Abstract
Stimulation of death receptors by agonists such as FasL and TNFα activates apoptotic cell death in apoptotic competent conditions or a type of necrotic cell death dependent on RIP1 kinase, termed necroptosis, in apoptotic deficient conditions. In a genome-wide siRNA screen for regulators of necroptosis, we identify a set of 432 genes that regulate necroptosis, a subset of 32 genes that act downstream and/or as regulators of RIP1 kinase, 32 genes required for death receptor mediated apoptosis, and 7 genes involved in both necroptosis and apoptosis. We show that the expression of subsets of the 432 genes are enriched in the immune and nervous systems, and cellular sensitivity to necroptosis is regulated by an extensive signaling network mediating innate immunity. Interestingly, Bmf, a BH3-only Bcl-2 family member, is required for death receptor-induced necroptosis. Our study defines a cellular signaling network that regulates necroptosis and the molecular bifurcation that controls apoptosis and necroptosis.
Cell death has been traditionally classified as apoptosis or necrosis. While apoptosis is known as a regulated cellular mechanism, necrosis is known as passive cell death caused by overwhelming stress. Necrosis is characterized by rapid loss of plasma membrane integrity, organelle swelling and mitochondrial dysfunction, and the lack of typical apoptotic features such as internucleosomal DNA cleavage and nuclear condensation. Although necrosis is known to occur under a variety of pathological conditions, little effort has been made to study necrosis due to the belief in its unregulated nature. Support for a regulated necrosis mechanism came from studies of the death receptors. Activation of the Fas and TNFR family of death receptors induces a “prototypic” apoptotic pathway through the recruitment of adaptor proteins such as FADD and upstream caspases such as caspase-8. Interestingly, it was discovered that in certain cell types, stimulation with FasL or TNFα under apoptosis deficient conditions could induce cell death with morphological features of necrosis (Kawahara et al., 1998; Vercammen et al., 1997). The fact that the activation of Fas/TNFα receptors may lead to cell death with features of either apoptosis or necrosis argues strongly for the existence of a regulated cellular necrosis mechanism, discrete from apoptosis, which we termed “necroptosis” (Degterev et al., 2005).
RIP1 is a death-domain containing kinase associated with the death receptors but its kinase activity is dispensable for the induction of death receptor mediated apoptosis (Grimm et al., 1996). In apoptosis deficient conditions, however, RIP1 kinase activity has been found to be required for the activation of necroptosis by death receptor agonists (Holler et al., 2000). In our previous studies, we have isolated multiple small molecule inhibitors of necroptosis, termed necrostatins (Necs) (Degterev et al., 2008; Degterev et al., 2005). Importantly, we have shown that Nec-1 is an allosteric inhibitor of RIP1 kinase activity (Degterev et al., 2008). Using Nec-1 as a tool, necroptosis has since been found to contribute to a wide range of pathologic cell death paradigms including ischemic brain injury, myocardial infarction, excitotoxicity and chemotherapy-induced cell death (Degterev et al., 2005; Han et al., 2007; Smith et al., 2007; Xu et al., 2007). Here, we have broadly explored the molecular mechanism and functional significance of necroptosis by carrying out a genome-wide siRNA screen for genes required for necroptosis. Our study defines a genetic profile for a cellular necrotic pathway, elucidates the connection between apoptosis and necroptosis, and implicates necroptosis as a critical regulatory pathway for innate immunity and suggests a potential role of necroptosis in human disease.
Results
A screen for genes required for necroptosis
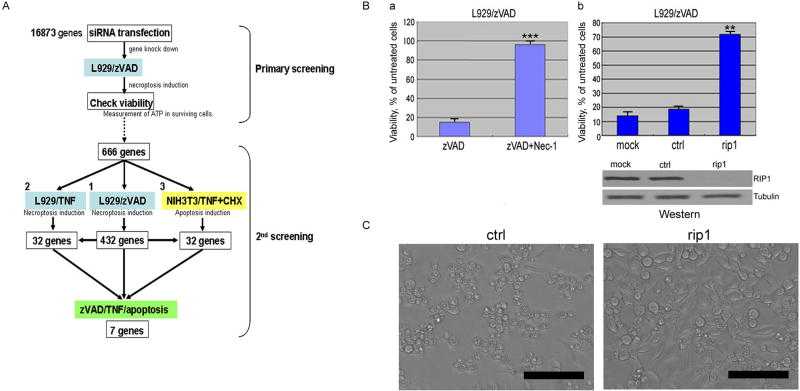
The treatment of L929 cells with zVAD.fmk has been shown to induce necroptosis, which can be inhibited by Nec-1 (Degterev et al., 2005). Using this model, we screened the Dharmacon siRNA library covering the entire mouse genome (16,873 genes) for genes required for necroptosis (Figure 1A). RIP1 siRNA was used as a positive control as knockdown of RIP1 efficiently blocked necroptosis induced by zVAD.fmk (Figure 1B). In the non-targeting siRNA (Dharmacon) transfected cells, the treatment of zVAD.fmk induced ∼80% cell death. An siRNA was scored as positive if its ATP level (a surrogate for cell survival) was >2SD above the mean ATP level of the plate. Using this criterion, 666 genes were scored as candidates required for zVAD.fmk-induced necroptosis in L929 cells. As expected, rip1 was scored as a hit in this assay, providing a validation for our approach (Figure 1B & C).
Figure 1. siRNA screen for genes required for necroptosis.
(A) A Schematic diagram of 1st and 2nd screens. (B) L929 cells, not-transfected (a), or transfected with either non-targeting siRNA(ctrl) or RIP1 siRNA at 50nM for 48hr (b), were treated with 20μM zVAD.fmk with or without 30μM Nec-1 for 18hrs. Lysates of siRNA transfected-L929 were analyzed by western blotting using anti-RIP1 or anti-Tubulin (bottom panel). (C) The morphological change of L929 cells transfected with indicated siRNAs after 18hr treatment of 20μM zVAD.fmk were shown by phase-contrast microscopic pictures. Scale bars: 100μm. Cellular viability was evaluated by measuring ATP levels in surviving cells using CellTiter-Glo. SD of cellular viability or cell death assays in all figures: *= p<0.05, **= p<0.01, ***= p<0.001, (n=4).
