |
| pRD105 | gfp-yeeV | This study |
| pRD106 | ykfH-ykfI | This study |
| pRD107 | gfp-ykfI | This study |
| pRD114 | relE | This study |
| pRD124 | gfp-ykfI-His6 | This study |
| pRD125 | ypjF-His6 | This study |
| pRD134 | UTR-yeeV | This study |
| pRD135 | yeeU-UTR-BamHI-yeeV | This study |
| pRD136 | Flag-ypjF | This study |
| pRD139 | yafW-BamHI-ykfI | This study |
| pRD140 | gfp-yeeV-His6 | This study |
| pRD141 | yeeU-BamHI-ykfI | This study |
| pRD142 | yafW BamHI-yeeV | This study |
| pBAD33 | None | 20 |
| pRD072a | yeeV | This study |
Low-copy-number pBAD33-based construct. All others described are high-copy pBAD18-based constructs.
FIG. 2.
Ectopic expression of six candidate toxin genes causes growth inhibition. Log-phase cultures (OD600 = 0.5) were diluted in LB plus ampicillin (100 μg/ml) liquid medium to OD600 = 0.01 and grown in the presence or absence of 0.2% arabinose. The known toxin genes relE and mazF were included as positive controls.
yeeV, ykfI, and ypjF define a novel toxin gene family.
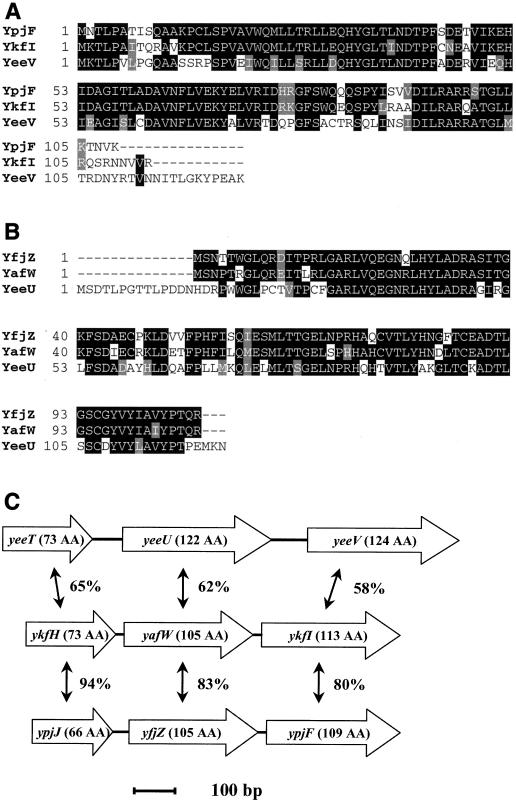
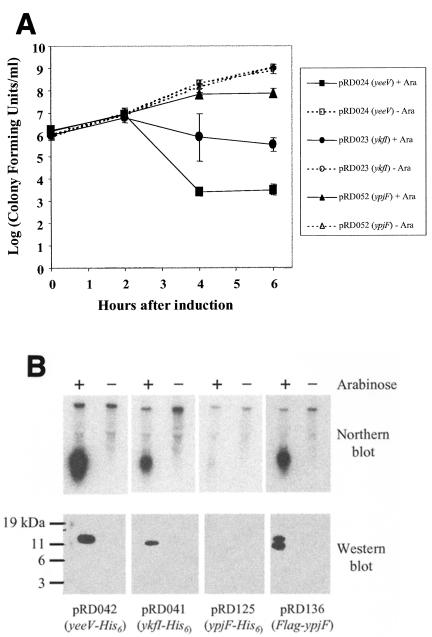
In this study, we initially focused our investigation on two of the six genes identified, yeeV and ykfI. These genes encode small proteins (124 and 113 aa, respectively) that share 58% aa sequence identity. Surprisingly, arabinose induction of a third highly similar gene, ypjF, did not cause growth inhibition in our assay, even though the YpjF protein is 80% identical to the YkfI protein (Fig. 3A and C). Quantitation of bacterial titers indicated that in addition to inhibiting growth, expression of yeeV or ykfI also caused a reduction in the number of CFU (Fig. 4A). After increasing at the same rate as the uninduced control culture for the first 2 h after induction, the viability of cells expressing ykfI decreased to approximately half of that at time zero, whereas the uninduced cells continued to grow exponentially. CFU of the yeeV-expressing strain decreased ∼500-fold from time zero.
