Abstract
LytF, LytE, and LytC are vegetative cell wall hydrolases in Bacillus subtilis. Immunofluorescence microscopy showed that an epitope-tagged LytF fusion protein (LytF-3xFLAG) in the wild-type background strain was localized at cell separation sites and one of the cell poles of rod-shaped cells during vegetative growth. However, in a mutant lacking both the cell surface protease WprA and the extracellular protease Epr, the fusion protein was observed at both cell poles in addition to cell separation sites. This suggests that LytF is potentially localized at cell separation sites and both cell poles during vegetative growth and that WprA and Epr are involved in LytF degradation. The localization pattern of LytE-3xFLAG was very similar to that of LytF-3xFLAG during vegetative growth. However, especially in the early vegetative growth phase, there was a remarkable difference between the shape of cells expressing LytE-3xFLAG and the shape of cells expressing LytF-3xFLAG. In the case of LytF-3xFLAG, it seemed that the signals in normal rod-shaped cells were stronger than those in long-chain cells. In contrast, the reverse was found in the case of LytE-3xFLAG. This difference may reflect the dependence on different sigma factors for gene expression. The results support and extend the previous finding that LytF and LytE are cell-separating enzymes. On the other hand, we observed that cells producing LytC-3xFLAG are uniformly coated with the fusion protein after the middle of the exponential growth phase, which supports the suggestion that LytC is a major autolysin that is not associated with cell separation.
Bacillus subtilis produces several murein hydrolases during vegetative growth (10, 38, 42). Among these enzymes there are two major vegetative autolysins, a 50-kDa N-acetylmuramoyl-l-alanine amidase (amidase or LytC [CwlB]) and a 90-kDa endo-β-N-acetylglucosaminidase (glucosaminidase or LytD [CwlG]), which have been cloned and characterized (20, 23, 29, 37). In addition to these major autolysins, minor 50- and 35-kDa autolysins, LytF (CwlE) and LytE (CwlF), respectively, were recently found in a vegetative cell surface extract (14, 32, 33, 34). Previous reports demonstrated that the C-terminal domain of LytF has dl-endopeptidase activity that breaks the linkage of d-γ-glutamyl-meso-diaminopimelic acid in a murein peptide (32, 34). The lytF gene is transcribed by EσD RNA polymerase, and the LytF protein consists of two domains, a putative cell wall binding (CWB) domain at its N terminus and a catalytic domain at its C terminus. Cells lacking the lytF gene form chains during the vegetative growth phase (34). In addition to LytF, B. subtilis produces another putative dl-endopeptidase, LytE, during vegetative growth (14, 32). The C-terminal domain of LytE also seems to have dl-endopeptidase activity because of the very high level of homology between LytF and LytE. The lytE gene is transcribed by EσA and EσH RNA polymerases, and the cells of a lytE mutant are slightly longer than those of the wild type in the early vegetative growth phase (14). Moreover, a lytF lytE double mutant forms extra long chains (34). Thus, we have presumed that the two dl-endopeptidases must play very important roles in cell separation during cell division. However, there has been no direct evidence concerning the localization of vegetative murein hydrolases in B. subtilis.
The N-terminal domains of LytF and LytE comprise five and three LysM domains, respectively, which are separated by serine-rich regions. There are 17 paralogs containing the LysM domain in B. subtilis (18; Pfam database [http://www.sanger.ac.uk/Software/Pfam/]). One of the LysM family proteins, membrane-bound lytic murein transglycosylase D (MltD) of Escherichia coli, has been well studied (4). The LysM motif, which has a βααβ secondary structure, is a widespread protein motif. This motif seems to function as a general peptidoglycan binding module. In addition to this suggestion, we propose that the LysM motifs of cell wall lytic enzymes in B. subtilis may play important roles in appropriate localization and/or recognition of substrates. The C-terminal catalytic domains of LytF and LytE are classified in the NlpC/p60 family (Pfam database), and they exhibit a high level of sequence homology (67.0%) with each other. In B. subtilis, there are five paralogs (YojL, YddH, YvcE, YkfC, and YwtD) of the catalytic domains of LytF and LytE (18; Pfam database). One of these paralogs, YojL, exhibits high levels of similarity to the complete LytF and LytE sequences, suggesting that it may be a third dl-endopeptidase in B. subtilis. In addition, the C-terminal domains of these molecules are very similar to those of the p60 and p45 proteins of Listeria monocytogenes (17, 40, 47) and the entire E. coli NlpC protein (SWISS-PROT protein sequence data bank [http://us.expasy.org/cgi-bin/get-sprot-entry?P23898]).
In contrast to LytE and LytF, LytC is an amidase that hydrolyzes the linkage of N-acetylmuramoyl-l-alanine in peptidoglycan. A lytC mutant strain did not form chains during vegetative growth, suggesting that LytC is not involved in the cell separation event (20, 30). LytC consists of a CWB domain in the N-terminal region and a catalytic domain in the C-terminal region. The CWB domain has three direct repeat sequences that are not similar to the LysM motifs in the N-terminal CWB domains of LytF and LytE. The CWB domain of LytC (CWBb) has been used as a CWB anchor of two artificial fusion proteins, CWBb-LipB and CWBb-CutL (15, 16, 44). These artificial lipases had both enzyme activity and CWB activity.
B. subtilis produces at least eight extracellular proteases (2). One of these enzymes, WprA, is the major cell surface protease (31). In a previous paper, it was reported that two artificial lipases fused to the C terminus of the CWBb domain accumulated more in a wprA mutant strain than in the wild type. Moreover, the fusion lipases were most stable in a wprA sigD double mutant (15). The results strongly suggest that there is another extracellular protease(s) regulated by SigD. Very recently, Bruckner et al. (5), Dixit et al. (7), and Sloma et al. (41) reported that the epr gene, encoding a minor extracellular serine protease, is transcribed by EσD RNA polymerase and that an epr mutation affects cell motility. In addition, it has been reported that Epr seems to play an important role in the processing of a major cell surface protein, WapA (2). Thus, we propose that not only WprA but also Epr might affect the stability of cell wall lytic enzymes.