To confirm the screening result, we re-screened the 666 primary siRNA hits using 4 individual siRNAs for each gene. In order to restrict our analysis to genes that have major impacts on cellular sensitivity to necroptosis, we required that at least 2 out of the 4 siRNAs increased cell survival for >3SD above that of cells transfected with non-targeting siRNA control, and showed at least 60% of the viability of cells expressing the positive control rip1 siRNA. Using these criteria, 432 genes were scored positive. RIP1 was again one of the validated hits, with all 4 siRNAs showing a protective effect against necroptosis induced by zVAD.fmk (data not shown).
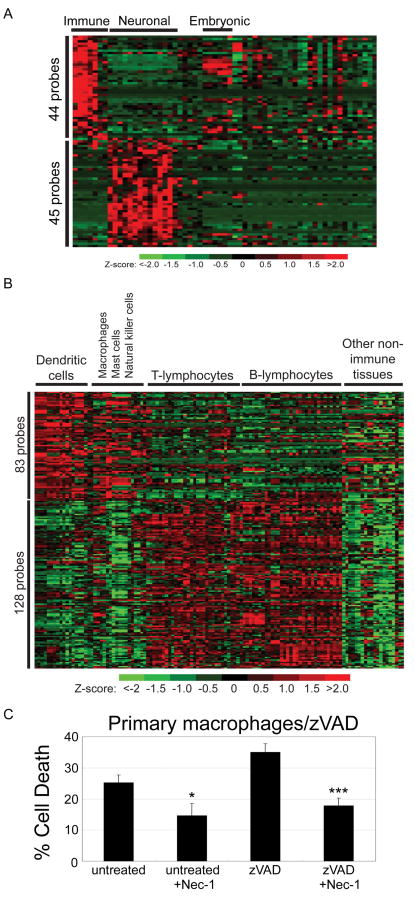
We examined the expression of genes identified in the zVAD.fmk screen across 61 normal mouse tissues using microarray data files from the Novartis GNF mouse expression atlas resource (Su et al., 2004) and found clusters of hits showing increased expression in immune and neuronal cells/tissues (FDR-adjusted p<0.05) (Figure 2A & Supplemental Table 1 & 2). To further define the immune cell types, we also analyzed a larger mouse immune microarray panel comprising gene expression profiles from 119 mouse cell/tissue samples of which 83% represent various types of primary immune cells (Hijikata et al., 2007). We observed a cluster of genes from the zVAD.fmk screen exhibiting increased expression in macrophages, dendritic and NK cells and another cluster showing enhanced expression in B- and T-lymphocytes (Figure 2B & Supplemental Table 3 & 4). The enriched expression in cells of the immune system also suggests a role of necroptosis in regulating immune function.
Figure 2. Expression patterns of the zVAD hits in mouse tissues and primary cells.
(A) Expression profiles of genes from the zVAD.fmk screen showing significantly higher expression in immune cells or neuronal cells (FDR-adjusted p<0.05) as observed in the Novartis GNF1m microarray data examining expression across 61 mouse tissues. (B) Expression profiles of genes from the zVAD.fmk screen exhibiting increased expression in mouse macrophages, dendritic cells and NK cells or in B- and T-lymphocytes, from a large microarray panel of 119 mouse cell/tissue samples obtained from the RIKEN resource. (C) Involvement of necroptosis in primary macrophage cell death induced by zVAD.fmk. Isolated peritoneal macrophages from mice stimulated by thioglycollate were untreated or treated with 100μM zVAD.fmk with or without 30μM Nec-1 for 24hr and cell death was measured by Sytox assay (Invitrogen).
To further confirm a functional role of necroptosis in the immune system, we tested the sensitivity of primary peritoneal macrophages to necrostatin-1. As shown in Figure 2C, spontaneous as well as zVAD.fmk induced cell death of macrophages was inhibited by Nec-1, suggesting that necroptosis plays a role in regulating the survival of activated primary macrophages, one of the key cell types involved in innate immunity. On the other hand, death receptor mediated cell death of epithelial cell lines, such as HeLa cells, 293 cells (Degterev et al., 2005) and MCF10A (data not shown) are not sensitive to the inhibition of Nec-1.
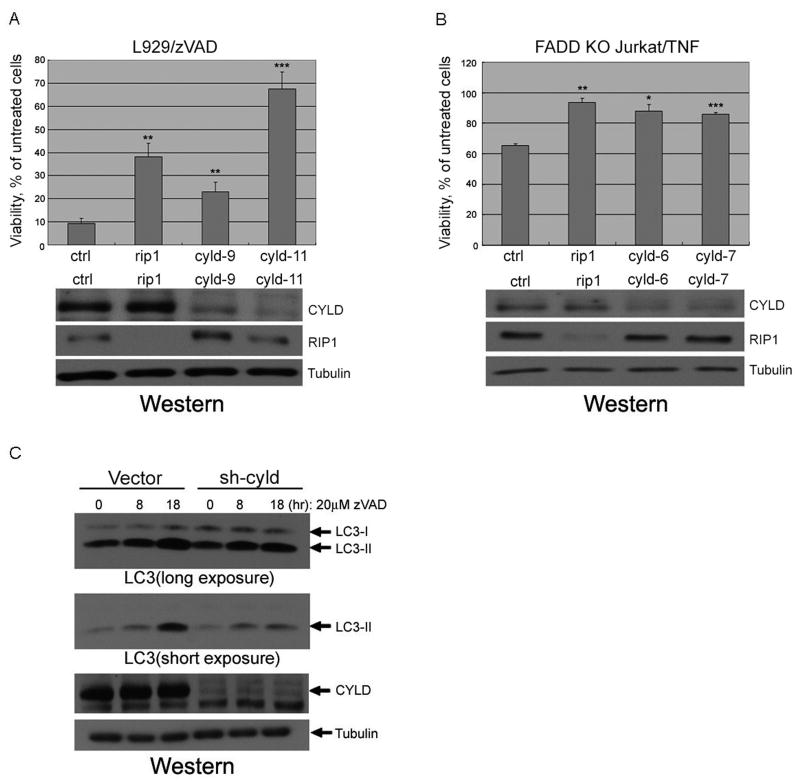
CYLD, a tumor suppressor and a de-ubiquitinating enzyme encoded by a gene that is mutated in familial cylindromatosis (Simonson et al., 2007), was identified in the microarray datasets as having increased gene expression in immune and neuronal cells/tissues (Supplemental Table 1 & 2). CYLD is known to be recruited to the TNFα receptor upon its activation, and furthermore, RIP1 has been identified as a CYLD substrate (Wright et al., 2007), suggesting that CYLD may represent a key regulatory factor in the necroptotic pathway. To confirm the role of CYLD in necroptosis, we compared the efficiency of cyld knockdown with necroptosis inhibition. As shown in Figure 3A, we found an excellent correlation between the efficiency of cyld knockdown by individual siRNAs and protection against zVAD.fmk induced necroptosis in L929 cells. To further confirm the role of cyld in necroptosis, we used siRNAs against human cyld to inhibit its expression in FADD-deficient Jurkat cells. We found that inhibition of cyld expression in Jurkat cells also attenuated necroptosis (Figure 3B). Execution of necroptosis is associated with activation of autophagy and increased formation of its marker, LC3II, which is efficiently inhibited by Nec-1 (Degterev et al., 2005). Consistently, knock down of cyld also inhibited LC3II induction in L929 cells treated with zVAD.fmk (Figure 3C).