FIG. 3.
YeeV, YkfI, and YpjF define a novel family of proteins. (A) Amino acid sequence alignment of the toxins YeeV, YkfI, and YpjF. Identical residues are highlighted in black, and chemically conserved residues are highlighted in gray. (B) Amino acid sequence alignment of the gene upstream of each toxin. (C) Chromosomal organization of yeeV, ykfI, ypjF, and the two proximal genes for each. The percent amino acid sequence identity for each homolog is indicated.
FIG. 4.
(A) Viability of strains expressing yeeV, ykfI, and ypjF. Growth in the presence (solid lines) or absence (dashed lines) of 0.2% arabinose was performed as described in the legend for Fig. 2. To quantitate CFU per milliliter, cells were diluted and plated on LB agar plus 100 μg of ampicillin/ml at the times indicated. (B) Analysis of mRNA and protein concentration for strains expressing yeeV-His, ykfI-His, ypjF-His, or FLAG-ypjF. Equivalent masses of total RNA and cellular protein isolated from log-phase cultures (OD600 = 0.5) shaken at 37°C for 30 min in the presence or absence of 0.2% arabinose were resolved by gel electrophoresis. mRNA was detected by hybridization with 32P-labeled DNA for each gene, and protein was detected using the appropriate antibody to each epitope tag.
We constructed C-terminal His6-tagged forms of each protein to allow analysis of protein expression. Addition of the epitope tag did not affect the phenotype of overexpression (Table 2). Examination of cellular protein concentration showed that in the presence of arabinose, YeeV-His6 and YkfI-His6 accumulate in the cell but YpjF-His6 does not, explaining the lack of toxicity (Fig. 4B). Northern blot analysis showed that the level of ypjF-His6 mRNA was significantly lower as well (Fig. 4B). Because all pBAD18 promoter and regulatory sequences were conserved between the expression constructs, we inferred that the difference in mRNA concentration may be an indirect effect. For example, if the translation rate for YpjF-His6 were low, mRNA stability could be decreased due to the tight linkage between bacterial transcription and translation. Inefficient translation is known to cause both premature termination of transcription (35) and a decrease in mRNA stability (34). Replacement of the His6 tag on YpjF with an N-terminal Flag tag resulted in both a large increase in cellular protein accumulation and growth inhibition (Fig. 4B; Table 2). Two slightly different forms of Flag-YpjF were detected by Western blotting.
TABLE 2.
Effect of epitope tagging on toxin induced growth inhibition
| Plasmid (gene expressed) | Arabinose | Growth at 6 ha |
|---|---|---|
| pBAD18 | + | +++ |
| pBAD18 | − | +++ |
| pRD052 (ypjF) | + | +++ |
| pRD125 (ypjF-His6) | + | +++ |
| pRD136 (Flag-ypjF) | + | − |
| pRD041 (ykfI-His6) | + | −b |
| pRD042 (yeeV-His6) | + | − |
Growth was monitored as described for Fig. 2. Optical density after 6 h of growth is represented as follows: −, OD600 < 0.5; +++, OD600 > 1.5.
In this experiment, OD600 = 0.45.
yeeU and yafW can prevent toxicity.
yeeV, ykfI, and ypjF are each preceded on the chromosome by two potential antitoxin genes that encode proteins of approximately the same size, the first of which is slightly smaller (Fig. 3C). In all three cases, none of the upstream genes inhibited growth when overexpressed individually (Table 3). Each of the ORFs was coexpressed with the appropriate toxins to test for the ability to prevent growth inhibition. In all previously characterized TA pairs, the antitoxin is immediately adjacent to the toxin. However, since yeeT and ykfH encode slightly smaller proteins than yeeV and ykfI, a common characteristic for antitoxins, these were also assayed for activity. Finally, the gene encoding the unrelated green fluorescent protein (GFP) was coexpressed as a negative control. ORFs that are adjacent in the genome were amplified as a pair from genomic DNA and subcloned into pBAD18 for expression. Others were constructed such that only the sequence of the proximal ORF varied; all regulatory and untranslated intergenic sequences were conserved. In both cases, coexpression of the adjacent gene (yeeU-yeeV and yafW-ykfI) restored the growth rate to the level of the uninduced strain (Table 3) and prevented a reduction in the number of CFU (data not shown). Growth of strains coexpressing the other pairs of genes (yeeT-yeeV, gfp-yeeV, ykfH-ykfI, or gfp-ykfI) was inhibited to approximately the same degree as growth of those expressing toxin alone (Table 3).