Information concerning protein localization is very important for predicting or understanding the functions of cell surface proteins. In Staphylococcus aureus, it has been reported that Atl, a bifunctional protein that has an amidase domain at the N terminus and a glucosaminidase domain at the C terminus (35), forms a ring structure on the cell surface at the septal region for the next cell division site (3, 49). Moreover, Loessner and colleagues have reported that two phage endolysins, Ply118 and Ply500, of Listeria monocytogenes exhibit different CWB patterns (28). A fusion between green fluorescent protein (GFP) and the CWB domain of Ply118 is predominantly localized at the septal regions and cell poles, whereas another GFP fusion protein derived from Ply500 binds to the entire cell surface. In B. subtilis, the subcellular localization of many intracellular and membrane proteins comprising the division apparatus has been determined (8). For example, Pedersen and colleagues found that penicillin binding protein 1, PonA, is localized at a cell division site by performing immunofluorescence microscopy (IFM) with cells producing a PonA-FLAG fusion protein (36). On the other hand, there is little information concerning cell surface and extracellular proteins, such as cell wall lytic enzymes in B. subtilis. In this study, we performed IFM with three vegetative cell wall lytic enzymes fused to a 3xFLAG epitope tag.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains of B. subtilis and E. coli and the plasmids used in this study are listed in Table 1. B. subtilis 168 was the parent strain throughout this study. B. subtilis strains were cultured in Luria-Bertani (LB) medium (39) at 37°C unless otherwise noted. When necessary, chloramphenicol, tetracycline, kanamycin, and erythromycin were added to final concentrations of 3, 5, 5, and 0.3 μg/ml, respectively. To culture conditional mutant strains with minC mutations (B. subtilis MINCp and WE1EMCp), isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.25 mM. E. coli strains were grown in LB medium at 37°C. If necessary, ampicillin was added to a final concentration of 100 μg/ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source or referencea |
|---|---|---|
| B. subtilis strains | ||
| 168 | trpC2 | S. D. Ehrlich |
| ED | trpC2 lytF::pM2HDD | 31 |
| EFD | trpC2 lytE::pM2cF | 14 |
| E3FL | trpC2 lytF::pCA3FLCE | pCA3FLCE→168b |
| F3FL | trpC2 lytE::pCA3FLCF | pCA3FLCF→168 |
| B3FL | trpC2 lytC::pCA3FLCB | pCA3FLCB→168 |
| MINCp | trpC2 minC::pM4MINC (Pspac-minCD) | pM4MINC→168 |
| E3FLFd | trpC2 lytF::pCA3FLCE lytE::pM2cF | EFD→E3FL |
| F3FLEd | trpC2 lytE::pCA3FLCF lytF::pM2HDD | ED→F3FL |
| B3FLEd | trpC2 lytC::pCA3FLCB lytF::pM2HDD | ED→B3FL |
| EPRTc | trpC2 epr::tet | pUCEPRTc→168 |
| EPRE3FL | trpC2 epr::tet lytF::pCA3FLCE | EPRTc→E3FL |
| EPRF3FL | trpC2 epr::tet lytE::pCA3FLCF | EPRTc→F3FL |
| EPRB3FL | trpC2 epr::tet lytC::pCA3FLCB | EPRTc→B3FL |
| WA | trpC2 wprA::kan | 15 |
| WAE3FL | trpC2 wprA::kan lytF::pCA3FLCE | WA→E3FL |
| WAF3FL | trpC2 wprA::kan lytE::pCA3FLCF | WA→F3FL |
| WAB3FL | trpC2 wprA::kan lytC::pCA3FLCB | WA→B3FL |
| WE1 | trpC2 epr::tet wprA::kan | EPRTc→WA |
| WEEd | trpC2 epr::tet wprA::kan lytF::pM2HDD | ED→WE1 |
| WE1E3FL | trpC2 epr::tet wprA::kan lytF::pCA3FLCE | E3FL→WE1 |
| WE1F3FL | trpC2 epr::tet wprA::kan lytE::pCA3FLCF | F3FL→WE1 |
| WE1B3FL | trpC2 epr::tet wprA::kan lytC::pCA3FLCB | B3FL→WE1 |
| WEB3FLEd | trpC2 epr::tet wprA::kan lytF::pM2HDD lytC::pCA3FLCB | ED→WE1B3FL |
| WE1EMCp | trpC2 epr::tet wprA::kan lytF::pCA3FLCE minC::pM4MINC (Pspac-minCD) | MINCp→WE1E3FL |
| E. coli strains | ||
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi-1 Δ(lac-proAB)/F′ [traD36 proAB+ lacIqlacZΔM15] | Takara |
| C600 | supE44 hsdR17 thi-1 thr-1 leuB6 lacY1 tonA21 | Laboratory stock |
| Plasmids | ||
| pCA3xFLAG | bla cat 3xFLAG | N. Ogasawara and K. Kobayashi |
| pCA3FLCE | bla cat lytF-3xFLAG | This study |
| pCA3FLCF | bla cat lytF-3xFLAG | This study |
| pCA3FLCB | bla cat lytC-3xFLAG | This study |
| pMUTIN4 | lacZ lacI bla erm | 44 |
| pM4MINC | pMUTIN4::ΔminC (containing minC Shine-Dalgarno sequence) | This study |
| pUCEPR | bla Δepr | 2 |
| pUCEPRTc | bla Δepr::tet | This study |
| pDG1513 | bla tet | BGSC |
| pUC19 | bla | Takara |
| pC194 | cat | 13 |
| pCA191 | bla cat | 48 |
| p3xFLAG-CMV-14 | bla neo 3xFLAG | Sigma |
BGSC, Bacillus Genetic Stock Center, Ohio State University.
The strains and plasmids before and after the arrows are the DNA donors and recipients for transformation, respectively.
Transformation of E. coli and B. subtilis.
E. coli transformation was performed as described by Sambrook et al. (39), and B. subtilis transformation was performed by the conventional transformation procedure (1).
Plasmid construction.
The pCA3xFLAG plasmid carrying the 3xFLAG and chloramphenicol resistance genes was obtained from N. Ogasawara and K. Kobayashi. According to the information provided by these workers for plasmid construction, the chloramphenicol resistance gene of pC194 (13) was digested with HpaII and ClaI. The fragment was inserted into the NarI site of pUC19 to obtain pCA191 (48). The 3xFLAG gene was amplified by using primers FLAG-F (5′-AAGAAGCTTGCGGCCGCGAATTC; the HindIII site is underlined) and FLAG-R (5′-CAACAATTGTCACTACTTGTCATCGTCATCC; the MunI site is underlined) with p3xFLAG-CMV-14 (Sigma) as a template. Then the amplified fragment was digested with MunI and HindIII and inserted into the EcoRI and HindIII sites of pCA191 to obtain pCA3xFLAG. To construct gene fusions between cell wall lytic enzyme and 3xFLAG genes, the 3′-terminal regions of lytF (cwlE; 243 bp), lytE (cwlF; 208 bp), and lytC (cwlB; 266 bp) just before their stop codons were amplified by PCR by using B. subtilis 168 DNA as the template and primers CwlEC-HF (5′-GCGCAAGCTTCTGAACAAAGTCACATCCG; the HindIII site is underlined) and CWLE-NSR (5′-GCGCCCCGGGGCTAGCGAAATATCGTTTTGCACCG; the NheI site is underlined) for lytF, primers CwlFC-HF (5′-GCGCAAGCTTCACTGCAGGATACTGGAG; the HindIII site is underlined) and CWLF-XR (5′-GCGCTCTAGAGAATCTTTTCGCACCGAG; the XbaI site is underlined) for lytE, and primers FGH-CwlB (5′-GCGCAAGCTTAGACGTACTACGATACAAC; the HindIII site is underlined) and RGX-CwlB (5′-CGGCTCTAGATCTGTAATAAGATACTGTGC; the XbaI site is underlined) for lytC. The amplified fragment of lytF was digested with HindIII and NheI, and the amplified fragments of lytE and lytC were digested with HindIII and XbaI. Then the digested fragments were ligated into the HindIII and XbaI sites of pCA3xFLAG. Then E. coli JM109 cells were transformed with the ligation mixtures to generate pCA3FLCE, pCA3FLCF, and pCA3FLCB. After the sequences were confirmed, the plasmids were used for transformation of E. coli C600 to produce concatemeric plasmid DNAs (6). For construction of a conditional minC mutant (MINCp), the 5′-terminal region including the ribosome binding site was amplified by using primers PspMinHF (5′-GCGCAAGCTTGTTGAGGTGAATATTGTG; the HindIII site is underlined) and MinC-BR (5′-CGGCGGATCCTTAGTAATGACTTCACTGTC; the BamHI site is underlined) with B. subtilis 168 chromosomal DNA as the template. The resultant fragment was digested with HindIII and BamHI, and this was followed by ligation into the corresponding sites of pMUTIN4 (45) to generate pM4MINC. After the nucleotide sequence of the insert was confirmed, the plasmid was used for transformation of E. coli C600 to obtain concatemeric DNAs. For construction of an epr mutant, a tetracycline resistance cassette derived by BamHI-ClaI digestion of pDG1515 was cloned into the corresponding sites of pUCEPR (2) to obtain pUCEPRTc.
Construction of mutants and 3xFLAG fusion strains.