Figure 3. CYLD knockdown inhibits necroptosis.
(A) L929 cells were transfected with 3 different siRNAs against cyld (cyld9 & cyld11), and rip1 (50nM) for 48hr, treated with 20μM zVAD.fmk for additional 18hr and the viability was measured as in Figure 1. Knockdown efficiency of cyld was confirmed by western blot using anti-CYLD, anti-RIP1 or anti-Tubulin antibodies. (B) Human FADD-deficient Jurkat cells were electroporated with siRNAs against human cyld (cyld6 & cyld7). Forty-eight hrs later, the cells were treated with 10ng/ml TNFα for additional 16hr and the viability was measured as in Figure 1. The cell lysates were analyzed by western blotting as in (A). (C) Inhibition of LC3-II induction in cyld-knockdown cells. L929 cells stably expressing shRNA for cyld(sh-cyld) or vector alone(vector) were treated with 20uM zVAD.fmk for indicated periods and the cell lysates were analyzed by western blotting using anti-LC3-II, anti-CYLD or anti-Tubulin antibodies.
Since CYLD regulates TNFα signaling (Wright et al., 2007), the requirement of cyld suggests that autocrine regulation of TNFα signaling might be involved in regulating cellular sensitivity to necroptosis induced by zVAD.fmk. Consistent with this possibility, TNFR1 (TNFRSF1a) was identified as one of the genes required for zVAD.fmk-induced necroptosis (Supplemental Table 6). Since inhibition of caspases by expression of crmA, a caspase inhibitor encoded by cowpox virus, or by multiple peptide caspase inhibitors, was found to sensitize L929 cells to TNFα induced necrosis (Vercammen et al., 1998), our results indicate that an autocrine regulation of TNFR1 contributes to necroptosis induced by zVAD.fmk.
Canonical pathways in necroptosis
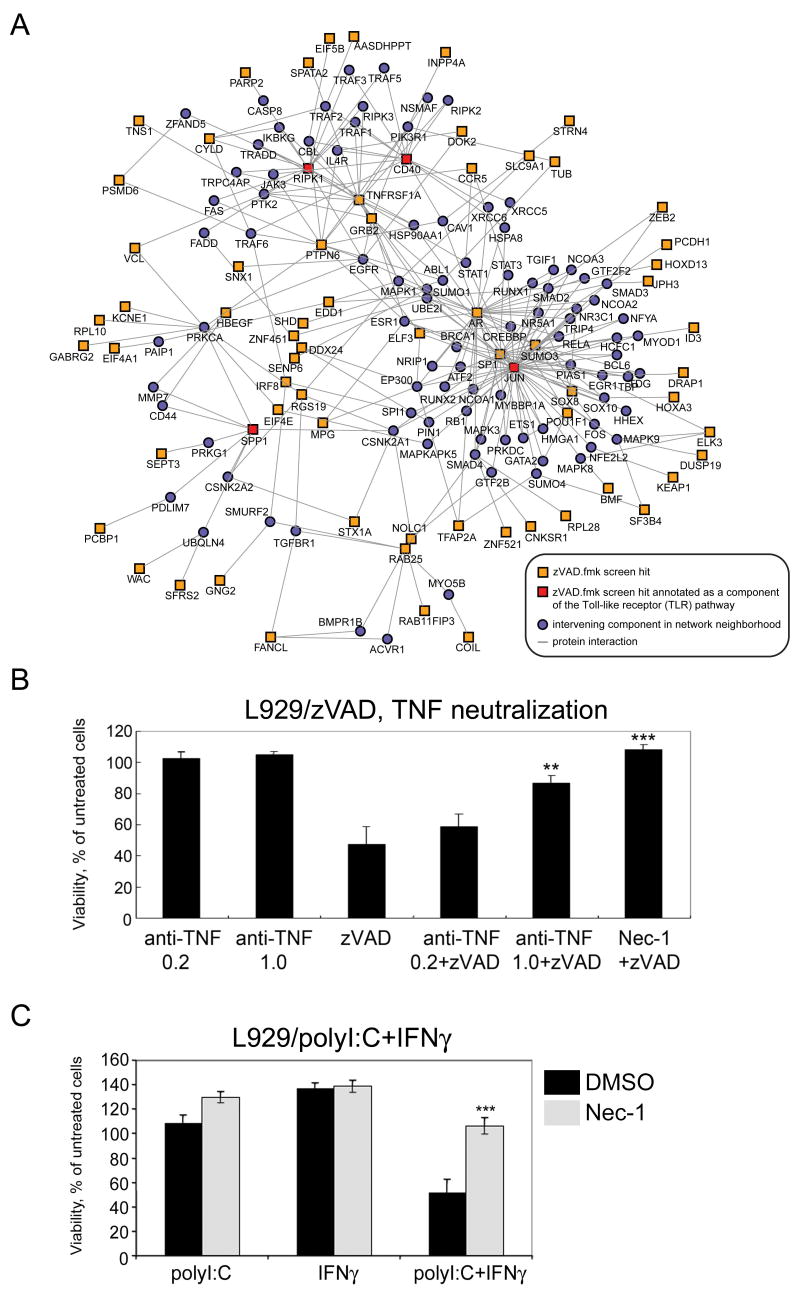
To gain insights into the function of genes scored in our siRNA screen, we used a gene-set approach to identify common pathways associated with these gene clusters. Using the Molecular Signatures Database (MSigDB) (Subramanian et al., 2005), which provides a catalog of some 3000 sets of genes divided into various annotation groupings, we interrogated human orthologs of the 432 genes involved in zVAD.fmk-induced necroptosis against curated gene sets (canonical pathways) and motif-based gene sets (transcription factor targets) from the MSigDB. We identified canonical pathways that were found to be significantly enriched (p<0.05) among zVAD.fmk hits compared to the full set of genes screened in the siRNA library to which human orthologs could be mapped. Involvement of the TNFR1 pathway is very clear as multiple hits in the zVAD.fmk screen, such as TNFR1 (TNFRSF1A), RIPK1, CYLD, JUN, and Grb2, have established roles in the TNF signaling. In addition, the Toll-like receptor (TLR) pathway, which was identified to be significantly enriched in the canonical pathway analysis, is also likely to be involved in necroptosis, as several zVAD.fmk hits, such as RIPK1, CYLD (Meylan et al., 2004; Yoshida et al., 2005), multiple members of the interferon family (IFNA1, IFNA7 and IFNA13) and an IFN induced gene, LRG47/Irgm1, are also known to have roles in the TLR pathway. Interestingly, our analysis identified that at least 70 out of 432 genes identified in the zVAD.fmk screen are connected through an extensive network surrounding innate immune pathways to TNFR1 and TLR signaling using pathway components, RIPK1, JUN, CD40 and SPP1, which are also zVAD.fmk hits, as anchor points (Figure 4A). An important role of necroptosis in innate immunity is consistent with the enriched expression of zVAD.fmk hits in the immune system. This network provides a framework for future explorations into the mechanisms by which cellular sensitivity to necroptosis is regulated in response to multiple TLR and death receptor family members.