TABLE 3.
Effect of coexpression of the adjacent gene, the distal gene, or gfp on the growth inhibition induced by yeeV and ykfI
| Plasmid (gene[s] expressed) | Arabinose | Growth at 6 ha |
|---|---|---|
| pRD069 (yeeT) | + | +++ |
| pRD030 (yeeU) | + | +++ |
| pRD024 (yeeV) | + | − |
| pRD024 (yeeV) | − | +++ |
| pRD104 (yeeT-yeeV) | + | − |
| pRD050 (yeeU-yeeV) | + | +++ |
| pRD105 (gfp-yeeV) | + | − |
| pRD068 (ykfH) | + | +++ |
| pRD029 (yafW) | + | +++ |
| pRD023 (ykfI) | + | − |
| pRD023 (ykfI) | − | +++ |
| pRD106 (ykfH-ykfI) | + | − |
| pRD049 (yafW-ykfI) | + | +++ |
| pRD107 (gfp-ykfI) | + | − |
| pRD103 (ypjJ) | + | +++ |
| pRD086 (yfjZ) | + | +++ |
| pRD052 (ypjF) | + | +++ |
Growth curves were performed as described for Fig. 2. Optical density after 6 h of growth is represented as follows: −, OD600 < 0.5; +++, OD600 > 1.5.
Most known antitoxins prevent toxicity by physically interacting with the toxin partner (16, 18). Therefore, we performed coimmunoprecipitation experiments to detect in vivo protein-protein interactions by using expression constructs in which both the toxin and antitoxin were epitope tagged. Addition of an epitope tag to the N terminus of the antitoxins YeeU and YafW did not affect their ability to prevent growth inhibition. However, although the tagged forms of each protein could be individually immunoprecipitated when expressed, demonstrating that sufficient amounts of proteins were produced for these studies, interactions between YeeU and YeeV or YafW and YkfI were not observed under a wide variety of experimental conditions (data not shown). Instead, Western blotting revealed that antitoxin coexpression resulted in almost a complete absence of His6-tagged toxin proteins in whole-cell extracts, whereas GFP did not (Table 4; Fig. 5). Because all transcriptional regulatory sequences were conserved among all of the expression constructs, it was anticipated that mRNA synthesis should be similar. Therefore, these results suggest that rather than binding to the toxins to prevent toxicity, YeeU and YafW may function as antitoxins by either preventing toxin protein translation or promoting toxin degradation. Both the degree of growth inhibition and the amount of toxin observed upon induction of pRD124 (gfp-ykfI-His6) were increased compared to that conferred by expression from pRD041 (ykfI-His6). The converse was true for cells transformed with pRD140 (gfp-yeeV-His6)—both the cellular toxin level and the degree of growth inhibition were decreased compared to that in a strain carrying pRD042 (yeeV-His6). These data, combined with those shown in Fig. 4, suggest that the degree of growth inhibition is directly related to the amount of toxin protein present in the cell. Therefore, the toxins may be titrating out an essential cellular component that may be present in limiting quantities.
TABLE 4.
The antitoxin activity of YeeU and YafW is not prevented by the presence of a C-terminal His tag on the YeeV or YkfI toxins
| Plasmid (gene[s] expressed) | Arabinose | Growth at 6 ha |
|---|---|---|
| pRD042 (yeeV-His6) | + | − |
| pRD042 (yeeV-His6) | − | +++ |
| pRD062 (yeeU-yeeV-His6) | + | +++ |
| pRD140 (gfp-yeeV-His6) | + | + |
| pRD041 (ykfI-His6) | + | +b |
| pRD041 (ykfI-His6) | − | +++ |
| pRD061 (yafW-ykfI-His6) | + | +++ |
| pRD124 (gfp-ykfI-His6) | + | − |
Growth curves were performed as described for Fig. 2. Optical density after 6 h of growth is represented as follows: −, OD600 < 0.5; +, 0.5 < OD600 < 1.0; +++, OD600 > 1.5.