The sources of donor DNAs and recipient cells used for B. subtilis mutant construction are listed in Table 1. For construction of an epr mutant strain, pUCEPRTc linearized with AatII was used. B. subtilis 168 was transformed with the linearized plasmid to generate B. subtilis EPRTc by double-crossover recombination. To construct the C-terminal lytF-3xFLAG, lytE-3xFLAG, and lytC-3xFLAG fusion strains, pCA3FLCE, pCA3FLCF, and pCA3FLCB were used for transformation of B. subtilis 168, respectively. The C-terminal regions of the fusion proteins included two types of short linker sequences (six amino acids; ARGSRA for LytF-3xFLAG and SRGSRA for LytE-3xFLAG and LytC-3xFLAG), followed by a 3xFLAG epitope tag sequence (22 amino acids; DYKDHDGDYKDHDIDYKDDDDK) derived from plasmid pCA3xFLAG. The resultant strains, B. subtilis E3FL, F3FL, and B3FL, integrated by single-crossover recombination, were selected with chloramphenicol. For zymography of LytC-3xFLAG, we also constructed a series of lytF mutants since LytF (CwlE; 49 kDa) and LytC (CwlB; 50 kDa) overlap on a sodium dodecyl sulfate (SDS)-polyacrylamide gel (34, 38). The chromosomal DNA of B. subtilis ED (34) was used for transformation of B. subtilis WE1, B3FL, and WE1B3FL to obtain B. subtilis WEEd, B3FLEd, and WEB3FLEd, respectively. For construction of B. subtilis MINCp, pM4MINC was used for transformation of B. subtilis 168. The resultant transformants were selected on LB agar plates containing erythromycin and IPTG. The chromosomal DNA extracted from B. subtilis MINCp was used for transformation of B. subtilis WE1E3FL to obtain B. subtilis WE1EMCp. All strains were confirmed by PCR.
Sample preparation for IFM observation.
B. subtilis strains were grown overnight at 25°C in LB medium. When sampling was to be carried out, parental strains without the 3xFLAG fusion gene were also cultured and prepared as a control. A culture having an optical density at 600 nm (OD600) of 0.5 was centrifuged, and the cells were suspended in 10 ml of LB medium and then cultured at 30°C with vigorous shaking. Samples used for IFM were prepared as described by Pedersen et al. (36), with some modifications, as described below. In order to detect the cell wall lytic enzyme-3xFLAG fusion proteins, cells at an OD600 of 0.5 were harvested and fixed for 15 min at room temperature in a solution containing 4.4% (wt/vol) (final concentration) paraformaldehyde, 30 mM (final concentration) Na2HPO4, and 30 mM (final concentration) NaH2PO4 (pH 7.0). After fixation, the cells were washed twice with 800 μl of phosphate-buffered saline (PBS) (80 mM Na2HPO4, 20 mM NaH2PO4, 100 mM NaCl; pH 7.5). Next, the cells were suspended in 400 μl of bovine serum albumin (BSA)-PBS (PBS containing 2% [wt/vol] BSA) and stored at −80°C, if necessary. The samples were incubated at 4°C for 20 min and then centrifuged. The cells were suspended in 400 μl of BSA-PBS containing 4 μg of anti-FLAG M2 monoclonal antibody (Sigma) per ml as a primary antibody and then incubated at 4°C for 1 h. After the cells were washed once with 800 μl of PBS, they were resuspended in 400 μl of a 1:400 dilution of anti-mouse immunoglobulin G-fluorescein isothiocyanate (FITC) conjugate antibody (Sigma) as a secondary antibody in BSA-PBS and then incubated at 4°C for 1 h. For cell wall staining, wheat germ agglutinin-tetramethylrhodamine conjugate (WGA-TMR) (Molecular Probes) was added at a final concentration of 1 μg/ml to the reaction mixtures with secondary antibody. Then the cells were washed with 800 μl of PBS and resuspended in 100 μl of PBS containing 0.025 mg of lysozyme per ml. Fifteen microliters of each cell suspension was immediately spotted onto a poly-l-lysine-coated microscope slide and incubated for 30 s at room temperature. Then the spots were washed once with 20 μl of PBS and completely dried. The slides were either stored at −20°C or equilibrated in Slow Fade (Molecular Probes) containing 1 μg of 4′,6-diamidino-2-phenylindole (DAPI) (Wako) per ml.
Phase-contrast and fluorescence microscopy.
Fluorescence microscopy of cells was performed as described previously (11) with an Olympus BX61 microscope equipped with a BX-UCB control unit, a UPPlan Apo Fluorite phase-contrast objective (magnification, ×100; numerical aperture, 1.3), and standard filter sets for visualizing DAPI, FITC, and rhodamine. The exposure times were 0.1 s for phase-contrast microscopy, 0.001 or 0.005 s for DAPI, 0.05 or 0.1 s for FITC, and 0.05 s for rhodamine. Under these conditions, we confirmed that no fluorescence of FITC was detected for the parent strains, B. subtilis 168 and WE1, without the 3xFLAG fusion genes (data not shown). Cells were photographed with a charge-coupled device camera (CoolSNAP HQ; Nippon Roper) driven by MetaMorph software (version 4.6r5; Universal Imaging). All images were processed with Adobe Photoshop software.
Preparation of cell surface proteins and cell walls.
For preparation of CWB proteins, we used an extraction method involving high concentrations of LiCl described previously (38). B. subtilis strains were cultured in 100 ml of LB medium at 37°C during the transition to the stationary phase. After centrifugation, the cells were washed twice with 10 mM Tris-HCl (pH 7.5). Then the cell pellets were resuspended in 2 ml of LiCl buffer (10 mM Tris-HCl [pH 7.5], 3 M LiCl) and stored on ice for 15 min. Then the cell suspensions were centrifuged at 20,000 × g for 5 min at 4°C to remove the cells. CWB proteins in the supernatants were precipitated on ice for 30 min by addition of 5% (wt/vol) (final concentration) trichloroacetic acid. After the mixtures had been centrifuged at 20,000 × g for 5 min at 4°C, the pellets were rinsed twice with 70% ethanol and then dissolved in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer (22). The mixtures were boiled for 5 min and then used for SDS-PAGE, Western blot analysis, and zymography. Cell walls of B. subtilis ATCC 6633 (Sigma) were prepared essentially as described previously (9, 19).
SDS-PAGE, Western blot analysis, and zymography.
SDS-PAGE was carried out as described by Laemmli (22). For Western blot analysis, the CWB proteins were separated by SDS-12% PAGE. After electrophoresis, the proteins in the gels were transferred to Hybond membranes (Amersham Biosciences) in transfer buffer (25 mM Tris-HCl [pH 8.3], 192 mM glycine, 20% [vol/vol] methanol, 0.1% [wt/vol] SDS) by using a semidry blotting system (Bio-Rad). Immunoblotting was carried out essentially as described in the instruction manual for the ECL Plus Western blotting starter core kit (Amersham Biosciences). For immunodetection, anti-FLAG M2 monoclonal antibody (Sigma) that was diluted 1:10,000 was used as the primary antibody. Zymography was performed as described by Leclerc and Asselin (24) by using SDS-10% polyacrylamide gels (for LytE-3xFLAG) or SDS-12% polyacrylamide gels (for LytC-3xFLAG and LytF-3xFLAG) containing 0.1% (wt/vol) B. subtilis cell walls. Renaturation was performed at 37°C in a renaturation solution (25 mM Tris-HCl [pH 7.2], 1% [vol/vol] Triton X-100) as described previously (38). The incubation times for the renaturation procedure were 1 h (LytF-3xFLAG), 3 h (LytE-3xFLAG), and 4 h (LytC-3xFLAG).
RESULTS
For localization analysis of vegetative cell wall lytic enzymes, we constructed three strains, E3FL, F3FL, and B3FL, carrying lytF-3xFLAG, lytE-3xFLAG, and lytC-3xFLAG fusion genes, respectively. We confirmed by PCR that the fusion genes were constructed at the original loci on the B. subtilis chromosome (data not shown). Therefore, these fusion genes were expressed with their original promoters and ribosome binding sites. To determine whether the fusion proteins were secreted and bound on the B. subtilis cell surface, cell surface extracts were prepared and used for Western blot analysis and zymography.
Effects of extracellular proteases on the stability of the LytF-3xFLAG, LytE-3xFLAG, and LytC-3xFLAG fusion proteins.