Figure 4. Network extension of the innate immune Toll-like receptor (TLR) signaling pathway and its participation in necroptosis.
(A) Network was constructed by anchoring on TLR pathway components (CD40, RIPK1, JUN and SPP1) that are also zVAD.fmk hits, using protein-protein interaction data curated from literature and high-throughput yeast-two-hybrid screens. (B) Autocrine production of TNFα is involved in zVAD-induced necroptosis. L929 cells were pretreated with indicated concentrations of anti-mouse TNFα antibody (μg/ml) for 1hr followed by treatment with or without 20μM zVAD.fmk or 30μM Nec-1 for 16hr. (C) Involvement of TLR pathway in necroptosis. L929 cells were treated with indicated chemicals for 19hr. Cellular viability was measured as described in Figure 1A. polyI:C; 25μg/ml, interferon gamma (IFNγ); 1000U/ml, Nec-1; 30μM.
Although it was not known that the treatment with zVAD.fmk would lead to the activation of some of the signaling processes involved in the TNFR1 and TLR pathways, the agonists of TNFR1 and TLR have been previously shown to induce caspase-independent necrosis that we termed necroptosis (Degterev et al., 2005; Kalai et al., 2002; Temkin et al., 2006; Vercammen et al., 1998; Xu et al., 2006). Consistent with a role of TNFR1 in zVAD induced necroptosis, neutralization of TNFα by antibody protected L929 cells from necroptosis induced by zVAD.fmk (Figure 4B). Furthermore, we found that L929 cells can be induced to die by co-treatment with IFNγ and polyI:C (polyinosinic:polycytidylic acid), a synthetic double-stranded RNA agonist of TLR3 commonly used to mimic viral infection, and the cell death was inhibited by Nec-1 (Figure 4C). Thus, consistent with the connection of necroptosis and innate immunity, the cell death induced by the activation of TLR3 in L929 cells in the presence of IFNγ requires RIP1 kinase activity to induce necroptosis.
The canonical pathway enrichment analysis also uncovered the involvement of the glutathione metabolic pathway, the glycosylphosphatidylinositol pathway, proteins involved in translation (including translation initiation factors) and ribosomal proteins (Supplemental Table 5). The protection offered by reducing the levels of glutathione peroxidase (GPX4) and glutathione-S-transferases (GSTA3 and GSTO2), which is expected to increase the availability of free glutathione, is consistent with the fact that RIP1 activation leads to increases in the cellular levels of free radical species which plays an important role in mediating necroptosis (Xu et al., 2006; Yu et al., 2006). Similarly, inhibition of glycosylphosphatidylinositol (GPI) anchor biosynthesis by knockdown of PGAP1, PIGL and PIGM is consistent with the requirement of GPI in TNFα signaling (Fukushima et al., 2004). Identification of these canonical pathways provides new insights into the molecular mechanism of necroptosis induced by zVAD.fmk as well as a validation for our screen.
Role of transcription and translation in necroptosis
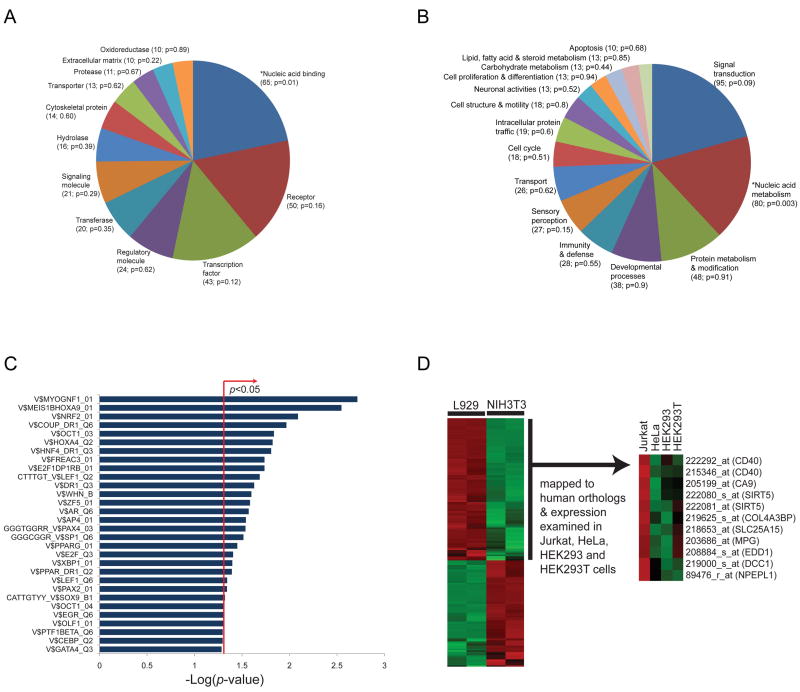
Of the 432 genes involved in zVAD.fmk-induced necroptosis, 291 (67%) and 281 (65%) could be classified into broad molecular function (Figure 5A and Supplemental Table 6) and biological process (Figure 5B and Supplemental Table 7) categories for mouse genes, respectively. There appears to be an enrichment trend for nucleic acid binding (unadjusted hypergeometric p=0.01) and nucleic acid metabolism (hypergeometric p=0.003) categories in the set of genes involved in zVAD.fmk-induced necroptosis relative to their representation in the global set of genes examined in the siRNA screen. None of the functional enrichments were significant at the 0.05 level after being adjusted using Benjamini and Hochberg's (BH) method, but this may be overly conservative since the functional categories are not independent. Interestingly, forty-three genes in the zVAD.fmk hit list encode transcription factors (Supplemental Table 6).