In this experiment, OD600 = 0.55.
FIG. 5.
Quantitation of His-tagged toxin protein. Cellular protein isolated from log-phase cultures (OD600 = 0.5) shaken at 37°C for 30 min in the presence or absence of 0.2% arabinose was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The mass of total protein loaded was normalized using the optical density of each culture. Detection was performed using an anti-His6 antibody.
YeeU antitoxin function requires the presence of the intergenic UTR.
Each of these novel toxin-antitoxin gene pairs is separated by a conserved 20-bp untranslated region (UTR). However, an additional 68 bp of untranslated mRNA is located immediately downstream of yeeU, but not yafW or ypjJ. To investigate whether the UTR plays a role in YeeU antitoxin function, we compared the coexpression constructs described previously (pRD049 and pRD050 [Table 3]) to pRD095 and pRD139, constructs in which each ORF was amplified separately and ligated together in pBAD18 using a BamHI restriction site (yeeU-BamHI-yeeV, yafW-BamHI-ykfI). In the absence of the UTR, YeeU could not prevent growth inhibition mediated by YeeV, whereas if the UTR were amplified along with the yeeU ORF before ligation with yeeV (yeeU-UTR-BamHI-yeeV), antitoxin activity was restored (Table 5). The presence of the UTR alone (UTR-yeeV) was not sufficient to prevent toxicity (Table 5). Growth of arabinose-induced cells carrying pRD142 (yafW-BamHI-ykfI) was similar to that of the uninduced control, indicating that the conserved 20-bp portion of the UTR is not required for antitoxin function (Table 5).
TABLE 5.
The unique 68-bp UTR between yeeU and yeeV is required for yeeU antitoxin function
| Plasmid (gene[s] expressed) | Arabinose | Growth at 6 ha |
|---|---|---|
| pRD024 (yeeV) | + | − |
| pRD024 (yeeV) | − | +++ |
| pRD095 (yeeU-BamHI-yeeV) | + | − |
| pRD134 (UTR-yeeV) | + | + |
| pRD135 (yeeU-UTR-BamHI-yeeV) | + | +++ |
| pRD050 (yeeU-yeeV) | + | +++ |
| pRD142 (yafW BamHI-yeeV) | + | +++ |
| pRD072 (yeeV) | + | − |
| pRD072 (yeeV) + pRD030 (yeeU) | + | − |
| pRD023 (ykfI) | + | − |
| pRD023 (ykfI) | − | +++ |
| pRD139 (yafW-BamHI-ykfI) | + | +++ |
| pRD049 (yafW-ykfI) | + | +++ |
| pRD141 (yeeU-BamHI-ykfI) | + | ++ |
Growth curves were performed as described for Fig. 2. Optical density after 6 h of growth is represented as follows: −, OD600 < 0.5; +, 0.5 < OD600 < 1.0; ++, 1.0 < OD600 < 1.5; +++, OD600 > 1.5.
We further probed the requirement of cis elements for the UTR in YeeU function by expressing yeeV and yeeU from separate compatible plasmids in the same cell. To do this, a strain containing both pRD072 (yeeV subcloned into the low-copy-number arabinose-inducible vector pBAD33) and pRD030 (yeeU subcloned into pBAD18) was constructed. We found that expression of yeeU was unable to prevent toxicity in trans, supporting the hypothesis that the UTR contains critical cis-acting regulatory elements (Table 5).
We constructed expression cassettes in which yeeU and yafW were exchanged to examine the cross-compatibility of antitoxin function. We found that YafW was able to partially block YeeV toxicity but was less effective than YeeU (with the UTR present) (Table 5). Finally, YeeU was able to prevent YkfI toxicity to a moderate degree, even in the absence of the 68-bp portion of the UTR, but was less effective than YafW (Table 5).