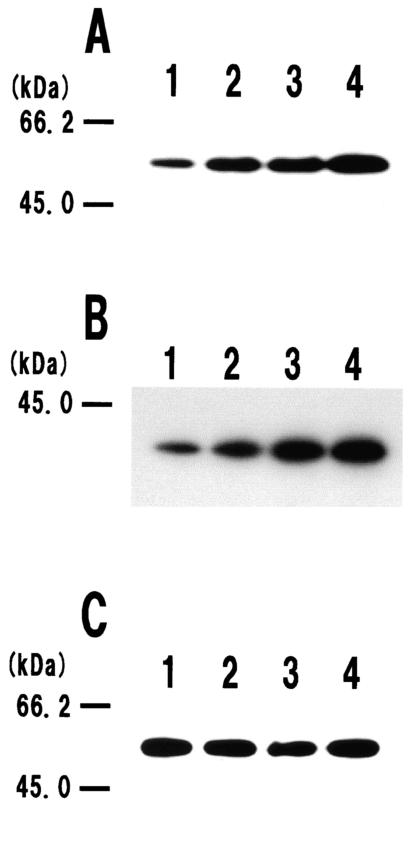
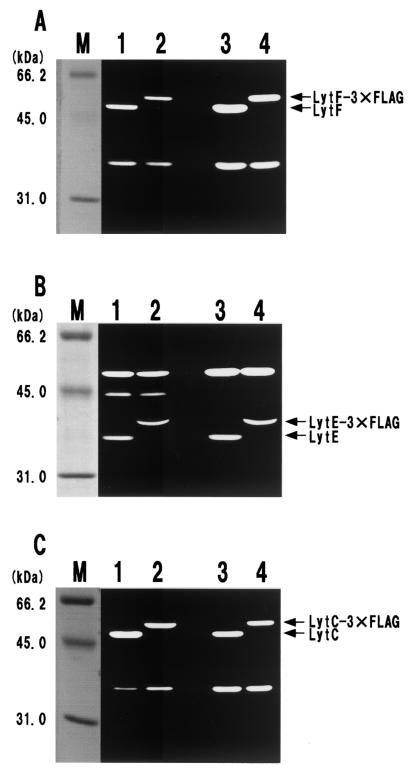
In a previous study, it was found that cell surface and extracellular proteases (WprA and Epr, respectively) seem to play important roles in the stability and processing of cell surface proteins in B. subtilis (2). Thus, we examined whether WprA and Epr cause the instability of vegetative cell wall lytic enzymes. To examine a fusion between LytF (CwlE; 462 amino acids without a putative signal peptide [32, 34, 43]; Mr, 48,819) and 3xFLAG (28 amino acids; Mr, 3,326), we constructed three mutant strains with mutations in the protease genes (wprA and/or epr). Western blot analysis was performed with the cell surface proteins that were extracted from mutant cells expressing LytF-3xFLAG (490 amino acids without the signal peptide; Mr, 52,145) at the transition phase. On an SDS-PAGE gel stained with Coomassie brilliant blue R250, the fusion protein band was not clear even for a protease-deficient double mutant, WE1E3FL (wprA epr lytF-3xFLAG) (data not shown). In contrast, we readily detected an approximately 52-kDa band corresponding to the LytF-3xFLAG fusion protein on a Western blot with the anti-FLAG M2 monoclonal antibody (Fig. 1A). The fusion protein band signals for EPRE3FL (epr lytF-3xFLAG) and WAE3FL (wprA lytF-3xFLAG) were stronger than the signal for the wild-type background strain, E3FL (lytF-3xFLAG), indicating that both WprA and Epr significantly affect the stability of LytF-3xFLAG. Moreover, the strongest signal was observed for a wprA epr double mutant, WE1E3FL (Fig. 1A, lane 4). On a zymographic gel, the original 49-kDa LytF band was observed for the wild-type strain, whereas a 52-kDa cell wall lytic band corresponding to the LytF-3xFLAG fusion protein was detected for the E3FL extract (Fig. 2A, lane 2). This result strongly indicates that the LytF-3xFLAG fusion protein retains the cell wall lytic activity. Moreover, more fusion protein accumulated in the WE1E3FL extract (Fig. 2A, lane 4), supporting the results of the Western blot analysis. These findings strongly suggest that LytF-3xFLAG is at least degraded by the cell surface and extracellular proteases WprA and Epr. In addition, the 3xFLAG-tagged peptide seemed to be quite stable on the B. subtilis cell surface, because no band corresponding to the original size of LytF (49 kDa) was observed for the cell surface extract from cells producing LytF-3xFLAG (Fig. 2A, lanes 2 and 4).
FIG.1.
Effects of extracellular proteases on degradation of the LytF-3xFLAG (A), LytE-3xFLAG (B), and LytC-3xFLAG (C) fusion proteins. Electrophoresis was performed on SDS-12% polyacrylamide gels. Cell surface proteins were prepared and subjected to Western blot analysis as described in Materials and Methods. The molecular masses of the protein standards (Bio-Rad) are indicated on the left. We confirmed the reproducibility of Western blot analysis in at least three independent experiments. (A) Western blot of the LytF-3xFLAG fusion protein. Lanes 1 to 4 contained cell surface extracts of the E3FL (lytF-3xFLAG), EPRE3FL (epr lytF-3xFLAG), WAE3FL (wprA lytF-3xFLAG), and WE1E3FL (wprA epr lytF-3xFLAG) strains, respectively. Cells were cultured in LB medium at 37°C and were harvested at the transition stage (OD600, 1.9). Equal amounts of cell surface proteins (equivalent to 0.15 OD600 unit) were applied to the lanes. (B) Western blot of the LytE-3xFLAG fusion protein. Lanes 1 to 4 contained cell surface extracts of the F3FL (lytE-3xFLAG), EPRF3FL (epr lytE-3xFLAG), WAF3FL (wprA lytE-3xFLAG), and WE1F3FL (wprA epr lytE-3xFLAG) strains, respectively. Cells were cultured in LB medium at 37°C and were harvested at the transition stage (OD600, 1.8). Equal amounts of cell surface proteins (equivalent to 0.1 OD600 unit) were applied to the lanes. (C) Western blot of the LytC-3xFLAG fusion protein. Lanes 1 to 4 contained cell surface extracts of the B3FL (lytC-3xFLAG), EPRB3FL (epr lytC-3xFLAG), WAB3FL (wprA lytC-3xFLAG), and WE1B3FL (wprA epr lytC-3xFLAG) strains, respectively. Cells were cultured in LB medium at 37°C and were harvested at the transition stage (OD600, 2.3). Equal amounts of cell surface proteins (equivalent to 0.1 OD600 unit) were applied to the lanes.
FIG.2.
Zymography of the 3xFLAG fusion proteins. Cell surface proteins from cells expressing the LytF-3xFLAG (A), LytE-3xFLAG (B), and LytC-3xFLAG (C) fusion proteins were extracted as described in Materials and Methods. Equal amounts of proteins (equivalent to 10 OD600 units) were applied to the lanes. We confirmed the reproducibility of zymography in at least three independent experiments. (A) Detection of the LytF-3xFLAG fusion protein. Cells were cultured in LB medium at 37°C and were harvested at the transition stage (OD600, 2.2). Lane M, size marker; lane 1, wild type; lane 2, E3FL (lytF-3xFLAG); lane 3, WE1 (wprA epr); lane 4, WE1E3FL (wprA epr lytF-3xFLAG). (B) Detection of the LytE-3xFLAG fusion protein. Cells were cultured in LB medium at 37°C and were harvested at the transition stage (OD600, 2.0). Lane M, size marker; lane 1, wild type; lane 2, F3FL (lytE-3xFLAG); lane 3, WE1 (wprA epr); lane 4, WE1F3FL (wprA epr lytE-3xFLAG). (C) Detection of the LytC-3xFLAG fusion protein. A lytF mutation was introduced into each strain used for zymography since LytF and LytC overlapped, as described in Materials and Methods. Cells were cultured in LB medium at 37°C and were harvested at the transition stage (OD600, 2.4). Lane M, size marker; lane 1, ED (lytF); lane 2, B3FLEd (lytF lytC-3xFLAG); lane 3, WEEd (wprA epr lytF); lane 4, WEB3FLEd (wprA epr lytF lytC-3xFLAG).