Figure 5. Enrichment of transcription factor binding sites and nucleic acid binding function of genes involved in necroptosis.
The 432 ‘hit’ genes from the secondary screen for zVAD.fmk-induced necroptosis were classified into (A) molecular function and (B) biological process categories for mouse genes according to the PANTHER classification system. Genes for which no annotations could be assigned were excluded from the analysis for both the hits and the global set (i.e. genes examined in the siRNA screen). Categories with at least 10 genes are displayed in the pie charts. The number of genes assigned to each category and enrichment p-values are shown in brackets. *denotes categories found enriched (unadjusted hypergeometric p<0.05) relative to the global set of genes examined in the screen. Lists of assigned genes grouped by molecular function or biological process categories are provided in Supplemental Tables 6 and 7, respectively. (C) Enrichment analysis of cis-regulatory elements, in particular transcription factor (TF) binding sites in the promoters of genes involved in zVAD.fmk-induced necroptosis (vertical axis of graph), using motif-based gene sets from the MSigDB and TF binding sites defined in the TRANSFAC database. TF-binding motifs were examined for enrichment among human orthologs of zVAD.fmk hits compared to the global set of genes screened in the siRNA library to which human orthologs could be mapped. The bar chart displays the negative log of the enrichment p-values for each pathway using the hypergeometric distribution. (D) Expression profiles of probes against zVAD.fmk screen hits showing elevated expression in necroptosis-sensitive L929 cells compared to necroptosis-resistant NIH3T3 cells were mapped to human orthologs and expression trends examined in four human cell lines: necroptosis-sensitive Jurkat cells, and necroptosis-resistant HeLa, HEK293 and HEK293T cells (from the GNF microarray collection). Expression values were z-score-transformed for each probe across arrays.
To begin to characterize the regulation of necroptosis, we explored whether the promoter regions of genes identified in our siRNA screen contain shared transcription factor binding sites defined in the TRANSFAC database (www.gene-regulation.com), we examined the MSigDB collection of transcription factor (TF) targets (i.e. motif-based gene sets) (Xie et al., 2005). We found an enrichment of binding sites (unadjusted hypergeometric p<0.05) for TFs such as MYOG/NF1, MEIS1B/HOXA9, NRF2, HNF4, LEF1, AR, PAX4 and PPARG in the promoters of genes involved in zVAD.fmk-induced necroptosis (Figure 5C), suggesting the involvement of these cis elements in the underlying transcriptional regulatory control of genes involved in zVAD.fmk-induced necroptosis. Consistent with this proposal, a binding site for the androgen receptor (AR) was found to be enriched in the promoters of genes involved in necroptosis, and AR was also a hit in the screen for genes involved in zVAD.fmk-induced necroptosis (Supplemental Table 6). This provides further evidence for the importance of transcriptional control of necroptosis.
In contrast to death receptor mediated apoptosis, which is sensitized by the inhibition of protein synthesis via inhibition of the NF-κB transcriptional response, inhibition of protein synthesis by CHX has been shown to inhibit necroptosis induced by zVAD.fmk (Yu et al., 2004) but not by TNFα (Supplemental Figure 1). Thus, the protection provided by the inhibition of some of the essential cellular machinery involved in protein translation, such as Eif3s10, Eif4a1, Eif4e, Eif4ebp2, Eif5b, Pcbp1 as well as multiple ribosomal proteins and proteins involved in mRNA splicing (Supplemental Table 6), is likely due to the inhibition of protein translation. Consistent with necroptosis being a cellular program of necrotic cell death, transcription and translation may be essential for the progression of necroptosis, at least under certain conditions. Furthermore, these data suggest an intriguing possibility that availability of the translational machinery might represent another factor determining cellular choice in executing cell death.
Gene expression profile of cells sensitive to necroptosis
To examine the transcriptional profile associated with cellular sensitivity to necroptosis, we performed microarray analysis on L929 cells, which are sensitive to necroptosis, and NIH3T3 cells, which we have shown previously to be unable to undergo necroptosis (Degterev et al., 2005). By comparing the expression profiles of the 432 hit genes from the zVAD.fmk screen in these two cell types, we identified 60 zVAD.fmk hits that were expressed at significantly higher levels in L929 cells relative to NIH3T3 cells (FDR-adjusted p<0.05, fold-change>1.5) (Supplemental Table 8).
To determine if this transcriptional profile is preserved in other cell lines known to be sensitive or resistant to necroptosis, we next examined gene expression data for four human cell lines (Jurkat, HeLa, HEK293, HEK293T) from the Genomics Institute of the Novartis Foundation (GNF) cell line collection. The trend appears to be consistent for the 9 genes that we examined (EDD1, MPG, CA9, SLC25A15, SIRT5, NPEPL1, DCC1, CD40 and COL4A3BP) (Figure 5D). These genes exhibited elevated expression in necroptosis-sensitive Jurkat cells relative to HeLa, HEK293 and HEK293T cells, which are resistant. These results suggest that increased expression of a subset of zVAD hit genes might convey cellular sensitivity to necroptosis.