DISCUSSION
We used a method that relied on protein size, rather than sequence homology, to search for novel bacterial TA pairs. Out of 32 genes tested, 6 caused growth inhibition when ectopically expressed in E. coli. We focused our attention on two of these chromosomal genes, yeeV and ykfI, that along with a third highly related gene, ypjF, define a novel family of toxins. Overproduction of each of these resulted in growth inhibition, although YpjF only accumulated to levels sufficient to inhibit growth when stabilized with an N-terminal FLAG tag. None of the toxin genes contained an unusual number of rare codons, which might be expected to lead to growth inhibition if the protein were overexpressed. Given that ypjF and ykfI share 80% sequence homology and the expression constructs are identical outside of the ORF, we suspect that specific sequences near the 5′ end of the ypjF gene are responsible for either inefficient translation or increased degradation and that these differences are masked by the presence of the FLAG tag. Coexpression of the adjacent, upstream gene of each toxin (yeeU-yeeV, yafW-ykfI) restored normal growth and viability. These genes encode proteins of approximately the same size as the toxin, which was somewhat surprising since our initial search was for pairs in which the antitoxin was slightly smaller. Indeed the antitoxin-encoding genes yeeU and yafW are preceded by the slightly smaller genes yeeT and ykfH, respectively. However, yeeU and yafW were not toxic when overexpressed, and yeeT and ykfH did not prevent toxicity when coexpressed with yeeV or ykfI. Additionally, we found that yeeU-mediated suppression of yeeV toxicity required the unique 68-bp intergenic UTR that was not present in the other two pairs. Cluster alignment analysis suggests the UTR may be derived from a duplication of sequence within the yeeU coding region (unpublished observation). This may explain why yafW, which does not have the extended downstream UTR, does not require it for antitoxin function. The fact that yeeU requires the presence of the downstream UTR to prevent growth inhibition caused by yeeV, but not ykfI, demonstrates that antitoxin function is not modular but relies on specific interactions with the paired toxin gene.
Identifying the mechanism by which yeeV, ykfI, and ypjF induce cell death is of primary importance. A general mode of action for antimicrobial agents can often be determined by assaying for the ability of the compound to inhibit a specific macromolecular synthesis pathway (27). However, in contrast to the immediate effect on cellular metabolism observed upon addition of inhibitor to a culture, the onset of toxicity caused by ectopic expression of YeeV or YkfI was relatively slow (Fig. 2 and 4A), presumably due to a lag in the accumulation of the toxin protein. These kinetics did not allow sufficient resolution to distinguish the primary macromolecular synthetic pathway inhibited by YeeV or YkfI. BLAST searches for related proteins also did not yield insight into the mechanism of action of YeeV-, YkfI-, or YpjF-mediated growth inhibition. These proteins do contain a moderate degree of chemical conservation to SIS domains (sugar isomerase), which are found in a number of proteins known to bind various phosphosugar metabolites (3). The most similar SIS domain to YeeV, YkfI, and YpjF is found in the E. coli protein LpcA (9 of 13 amino acids thought to be conserved in SIS domains [3] are chemically related). Deletion of the lpcA gene results in the production of a shortened lipopolysaccharide core and supersensitivity to novobiocin (10). However, we did not observe increased novobiocin sensitivity upon induction of yeeV or ykfI, nor did we observe synergy with several other antimicrobial agents whose mechanism of action is known (unpublished data). Our observation that there is a positive correlation between the cellular concentration of toxin protein and the severity of growth inhibition suggests that the toxins may act by binding to and titrating out an essential protein or metabolite.
These novel TA pairs varied from previously characterized pairs in that we could not detect any physical interaction between the proteins. Instead, we found that expression of the antitoxins resulted in a decrease in the level of toxin protein in the cell. The mechanism by which the antitoxins affect this regulation requires further investigation but could involve preventing the translation of toxin mRNA or increasing the efficiency of degradation of either toxin mRNA or protein. The requirement of the downstream UTR for yeeU activity suggests binding of antitoxin protein to the mRNA transcript may be important.
The physiological role of these new TA pairs is currently unclear. If it were true that the toxicity associated with this family of proteins results from the titration of an essential metabolite or enzyme, this would be consistent with the model proposing a role for TA pairs in the regulation of specific metabolic pathways (18). However, a role in programmed cell death cannot be ruled out. For instance, if a pool of untranslated toxin mRNA were present in the cytoplasm, cell death could be triggered by inactivation of the antitoxin. A detailed examination of the in vivo regulation of the endogenous genes and proteins will be required to distinguish between these and other possibilities.