In order to determine the stability and dl-endopeptidase activity of LytE-3xFLAG (337 amino acids without the signal peptide; Mr, 36,327), we carried out the same experiments with cell surface proteins extracted from cells expressing the LytE-3xFLAG fusion protein. The results of Western blot analysis and zymography of LytE-3xFLAG were very similar to the results obtained for LytF-3xFLAG. On the Western blot, a 36-kDa signal for EPRF3FL (epr lytE-3xFLAG) was stronger than the signal for E3FL (lytE-3xFLAG), and the signal for WAF3FL (wprA lytE-3xFLAG) was stronger than the signal for EPRF3FL (Fig. 1B). This suggests that WprA is more effective than Epr for specific degradation of LytE. Moreover, LytE-3xFLAG accumulated most in WE1F3FL (wprA epr lytE-3xFLAG). As shown in Fig. 2B, zymography of LytE-3xFLAG revealed that the fusion protein had cell wall lytic activity and was more stable in the wprA epr mutant strain. In addition, the results indicated that degradation of LytE-3xFLAG seemed to occur mainly in the LytE region and not in the 3xFLAG region, since no cell wall lytic band corresponding to the original size (35 kDa) of LytE (14) was observed (Fig. 2B, lanes 2 and 4). The approximately 45-kDa band in lanes 1 and 2 in Fig. 2B may be a degradation product derived from LytF, and it became visible after 3 h of incubation in the renaturation buffer.
As shown in Fig. 1C, the LytC-3xFLAG bands (500 amino acids without the signal peptide; Mr, 53,228) extracted from B3FL, EPRB3FL, WAB3FL, and WE1B3FL were detected as approximately 53-kDa signals. Interestingly, the fusion protein bands for the four strains were very similar on Western blots. These results are considerably different from the results obtained for LytF-3xFLAG or LytE-3xFLAG, as described above. The LytF and LytC proteins have very similar molecular masses, as determined by SDS-PAGE (34, 38). Therefore, we introduced a lytF mutation into B3FL and WE1B3FL in order to obtain B3FLEd and WEB3FLEd, respectively. Then the cell surface proteins were extracted and loaded onto an SDS-12% polyacrylamide gel containing B. subtilis cell walls as the substrate (Fig. 2C). The results of zymography indicated that LytC-3xFLAG had murein-hydrolyzing activity, like LytC, and that there was not a significant difference in the cell wall lytic bands corresponding to the LytC-3xFLAG fusion proteins of B3FLEd and WEB3FLEd (Fig. 2C). These findings strongly suggest that LytC-3xFLAG is degraded little by the cell surface (WprA) and extracellular (Epr) proteases.
Effects of cell surface and extracellular proteases on localization of the LytF-3xFLAG fusion protein.
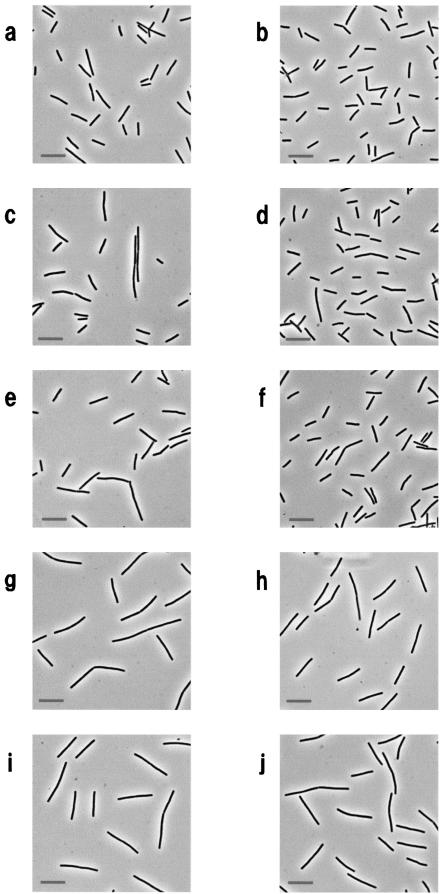
As described above, we found that LytF-3xFLAG had cell wall lytic activity and accumulated most on the cell surface of the double mutant lacking cell surface (WprA) and extracellular (Epr) proteases (Fig. 1A and 2A). To confirm these results, we carried out IFM with two strains producing the LytF-3xFLAG fusion protein. One of these strains was a wild-type background strain, E3FL, and the other was a double-mutant background strain, WE1E3FL. These two strains were cultured at 30°C in LB medium. The cells were fixed and then treated with primary and secondary antibodies. We did not observe any significant difference in cell shape between EFD (lytE::pM2cF) and E3FLFd (lytF-3xFLAG lytE::pM2cF) (Fig. 3), suggesting that the LytF-3xFLAG fusion protein functions like LytF on the B. subtilis cell surface. In addition, the growth rate of WE1 was similar to that of the wild-type strain (data not shown). Moreover, for the control strains, B. subtilis 168 and WE1, we did not detect any fluorescence of FITC under the same conditions, as determined by IFM observation.
FIG.3.
Phase-contrast microscopy of B. subtilis 168 (wild type) (a and b), EFD (lytE) (c and d), E3FLFd (lytE lytF-3xFLAG) (e and f), ED (lytF) (g and h), and F3FLEd (lytF lytE-3xFLAG) (i and j). All strains were cultured in LB medium at 30°C. Cells were harvested and fixed as described in Materials and Methods at the early exponential phase (OD600, 0.5) (a, c, e, g, and i) and the mid-exponential phase (OD600, 1.5) (b, d, f, h, and j). Bars = 10 μm.
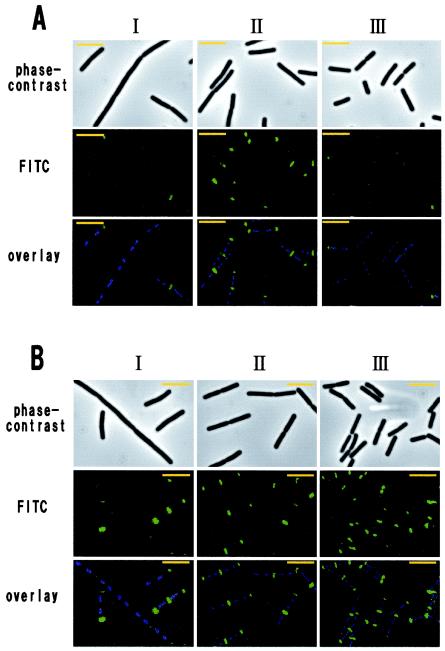
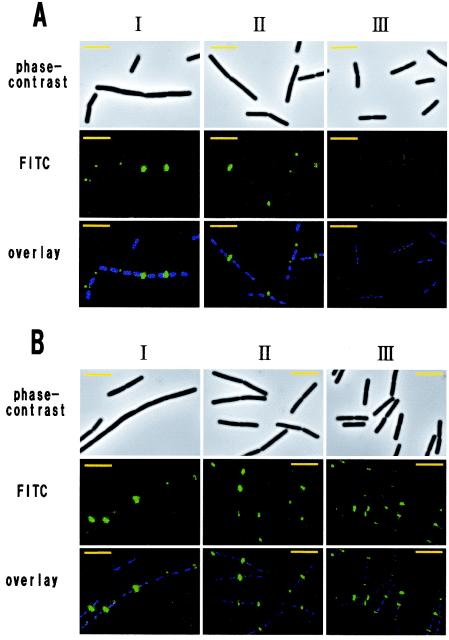
Since the lytF gene is transcribed by EσD (32, 34), the LytF-3xFLAG fusion protein is efficiently produced after the middle of the vegetative growth phase. Indeed, time course IFM of the wild-type background strain, E3FL, indicated that the percentage of fluorescent cells was maximal at the middle of the vegetative growth phase (207 of 219 cells [94%]) and then immediately decreased in the stationary phase (66 of 257 cells [26%]) (Fig. 4A). In the early vegetative phase, we observed two types of cells; some cells were normal and rod shaped, and other cells were slightly longer and in chains. The LytF-3xFLAG signals at this time were strong in the former cells but weak in the latter cells (Fig. 4A, panels I), suggesting that EσD-driven transcription may not have been activated in the latter cells.