Core components of the necroptotic pathway
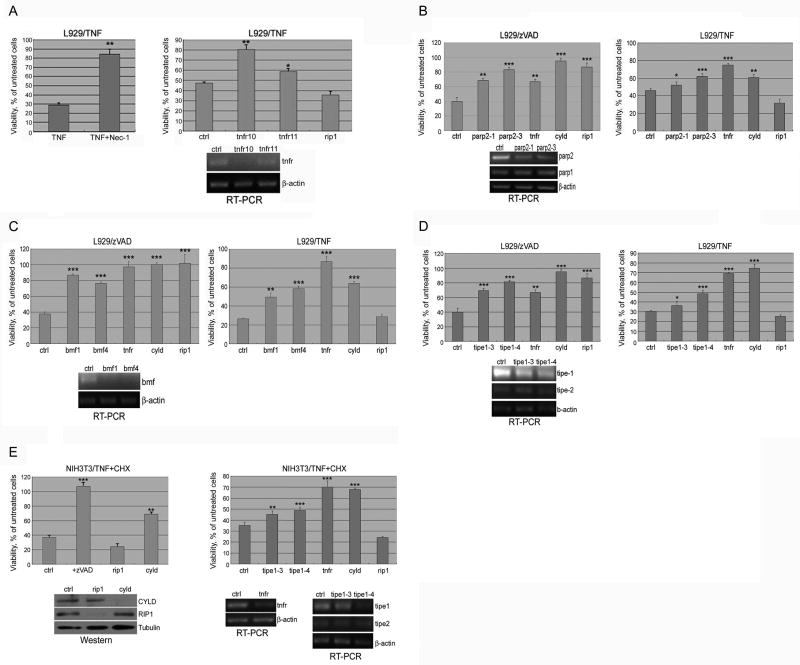
Treatment of L929 cells with TNFα strongly induces necroptosis (Fiers et al., 1995). TNFα, however, also induces the activation of NF-κB, which requires the intermediate domain of RIP1 (Ting et al., 1996). Since activation of NF-κB is pro-survival, necroptosis of L929 cells induced by TNFα cannot be inhibited by RIP1 siRNA (Figure 6A). On the other hand, since Nec-1 targets RIP1 kinase activity specifically and does not affect the activation of NF-κB, Nec-1 inhibits TNFα-induced necroptosis of L929 cells (Degterev et al., 2005) (Figure 6A). Since RIP1 is specifically recruited to the TNFα receptor in the activation of necroptosis (Zheng et al., 2006), the genes required for TNFα-induced necroptosis are predicted to be specifically downstream and/or regulators of RIP1 kinase activity.
Figure 6. The Genes regulate necroptosis and apoptosis.
(A) L929 cells were treated with 40ng/ml TNFα for 20hr with or without co-treatment of 30μM Nec-1. L929 cells, transfected with indicated siRNAs (tnfr; tnf receptor, ctrl; non-targeting siRNA), were treated with 10ng/ml TNFα for 18hr. Knockdown efficiency of TNFα receptor in L929 cells was determined by RT-PCR (right bottom panel). (B) Knockdown of Parp-2 inhibits necroptosis. L929 cells transfected with indicated siRNAs, treated with 20mM zVAD.fmk (left panel) for 18hr or with 10ng/ml TNFα for 20hr (right panel). The knockdown efficiency was determined by RT-PCR using Parp-1 as a control (left bottom panel). (C) Knockdown of Bmf inhibits necroptosis. L929 cells tranfected with indicated siRNAs (bmf, ctrl, cyld, rip1) were treated as in B. Knockdown efficiency of Bmf was determined by RT-PCR (left bottom panel) (D) Knockdown of Tipe1 inhibits necroptosis and apoptosis. L929 cells transfected with indicated siRNAs (tipe1, tnfr, cyld, rip1) were treated as in B. (E) NIH3T3 cell transfected with indicated siRNAs (tipe1, tnfr, rip1, cyld) (40nM) were treated with 10ng/ml TNFα+1.0μg/ml CHX for 12hr with or without co-treatment of 100μM zVAD,fmk (left bottom panel). Knockdown efficiency was determined by RT-PCR for Tipe1 using Tipe2 as a control (left upper and right bottom panels) or western blotting for RIP1, CYLD (left bottom panel). Cellular viability was measured as described in Figure1.
We screened the 666 primary siRNA hits from the zVAD.fmk screen for genes required for necroptosis induced by TNFα (Figure 1A). In non-targeting siRNA-transfected cells, TNFα caused ∼80% cell death. tnfr1 siRNA, which provided a strong protection against necroptosis induced by TNFα, was used as a positive control. A gene was scored as positive if at least 2 out of 4 of its siRNAs increased cell survival to at least 50% of the level provided by tnfr1 siRNA. This screen identified 32 genes required for necroptosis induced by TNFα (Table 1). Since these 32 genes are also required for zVAD.fmk-induced necroptosis, we hypothesize that these 32 genes represent potential core components of the necroptotic pathway. Consistent with this proposal, CYLD, which has been shown previously to interact with RIP1 and regulate RIP1 ubiquitination (Wright et al., 2007), is required for necroptosis induced by either zVAD.fmk or TNFα. Importantly, PARP-2, a member of poly-ADP ribose polymerase family (Menissier de Murcia et al., 2003), and Bmf, a member of the Bcl-2 family, is also required for necroptosis induced by either zVAD.fmk or TNFα (Figure 6B & C). In addition, TIPE1 is also required for both zVAD.fmk and TNFα induced necroptosis (Figure 6D). TIPE1 is a close homolog of TIPE2 which has been shown to play an important role in mediating death receptor mediated apoptosis and innate immunity (Sun et al., 2008).
Table 1. The genes regulate necroptosis and apoptosis.
An siRNA library against 16873 genes in the mouse genome was screened for ability to protect against zVAD.fmk-induced necroptosis in L929 cells. 666 primary hits were selected for having achieved viability more than 2SD above the mean plate viability. 432 genes were scored positive in secondary confirmatory screen with zVAD.fmk induced necroptosis. The siRNAs against 666 genes were screened against TNFα-induced necroptosis in L929 cells and TNFα-CHX induced apoptosis in NIH3T3 cells, respectively. A set of 32 genes was scored positive (zVAD/TNF box) in TNFα induced necroptosis. Another set of 32 genes was scored positive (zVAD/apoptosis box) in TNFα+CHX induced apoptosis. A set of 7 genes (in both boxes) was scored positive in all 3 screens (zVAD.fmk or TNFα induced necroptosis and TNFα/CHX induced apoptosis).