Of the six toxin genes described in this report, only pspC has been previously studied (9, 30). This gene is part of an operon that is induced by phage infection, as well as a variety of other stresses (5, 8, 30). Coexpression of pspB with pspC restored normal growth rates (unpublished data). Although these proteins have not previously been described as a TA pair, they do possess additional characteristics of TA pairs, including physical interaction (1) and autoregulation of transcription (37, 38). PspC positively regulates the transcription of the psp operon, for which the only currently known function involves maintaining the proton motive force of the inner membrane under stress conditions (23). Overexpression of PspA, which is thought to be the effector protein in this process, does not inhibit growth (unpublished data). Interestingly, the Yersinia enterocolitica homolog of pspC was shown to be required for virulence and is likely to regulate additional genes outside the psp locus (13, 14).
Another particularly intriguing finding was the toxicity associated with the overexpression of yafO. This gene is located just downstream of yafN, which has been described as an “orphan” antitoxin gene (19). It has some sequence similarity to relB but lacks a transcriptionally linked relE homolog. Our results show that yafN is indeed directly upstream of a toxin-encoding gene, but one that is unexpectedly divergent from relE. YafN antitoxin activity against YafO has not yet been demonstrated.
Acknowledgments
We thank Gary Schoenhals for use of the E. coli ORF database and members of the Infectious Disease team for valuable discussions throughout the course of this work.
REFERENCES
- 1.Adams, H., W. Teertstra, J. Demmers, R. Boesten, and J. Tommassen. 2003. Interactions between phage-shock proteins in Escherichia coli. J. Bacteriol. 185:1174-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′, 5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bateman, A. 1999. The SIS domain: a phosphosugar-binding domain. Trends Biochem. Sci. 24:94-95. [DOI] [PubMed] [Google Scholar]
- 4.Bech, F. W., S. T. Jorgensen, B. Diderichsen, and O. H. Karlstrom. 1985. Sequence of the relB transcription unit from Escherichia coli and identification of the relB gene. EMBO J. 4:1059-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergler, H., D. Abraham, H. Aschauer, and F. Turnowsky. 1994. Inhibition of lipid biosynthesis induces the expression of the pspA gene. Microbiology 140(Pt. 8):1937-1944. [DOI] [PubMed] [Google Scholar]
- 6.Bernard, P., and M. Couturier. 1992. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J. Mol. Biol. 226:735-745. [DOI] [PubMed] [Google Scholar]
- 7.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 8.Brissette, J. L., M. Russel, L. Weiner, and P. Model. 1990. Phage shock protein, a stress protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 87:862-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brissette, J. L., L. Weiner, T. L. Ripmaster, and P. Model. 1991. Characterization and sequence of the Escherichia coli stress-induced psp operon. J. Mol. Biol. 220:35-48. [DOI] [PubMed] [Google Scholar]
- 10.Brooke, J. S., and M. A. Valvano. 1996. Biosynthesis of inner core lipopolysaccharide in enteric bacteria identification and characterization of a conserved phosphoheptose isomerase. J. Biol. Chem. 271:3608-3614. [DOI] [PubMed] [Google Scholar]
- 11.Cashel, M., D. R. Gentry, V. Z. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington D.C.