FIG. 4.
Localization pattern of LytF-3xFLAG on the vegetative cell surface. Time course IFM was performed with E3FL (lytF-3xFLAG) (A) and WE1E3FL (wprA epr lytF-3xFLAG) (B). These strains were grown in LB medium at 30°C. The OD600 values at the sampling times for panels I, II, and III were 0.4 (early exponential phase), 1.2 (mid-exponential phase), and 2.4 (transition phase), respectively (A), and 0.4, 1.1, and 2.3, respectively (B). Each overlay image is a DAPI-stained fluorescence image (blue) superimposed on an FITC-stained image (green). The exposure times were 0.1 s for phase-contrast microscopy, 0.001 s for DAPI, and 0.05 s for FITC. Bars = 5 μm.
The fusion protein was observed at cell separation sites in the E3FL strain (Fig. 4A). At the cell poles, however, the fluorescence was very weak and frequently asymmetric, and there was fluorescence only on one side (199 of 219 cells [91%] in the mid-exponential phase). It seemed that a newly formed pole was still fluorescent, whereas an older pole was weakly fluorescent or nonfluorescent. We assumed that this difference at the cell poles might have resulted mainly from proteolysis by cell surface and/or extracellular proteases and might have depended on the order of the periods in which cell separation occurred. In support of this idea, the LytF-3xFLAG fusion protein in the WE1 background strain, WE1E3FL, was frequently observed at both cell poles (59 of 229 cells [26%] in the mid-exponential phase and 246 of 266 cells [92%] in the transition phase) (Fig. 4B). Moreover, the LytF-3xFLAG signals for WE1E3FL were stronger than those for E3FL, especially at the cell poles, and we readily detected the signals even in the transition phase (Fig. 4, panels III). These findings indicate that LytF potentially can be localized at not only cell separation sites but also cell poles and that the LytF remaining at cell poles is promptly degraded by cell surface and extracellular proteases (WprA and Epr).
LytF-3xFLAG localization in MinCD-depleted cells.
It has been reported previously that the B. subtilis minC and minD genes are in the mreBCD-minCD operon and that mutation of the min genes causes frequent misplacement of the division septum, resulting in minicells (8, 25, 26, 27, 46). To determine whether LytF-3xFLAG is localized at minicell separation sites and cell poles even in a minCD mutant, a depletion experiment with both MinC and MinD was performed. For this purpose, we constructed a conditional mutant strain with deficiencies in the two proteases, WE1EMCp (wprA epr lytF-3xFLAG Pspac-minCD), whose minCD operon was under control of an IPTG-dependent Pspac promoter. Cells began to form minicells at cell poles frequently after 2 h of incubation without IPTG, and the LytF-3xFLAG signals were observed not only at both poles but also at asymmetric minicell separation sites (Fig. 5). Moreover, cell wall staining with WGA-TMR indicated that the LytF-3xFLAG targeting sites completely overlapped the septal positions in the minicell-producing cells (Fig. 5).
FIG. 5.
LytF-3xFLAG localization in MinCD-depleted cells. B. subtilis WE1EMCp (wprA epr lytF-3xFLAG Pspac-minCD) was first grown in LB medium with 0.2 mM IPTG at 37°C to an OD600 of 0.5 to 0.7. Cells were harvested and inoculated at an OD600 of 0.08 into 5 ml of fresh LB medium without IPTG. After 2 h of incubation, cells were harvested and fixed as described in Materials and Methods. Cell wall staining (red) was performed with WGA-TMR at a final concentration of 1 μg/ml. The overlay was obtained by superimposing a DAPI-stained fluorescence image (blue), an FITC-stained image (green), and a WGA-TMR-stained image (red). The exposure times were 0.1 s for phase-contrast microscopy, 0.005 s for DAPI, 0.1 s for FITC, and 0.05 s for rhodamine. Bars = 5 μm.
Localization of the LytE-3xFLAG and LytC-3xFLAG fusion proteins.
A previous study revealed that the lytE (cwlF) gene was transcribed by EσA (major) and EσH (minor) and that the cells of a lytE mutant strain were slightly longer than those of the wild-type strain, especially in the early vegetative growth phase (14). Phase-contrast microscopy indicated that there was not a significant difference in cell shape between ED (lytF::pM2HDD) and F3FLEd (lytE-3xFLAG lytF::pM2HDD) (Fig. 3), suggesting that the LytE-3xFLAG fusion protein has the same function as LytE. The localization pattern of the LytE-3xFLAG fusion protein was observed by time course IFM (Fig. 6). Supporting the previous observations concerning the morphology of lytE mutant cells, the results indicated that the fusion protein was specifically localized at cell separation sites and cell poles. The localization patterns of LytE-3xFLAG in F3FL and WE1F3FL were quite like those of LytF-3xFLAG (Fig. 4). However, the LytE-3xFLAG signals were strong in long-chain cells but weak in normal rod-shaped cells, especially during the early vegetative growth phase (Fig. 6A, panels I). This is clearly different from the results obtained for LytF-3xFLAG, as described above. The difference seemed to result from the main promoter activities, EσD for LytF and EσA for LytE. Moreover, when cells entered the stationary phase, the specific signals for WE1F3FL (250 of 258 cells [97%]) were readily observed at cell separation sites and cell poles, as they were for LytF (Fig. 4B, panels III, and 6B, panels III). However, the LytE signals at the same phase were very weak in the wild-type background strain, F3FL (39 of 218 cells [18%]) (Fig. 6A, panels III). This difference seemed to result from degradation by WprA and Epr. Consequently, these results strongly suggest that LytE also functions in cell separation of growing cells like LytF.
FIG. 6.
Localization pattern of LytE-3xFLAG on the vegetative cell surface. Time course IFM was performed with F3FL (lytE-3xFLAG) (A) and WE1F3FL (wprA epr lytE-3xFLAG) (B). These strains were grown in LB medium at 30°C. The OD600 values at the sampling times for panels I, II, and III, were 0.4 (early exponential phase), 1.0 (mid-exponential phase), and 2.3 (transition phase), respectively (A), and 0.4, 1.0, and 2.3, respectively (B). Each overlay is a DAPI-stained fluorescence image (blue) superimposed on an FITC-stained image (green). The exposure times were 0.1 s for phase-contrast microscopy, 0.001 s for DAPI, and 0.05 s for FITC. Bars = 5 μm.
It has been reported that LytC is an N-acetylmuramoyl-l-alanine amidase and that the cell morphology of a lytC mutant is not different from that of the wild-type (20, 30). Therefore, we presumed that the LytC protein is not required for cell separation. Since lytC is transcribed by EσA (minor) and EσD (major) (21, 23), IFM of B3FL producing LytC-3xFLAG indicated that signals for the fusion protein were observed during the late vegetative growth phase (194 of 212 cells [92%]) (Fig. 7A, panels III). The LytC-3xFLAG fusion protein was localized on the entire cell surface, indicating that the localization pattern of LytC is clearly different from the localization patterns of LytF and LytE. Moreover, we did not observe any difference in fluorescence strength and frequency between B3FL (194 of 212 cells [92%]) and WE1B3FL (189 of 201 cells [94%]) in the transition phase, suggesting that LytC may not be a target of WprA and Epr (Fig. 7). These observations are consistent with the results of Western blotting and zymography (Fig. 1C and 2C).
FIG. 7.
Localization pattern of LytC-3xFLAG. Time course IFM was performed with B3FL (lytC-3xFLAG) (A) and WE1B3FL (wprA epr lytC-3xFLAG) (B). These strains were grown in LB medium at 30°C. The OD600 values at the sampling times for panels I, II, and III were 0.5 (early exponential phase), 1.9 (mid-exponential phase), and 2.7 (transition phase), respectively (A), and 0.5, 1.9, and 2.6, respectively (B). The exposure times were 0.1 s for phase-contrast microscopy and 0.05 s for FITC (green). Bars = 5 μm.