| SYMBOL | ID | NAME | ||
|---|---|---|---|---|
| Tmem57 | 66146 | transmembrane protein 57 | ||
| Grb2 | 14784 | growth factor receptor bound protein 2 | ||
| Pvr | 52118 | poliovirus receptor | ||
| Foxi1 | 14233 | forkhead box I1 | ||
| Jph3 | 57340 | junctophilin 3 | ||
| Rab25 | 53868 | RAB25, member RAS oncogene family | ||
| S100a7a | 381493 | S100 calcium binding protein A7A | ||
| 4933439F11Rik | 66784 | RIKEN cDNA 4933439F11 gene | ||
| 4732429D16Rik | 217305 | RIKEN cDNA 4732429D16 gene | ||
| 9430015G10Rik | 230996 | RIKEN cDNA 9430015G10 gene | ||
| Bmf | 171543 | Bcl2 modifying factor | ||
| Defb1 | 13214 | defensin beta 1 | ||
| Eif5b | 226982 | eukaryotic translation initiation factor 5B | ||
| Dpysl4 | 26757 | dihydropyrimidinase-like 4 | ||
| Commd4 | 66199 | COMM domain containing 4 | zVAD/TNF | |
| Txnl4b | 234723 | thioredoxin-like 4B | ||
| F730015K02Rik | 319526 | RIKEN cDNA F730015K02 gene | ||
| Hspbap1 | 66667 | Hspb associated protein 1 | ||
| Mag | 17136 | myelin-associated glycoprotein | ||
| Mrcl | 353287 | mannose receptor-like precursor | ||
| Hoxa3 | 15400 | homeo box A3 | ||
| Kcnip1 | 70357 | Kv channel-interacting protein 1 | ||
| Galnt5 | 241391 | UDP-N-acetyl-alpha-D-galactosamine: polypeptide N–acetylgalactosaminyltransferase 5 | ||
| Olfr1404 | 258881 | olfactory receptor 1404 | ||
| Parp2 | 11546 | poly (ADP-ribose) polymerase family, member 2 | ||
| Tnfrsf1a | 21937 | tumor necrosis factor receptor superfamily, member 1a |
|
zVAD/TNF/apoptosis |
| Sycp2 | 320558 | synaptonemal complex protein 2 | ||
| Atp6v1g2 | 66237 | ATPase, H+ transporting, lysosomal V1 subunit G2 | ||
| Nudt13 | 67725 | nudix (nucleoside diphosphate linked moiety X)-type motif 13 | ||
| Spata2 | 263876 | spermatogenesis associated 2 | ||
| cyld | 74256 | cylindromatosis 1 | ||
| Tnfaip8l1 | 66443 | tumor necrosis factor, alpha-induced protein 8-like 1 | ||
| Grid2 | 14804 | glutamate receptor, ionotropic, delta 2 | ||
| Ephx1 | 13849 | epoxide hydrolase 1, microsomal | ||
| Insm2 | 56856 | insulinoma-associated 2 | ||
| Stx1a | 20907 | syntaxin 1A (brain) | ||
| Rasa4 | 54153 | RAS p21 protein activator 4 | ||
| Plrg1 | 53317 | pleiotropic regulator 1, PRL1 homolog (Arabidopsis) | ||
| Sfrs2 | 20382 | splicing factor, arginine/serine-rich 2 (SC-35) | ||
| Rpl37 | 67281 | ribosomal protein L37 | ||
| Trmt11 | 73681 | tRNA methyltransferase 11 homolog (S. cerevisiae) | ||
| 0610009D07Rik | 66055 | RIKEN cDNA 0610009D07 gene | ||
| Tmem107 | 66910 | transmembrane protein 107 | ||
| Sft2d2 | 108735 | SFT2 domain containing 2 | ||
| Srcap | 546001 | Snf2-related CREBBP activator protein | zVAD/apoptosis | |
| Psmd6 | 66413 | proteasome (prosome, macropain) 26S subunit, non–ATPase, 6 | ||
| Cwc15 | 66070 | CWC15 homolog (S. cerevisiae) | ||
| Adam26a | 13525 | a disintegrin and metallopeptidase domain 26A (testase 3) | ||
| A130070M06 | 230050 | hypothetical protein A130070M06 | ||
| Atp6ap2 | 70495 | ATPase, H+ transporting, lysosomal accessory protein 2 | ||
| Abce1 | 24015 | ATP-binding cassette, sub-family E (OABP), member 1 | ||
| A530021J07Rik | 330578 | Riken cDNA A530021J07 gene | ||
| Olfr380 | 259027 | olfactory receptor 380 | ||
| Rai14 | 75646 | retinoic acid induced 14 | ||
| Rpl10 | 110954 | ribosomal protein 10 | ||
| Crebl2 | 232430 | cAMP responsive element binding protein-like 2 | ||
| Gpr152 | 269053 | G protein-coupled receptor 152 |
Common regulators of apoptosis and necroptosis
Because multiple hits in our screen, such as CYLD and Bmf, have been previously shown to contribute to apoptosis, we hypothesized that apoptosis and necroptosis may share certain common regulators. To ask if any of the hits from the zVAD.fmk screen may also be required for apoptosis, we screened 666 primary hits from the zVAD.fmk screen against apoptosis induced by TNFα and CHX in NIH3T3 cells (Figure 1A). We chose this system because we have previously found that cell death induced by TNFα and CHX in NIH3T3 cells cannot be inhibited by Nec-1, but is efficiently prevented by zVAD.fmk, as is characteristic for apoptosis (Degterev et al., 2005). In control non-targeting siRNA transfected cells, the treatment with TNFα induced at least 60-70% cell death, which is blocked by knockdown of cyld and tnfr1 but not by that of rip1 (Figure 6E). tnfr1 siRNA was again used as a positive control. A gene was scored as positive if at least 2 out of 4 of its siRNAs increased cell survival to at least 80% of that positive control cells expressing tnfr1 siRNA on the same plate. This screen identified 32 genes required for both apoptosis of NIH3T3 cells induced by TNFα and CHX and necroptosis of L929 cells induced by zVAD.fmk (Table 1). The expression of these 32 genes may have a general impact on cellular sensitivity to multiple cell death stimuli.
Overall, comparing the lists of genes required for necroptosis of L929 cells induced by zVAD.fmk and TNFα and apoptosis of NIH3T3 cells induced by TNFα and CHX, we identified 7 genes that are involved in all 3 cell death paradigms (Table 1). Consistent with the role of TNFα in all 3 cell death paradigms, 3 out of 7 factors have been previously shown to regulate TNFα signaling: TNFR1, CYLD, which directly interacts the TNFR1 complex, and TIPE1, which is also very likely to be a component of the apical death receptor signaling complexes.