- 12.Christensen, S. K., M. Mikkelsen, K. Pedersen, and K. Gerdes. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 98:14328-14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darwin, A. J., and V. L. Miller. 1999. Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol. Microbiol. 32:51-62. [DOI] [PubMed] [Google Scholar]
- 14.Darwin, A. J., and V. L. Miller. 2001. The psp locus of Yersinia enterocolitica is required for virulence and for growth in vitro when the Ysc type III secretion system is produced. Mol. Microbiol. 39:429-444. [DOI] [PubMed] [Google Scholar]
- 15.Diderichsen, B., N. P. Fiil, and R. Lavalle. 1977. Genetics of the relB locus in Escherichia coli. J. Bacteriol. 131:30-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engelberg-Kulka, H., and G. Glaser. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53:43-70. [DOI] [PubMed] [Google Scholar]
- 17.Galvani, C., J. Terry, and E. E. Ishiguro. 2001. Purification of the RelB and RelE proteins of Escherichia coli: RelE binds to RelB and to ribosomes. J. Bacteriol. 183:2700-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerdes, K. 2000. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 182:561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gotfredsen, M., and K. Gerdes. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29:1065-1076. [DOI] [PubMed] [Google Scholar]
- 20.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayes, F. 1998. A family of stability determinants in pathogenic bacteria. J. Bacteriol. 180:6415-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horton, R. M., Z. L. Cai, S. N. Ho, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8:528-535. [PubMed] [Google Scholar]
- 23.Kleerebezem, M., W. Crielaard, and J. Tommassen. 1996. Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the proton-motive force under stress conditions. EMBO J. 15:162-171. [PMC free article] [PubMed] [Google Scholar]
- 24.Lavalle, R. 1965. New mutants for regulation of RNA synthesis. Bull. Soc. Chim. Biol. (Paris) 47:1567-1570. (In French.) [PubMed] [Google Scholar]
- 25.Lewis, K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuda, Y., K. Miyakawa, Y. Nishimura, and E. Ohtsubo. 1993. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol. 175:6850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNicholas, P. M., D. J. Najarian, P. A. Mann, D. Hesk, R. S. Hare, K. J. Shaw, and T. A. Black. 2000. Evernimicin binds exclusively to the 50S ribosomal subunit and inhibits translation in cell-free systems derived from both gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 44:1121-1126. [DOI] [PMC free article] [PubMed]
- 28.Miki, T., J. A. Park, K. Nagao, N. Murayama, and T. Horiuchi. 1992. Control of segregation of chromosomal DNA by sex factor F in Escherichia coli. Mutants of DNA gyrase subunit A suppress letD (ccdB) product growth inhibition. J. Mol. Biol. 225:39-52. [DOI] [PubMed] [Google Scholar]
- 29.Mittenhuber, G. 1999. Occurrence of mazEF-like antitoxin/toxin systems in bacteria. J. Mol. Microbiol. Biotechnol. 1:295-302. [PubMed] [Google Scholar]
- 30.Model, P., G. Jovanovic, and J. Dworkin. 1997. The Escherichia coli phage-shock-protein (psp) operon. Mol. Microbiol. 24:255-261. [DOI] [PubMed] [Google Scholar]
- 31.Pedersen, K., S. K. Christensen, and K. Gerdes. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45:501-510. [DOI] [PubMed] [Google Scholar]
- 32.Pedersen, K., A. V. Zavialov, M. Y. Pavlov, J. Elf, K. Gerdes, and M. Ehrenberg. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112:131-140. [DOI] [PubMed] [Google Scholar]
- 33.Ruiz-Echevarria, M. J., G. Gimenez-Gallego, R. Sabariegos-Jareno, and R. Diaz-Orejas. 1995. Kid, a small protein of the parD stability system of plasmid R1, is an inhibitor of DNA replication acting at the initiation of DNA synthesis. J. Mol. Biol. 247:568-577. [DOI] [PubMed] [Google Scholar]
- 34.Schneider, E., M. Blundell, and D. Kennell. 1978. Translation and mRNA decay. Mol. Gen. Genet. 160:121-129. [DOI] [PubMed] [Google Scholar]
- 35.Stanssens, P., E. Remaut, and W. Fiers. 1986. Inefficient translation initiation causes premature transcription termination in the lacZ gene. Cell 44:711-718. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiner, L., J. L. Brissette, and P. Model. 1991. Stress-induced expression of the Escherichia coli phage shock protein operon is dependent on sigma 54 and modulated by positive and negative feedback mechanisms. Genes Dev. 5:1912-1923. [DOI] [PubMed] [Google Scholar]
- 38.Weiner, L., J. L. Brissette, N. Ramani, and P. Model. 1995. Analysis of the proteins and cis-acting elements regulating the stress-induced phage shock protein operon. Nucleic Acids Res. 23:2030-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zielenkiewicz, U., and P. Ceglowski. 2001. Mechanisms of plasmid stable maintenance with special focus on plasmid addiction systems. Acta Biochim. Pol. 48:1003-1023. [PubMed] [Google Scholar]