DISCUSSION
To investigate the localization of cell wall lytic enzymes in vivo, we chose an epitope-tagging method in which a 3xFLAG polypeptide was used. We initially used a GFP fusion, but we did not detect any signals for the WE1 background strain because of proteolysis, mainly at the site of linkage between autolysin and the fused GFP domain (data not shown). Next, we attempted to localize a LytF-FLAG fusion protein by the method successfully employed by Pedersen et al. for penicillin binding protein 1 (36). However, no specific signals were observed (data not shown). Thus, we finally selected a 3xFLAG epitope that exhibits approximately 10-fold-higher affinity to an anti-FLAG monoclonal antibody than the FLAG epitope exhibits. The advantage of this method is that the fusion genes are transcribed from the original promoters and expressed with their own ribosome binding sites. Therefore, the information for the 3xFLAG fusion protein seems to be very similar to that for the original protein. In fact, zymography and Western blot analyses indicated that the LytF-3xFLAG, LytE-3xFLAG, and LytC-3xFLAG fusion proteins are normally secreted on the vegetative cell surface, have cell wall lytic activities, and are quite stable on the B. subtilis cell surface (Fig. 1 and 2). In addition, cells expressing these fusion proteins exhibited no significant difference in morphology compared with cells of the parental strains (data not shown).
In a previous report, cells lacking lytF and/or lytE were inhibited only in cell separation and were not inhibited in cell division, since normal septa seemed to be formed even in the chains of cells lacking both lytF and lytE (34). Therefore, we presumed that the two dl-endopeptidases might play important roles in cell separation in the final stage of cell division. In this study, we demonstrated that two cell wall hydrolases, LytF and LytE, are potentially localized at the cell separation sites and cell poles of vegetative cells (Fig. 4 and 6). These findings are consistent with the phenotypes of lytF (cwlE) and/or lytE (cwlF) mutant cells (14, 34). In addition, a MinCD depletion experiment revealed that LytF-3xFLAG is localized at minicell separation sites in addition to cell poles (Fig. 5). This finding strongly suggests that LytF targets the septum-forming sites and that depletion of MinCD may not affect LytF localization.
It has been reported that WprA is a major CWB protease (31), whereas Epr is a minor extracellular protease (41). In a previous proteome analysis, a processing product of Epr was found in the extracellular fraction but not in the cell surface fraction (2). However, Western blot analysis revealed that the LytF-3xFLAG fusion protein was more abundant in cell surface extracts of a wprA mutant and an epr mutant than in the extract of the wild-type strain and that there were equivalent amounts in the two mutants (Fig. 1A). The greatest accumulation of the fusion protein was in the wprA epr double mutant, WE1E3FL (Fig. 1A). We also observed that the LytE-3xFLAG fusion protein was degraded by WprA and Epr (Fig. 1B). These findings strongly suggest that LytF and LytE are attacked by at least cell surface and extracellular proteases (WprA and Epr) in the transition phase. Moreover, we demonstrated using IFM that the LytF-3xFLAG and LytE-3xFLAG fusion proteins are localized not only at the medial cell separation sites but also at cell poles of vegetative cells in double-mutant background strains WE1E3FL and WE1F3FL (Fig. 4B and 6B). On the other hand, the specific signals in the wild-type background strains were reduced, especially at the cell poles (Fig. 4A and 6A). It seemed that this reduction mainly resulted from degradation of LytF and LytE, suggesting that the two dl-endopeptidases are not reused or reassembled at the next cell separation site. Interestingly, since the epr gene was transcribed by EσD, as was lytF (7), it may be important for the growth of cells to maintain the balance of production and degradation of the dl-endopeptidases LytF and LytE. Consequently, one of the important roles of WprA and Epr may be specific degradation of unnecessary LytF and LytE at the cell poles. In support of this idea, we readily observed the LytF-3xFLAG- and LytE-3xFLAG-specific signals even in the transition state in the WE1 background cells, but not in the wild-type cells (Fig. 4 and 6, panels III).
In early vegetative growth, the LytF-3xFLAG signals were stronger in normal rod-shaped cells than in long-chain cells (Fig. 4), suggesting that EσD-specific transcription may not be activated in the latter cells. This suggestion is supported by the fact that either a sigD or lytF mutation results in a typical chain phenotype during vegetative growth (12, 34). In contrast to LytF-3xFLAG signals, the LytE-3xFLAG signals were strong in long-chain cells but weak in normal rod-shaped cells, especially in the early vegetative growth phase (Fig. 6, panels I). This difference in localization pattern between LytF and LytE seemed to result from the main promoters, EσD for lytF and EσA for lytE (14, 32, 33, 34). Moreover, the localization pattern of LytE-3xFLAG was consistent with chain formation by lytE mutant cells in this phase (32, 34).
In the case of LytC-3xFLAG, we observed that the fusion protein was localized on the entire cell surface after the middle of the exponential growth phase and that there was no significant difference in fluorescence strength between wild-type and WE1 background cells (Fig. 7). In support of these observations, the results of zymography and Western blotting revealed that LytC is not a target of the cell surface and extracellular proteases, WprA and Epr (Fig. 1C and 2C). These findings led us propose two possibilities. First, LytC may be potentially resistant to these proteases, and second, WprA and Epr may also be localized at cell separation sites and cell poles for the specific degradation of dl-endopeptidases. To examine these possibilities, systematic localization analysis of the B. subtilis cell surface proteins, including WprA and Epr, is under way. In addition, further studies are in progress to identify a factor(s) that allows LytF and LytE to be localized at cell separation sites.
Acknowledgments
We thank N. Ogasawara and K. Kobayashi for kindly providing plasmid pCA3xFLAG.
This research was supported by Grants-in-Aid for Scientific Research on Priority Areas Genome Biology (2) (grant 12206005), the 21st Century COE Program and Scientific Research (B) (grant 13460037) (to J.S.), and Young Scientists (B) (grant 14760046) (to H.Y.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antelmann, H., H. Yamamoto, J. Sekiguchi, and M. Hecker. 2002. Stabilization of cell wall proteins in Bacillus subtilis: a proteomic approach. Proteomics 2:591-602. [DOI] [PubMed] [Google Scholar]
- 3.Baba, T., and O. Schneewind. 1998. Targeting of muralytic enzymes to the cell division site of Gram-positive bacteria: repeat domains direct autolysin to the equatorial surface ring of Staphylococcus aureus. EMBO J. 17:4639-4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bateman, A., and M. Bycroft. 2000. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299:1113-1119. [DOI] [PubMed] [Google Scholar]
- 5.Bruckner, R., O. Shosheyov, and R. H. Doi. 1990. Multiple active forms of a novel serine protease from Bacillus subtilis. Mol. Gen. Genet. 221:486-490. [DOI] [PubMed] [Google Scholar]
- 6.Canosi, U., G. Morelli, and T. A. Trautner. 1978. The relationship between molecular structure and transformation efficiency of some Streptococcus aureus plasmids isolated from Bacillus subtilis. Mol. Gen. Genet. 166:259-267. [DOI] [PubMed] [Google Scholar]
- 7.Dixit, M., C. S. Murudkar, and K. Krishnamurthy Rao. 2002. epr is transcribed from a σD promoter and is involved in swarming of Bacillus subtilis. J. Bacteriol. 184:596-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Errington, J., and R. A. Daniel. 2002. Cell division during growth and sporulation, p. 97-109. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 9.Fein, J. E., and H. J. Rogers. 1976. Autolytic enzyme-deficient mutants of Bacillus subtilis 168. J. Bacteriol. 127:1427-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster, S. J., and D. L. Popham. 2002. Structure and synthesis of cell wall, spore cortex, teichoic acids, S-layers, and capsules, p. 21-41. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 11.Fukushima, T., H. Yamamoto, A. Atrih, S. J. Foster, and J. Sekiguchi. 2002. A polysaccharide deacetylase gene (pdaA) is required for germination and for production of muramic δ-lactam residues in the spore cortex of Bacillus subtilis. J. Bacteriol. 184:6007-6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helmann, J. D., L. M. Marquez, and M. J. Chamberlin. 1988. Cloning, sequencing, and disruption of the Bacillus subtilis sigma 28 gene. J. Bacteriol. 170:1568-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol. 150:815-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishikawa, S., Y. Hara, R. Ohnishi, and J. Sekiguchi. 1998. Regulation of a new cell wall hydrolase gene, cwlF, which affects cell separation in Bacillus subtilis. J. Bacteriol. 180:2549-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi, G., J. Toida, T. Akamatsu, H. Yamamoto, T. Shida, and J. Sekiguchi. 2000. Accumulation of an artificial cell wall-binding lipase by Bacillus subtilis wprA and/or sigD mutants. FEMS Microbiol. Lett. 188:165-169. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi, G., J. Toida, T. Akamatsu, H. Yamamoto, T. Shida, and J. Sekiguchi. 2000. Accumulation of a recombinant Aspergillus oryzae lipase artificially localized on the Bacillus subtilis cell surface. J. Biosci. Bioeng. 90:422-425. [PubMed] [Google Scholar]
- 17.Köhler, S., M. Leimeister-Wächter, T. Chakraborty, F. Lottspeich, and W. Goebel. 1990. The gene coding for protein p60 of Listeria monocytogenes and its use as a specific probe for Listeria monocytogenes. Infect. Immun. 58:1943-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 19.Kuroda, A., and J. Sekiguchi. 1990. Cloning, sequencing and genetic mapping of a Bacillus subtilis cell wall hydrolase gene. J. Gen. Microbiol. 136:2209-2216. [DOI] [PubMed] [Google Scholar]
- 20.Kuroda, A., and J. Sekiguchi. 1991. Molecular cloning and sequencing of a major Bacillus subtilis autolysin gene. J. Bacteriol. 173:7304-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuroda, A., and J. Sekiguchi. 1993. High-level transcription of the major Bacillus subtilis autolysin operon depends on expression of the sigma D gene and is affected by a sin (flaD) mutation. J. Bacteriol. 175:795-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Lazarevic, V., P. Margot, B. Soldo, and D. Karamata. 1992. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-l-alanine amidase and its modifier. J. Gen. Microbiol. 138:1949-1961. [DOI] [PubMed] [Google Scholar]
- 24.Leclerc, D., and A. Asselin. 1989. Detection of bacterial cell wall hydrolases after denaturing polyacrylamide gel electrophoresis. Can. J. Microbiol. 35:749-753. [DOI] [PubMed] [Google Scholar]
- 25.Lee, S., and C. W. Price. 1993. The minCD locus of Bacillus subtilis lacks the minE determinant that provides topological specificity to cell division. Mol. Microbiol. 7:601-610. [DOI] [PubMed] [Google Scholar]
- 26.Levin, P. A., J. J. Shim, and A. D. Grossman. 1998. Effect of minCD on FtsZ ring position and polar septation in Bacillus subtilis. J. Bacteriol. 180:6048-6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin, P. A., P. S. Margolis, P. Setlow, R. Losick, and D. Sun. 1992. Identification of Bacillus subtilis genes for septum placement and shape determination. J. Bacteriol. 174:6717-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loessner, M. J., K. Kramer, F. Ebel, and S. Scherer. 2002. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44:335-349. [DOI] [PubMed] [Google Scholar]
- 29.Margot, P., C. Mauël, and D. Karamata. 1994. The gene of the N-acetylglucosaminidase, a Bacillus subtilis cell wall hydrolase not involved in vegetative cell autolysis. Mol. Microbiol. 12:535-545. [DOI] [PubMed] [Google Scholar]
- 30.Margot, P., and D. Karamata. 1992. Identification of the structural genes for N-acetylmuramoyl-l-alanine amidase and its modifier in Bacillus subtilis 168: inactivation of these genes by insertional mutagenesis has no effect on growth or cell separation. Mol. Gen. Genet. 232:359-366. [DOI] [PubMed] [Google Scholar]
- 31.Margot, P., and D. Karamata. 1996. The wprA gene of Bacillus subtilis 168, expressed during exponential growth, encodes a cell-wall-associated protease. Microbiology 142:3437-3444. [DOI] [PubMed] [Google Scholar]
- 32.Margot, P., M. Pagni, and D. Karamata. 1999. Bacillus subtilis 168 gene lytF encodes a γ-d-glutamate-meso-diaminopimelate muropeptidase expressed by the alternative vegetative sigma factor, σD. Microbiology 145:57-65. [DOI] [PubMed] [Google Scholar]
- 33.Margot, P., M. Wahlen, A. Gholamhuseinian, P. Piggot, and D. Karamata. 1998. The lytE gene of Bacillus subtilis 168 encodes a cell wall hydrolase. J. Bacteriol. 180:749-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohnishi, R., S. Ishikawa, and J. Sekiguchi. 1999. Peptidoglycan hydrolase LytF plays a role in cell separation with LytE during vegetative growth of Bacillus subtilis. J. Bacteriol. 181:3178-3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oshida, T., M. Sugai, H. Komatsuzawa, Y.-M. Hong, H. Suginaka, and A. Tomasz. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-l-alanine amidase domain and an endo-β-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. USA 92:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedersen, L. B., E. R. Angert, and P. Setlow. 1999. Septal localization of penicillin-binding protein 1 in Bacillus subtilis. J. Bacteriol. 181:3201-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rashid, M. H., M. Mori, and J. Sekiguchi. 1995. Glucosaminidase of Bacillus subtilis: cloning, regulation, primary structure and biochemical characterization. Microbiology 141:2391-2404. [DOI] [PubMed] [Google Scholar]
- 38.Rashid, M. H., N. Sato, and J. Sekiguchi. 1995. Analysis of the minor autolysins of Bacillus subtilis during vegetative growth by zymography. FEMS Microbiol. Lett. 132:131-137. [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 40.Schubert, K., A. M. Bichmaier, E. Mager, K. Wolff, G. Ruhland, and F. Fiedler. 2000. P45, an extracellular 45 kDa protein of Listeria monocytogenes with similarity to protein p60 and exhibiting peptidoglycan lytic activity. Arch. Microbiol. 173:21-28. [DOI] [PubMed] [Google Scholar]
- 41.Sloma, A., A. Ally, D. Ally, and J. Pero. 1988. Gene encoding a minor extracellular protease in Bacillus subtilis. J. Bacteriol. 170:5557-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, T. J., S. A. Blackman, and S. J. Foster. 2000. Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249-262. [DOI] [PubMed] [Google Scholar]
- 43.Tjalsma, H., A. Bolhuis, J. D. H. Jongbloed, S. Bron, and J. M. van Dijl. 2000. Signal peptide-dependent protein transport in Bacillus subtilis: a genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 64:515-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuchiya, A., G. Kobayashi, H. Yamamoto, and J. Sekiguchi. 1999. Production of a recombinant lipase artificially localized on the Bacillus subtilis cell surface. FEMS Microbiol. Lett. 176:373-378. [Google Scholar]
- 45.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 46.Varley, A. W., and G. C. Stewart. 1992. The divIVB region of the Bacillus subtilis chromosome encodes homologs of Escherichia coli septum placement (MinCD) and cell shape (MreBCD) determinants. J. Bacteriol. 174:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wuenscher, M. D., S. Köhler, A. Bubert, U. Gerike, and W. Goebel. 1993. The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J. Bacteriol. 175:3491-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamada, K. 1989. Ph.D. thesis. Hiroshima University, Hiroshima, Japan.
- 49.Yamada, S., M. Sugai, H. Komatsuzawa, S. Nakashima, T. Oshida, A. Matsumoto, and H. Suginaka. 1996. An autolysin ring associated with cell separation of Staphylococcus aureus. J. Bacteriol. 178:1565-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]