Potential roles of necroptosis in human disease
To further explore the significance of necroptosis in human disease, we analyzed the 432 genes from zVAD.fmk screen against a database of known human diseases genes. Interestingly, we found 33 genes from the zVAD.fmk screen that are also implicated in human diseases (Supplemental Table 9). For example, junctophilin-3 (JPH3) has been implicated in Huntington's disease-like 2 (HDL2). HDL2 is an autosomal dominant, progressive, adult-onset neurodegenerative disorder similar to Huntington's disease pathologically. Although the mechanism of neurodegeneration of HDL2 is still not yet clear, a CAG/CTG expansion mutation was found in a variably spliced exon of JPH3 that is responsible for the disease (Holmes et al., 2001). Such connections of genes involved in necroptosis and human diseases suggest the possible involvement of necroptosis in these human diseases that needs be examined in future.
Discussion
In this study we show an extensive signaling network that regulates necroptosis, a cellular caspase-independent necrotic cell death pathway. The enriched expression of the genes required for necroptosis in the immune and nervous systems suggests that necroptosis may function as a physiological cell death mechanism in these two compartments. Interestingly, cell death with necrotic features, termed “type III cell death”, has been described to occur during normal development of nervous system (Clarke, 1990). We propose that necroptosis may represent a type of “type III” cell death. On the other hand, the ability of Nec-1 to extend the life span of activated primary macrophages suggests that necroptosis could function as a cellular mechanism to limit and regulate the numbers of active macrophages during infection. The sensitivity of cells involved in innate immunity to treatment with zVAD.fmk suggests that caspase inhibition might represent a cellular signal of being invaded by virus and therefore, such cells must be destroyed by activating a cellular suicide mechanism such as necroptosis. Thus, necroptosis might serve as a cellular defense mechanism in protecting mammals against the invasion of foreign organisms.
Core regulators of necroptosis
Our analysis identified a group of 25 genes, including parp2, bmf, pvr, rab25, jph3, mag and foxi1 (Table 1), which are required for necroptosis induced by both zVAD.fmk and TNFα, but not for apoptosis. These results provide a first insight into the composition of the “core” necroptotic machinery downstream and/or regulators of RIP1. Regulation of necrotic cell death is a new function for these genes, emphasizing the notion that necroptosis is a separate mechanism of regulated cell death, distinct from apoptosis.
Interestingly, this screen identified Grb2, an SH2 and SH3 domain containing adaptor molecule that has been known to bind to TNFR1 mediating TNFα-dependent activation of c-Raf-1 kinase (Hildt and Oess, 1999), as required for both zVAD.fmk and TNFα induced necroptosis. The involvement of Grb2 suggests a role of MAP kinases in the signaling of necroptosis. The identification of multiple known targets of MAP kinase, such as EDD, SP1, SLC9A1, RGS19, is consistent with this possibility.
Our screen uncovered a role of PARP-2 as required for zVAD.fmk and TNFα induced necroptosis. PARP-2 is the closest homolog of PARP-1 in PARP family, sharing 60% identity in the catalytic domain. Previous studies have implicated PARP-1 in necrosis induced by alkylating agent MNNG (N-methyl-N′-nitrosoguanidine), which requires RIP1 (Xu et al., 2006). PARP-2 and PARP-1 can heterodimerize and have partially redundant functions as indicated by the embryonic lethality of parp1-/-; parp2-/-double but not singly mutant mice (Menissier de Murcia et al., 2003). Oligomerization of Parp1 and Parp2 has been shown to stimulate PARP catalytic activity in assisting the base excision repair after DNA nicks. PARP-2 is also cleaved in apoptosis, albeit with a delayed time course compared to that of PARP-1, suggesting that PARP-2 is not an ideal substrate for caspases as is PARP-1. Interestingly, expression of a caspase-cleavage resistant form of PARP-1 (D214A) has been shown to lead to necrosis (Kim et al., 2000). The activation of PARP1 that catalyzes the hydrolysis of NAD+ into nicotinamide and poly-ADP ribose has been proposed to cause depletion of NAD+, which in turn may contribute to the energy failure in necrosis (Zong et al., 2004). Thus, it is possible that persistent elevated levels of PARP-2 activity resistant to caspase cleavage may contribute to the execution of necroptosis.
Transcriptional factor FOXI1 is also required for both zVAD.fmk and TNFα induced necroptosis. Mutant mice with a targeted disruption of FOXI1 display tubular acidosis resulting from a defect in cell differentiation characterized by the absence of vacuolar H(+)-ATPase expression (Kurth et al., 2006). Interestingly, an H(+)-ATPase, Atp6v1g2, was also a hit required for necroptosis induced by both zVAD.fmk and TNFα. The vacuolar H(+)-ATPase (V-ATPase) is a ubiquitous multi-subunit pump responsible for the acidification of intracellular organelles. Inhibition of V-ATPase is expected to increase the pH in acidic intracellular organelles such as lysosomes, which in turn obligates the activities of lysosomal hydrolytic enzymes.
Role of Bmf in necroptosis
We identified a member of the Bcl-2 family, Bmf, to be required for necroptosis induced by either zVAD.fmk or TNFα. Bmf is a member of the pro-death BH3-only subgroup of Bcl-2 family proteins (Puthalakath et al., 2001). BH3-only proteins serve as integrators of upstream signaling in both apoptosis and autophagy. Our result suggests that Bmf might play a similar role in necroptotic signaling. Overexpression of Bmf has been shown to reduce the colony formation of L929 cells (Puthalakath et al., 2001); however, the mechanism by which Bmf induces the death of L929 cells has not been not directly examined. Bmf-/- mice develop a B cell-restricted lymphadenopathy caused by the abnormal resistance of these cells to a range of apoptotic stimuli and have accelerated development of γ-irradiation-induced thymic lymphomas (Labi et al., 2008). The phenotype of Bmf-/- mice is consistent with a role of necroptosis in mediating homeostasis of immune system. We propose that Bmf may function as a tumor suppressor by mediating the execution of both apoptosis and necroptosis.
Interestingly, knockdown of Bmf blocked necroptosis induced by zVAD.fmk and TNFα but not apoptosis of TNFα/CHX-treated NIH3T3 cells. Although Bmf has been implicated as a pro-apoptotic molecule, this result suggests that at least in the death receptor signaling pathway, Bmf is primarily involved in mediating necroptosis but not apoptosis. It is possible that the activation of Bmf may induce either apoptosis or necroptosis in a stimulus and cellular context dependent manner.
Necroptosis in tumorigenesis?
The role of 2 tumor suppressor genes including cyld and edd1, and 4 Ras-related proteins, Rab25, Rasa4, Rassf7 and Rassf8, in regulating necroptosis suggests a possible function of necroptosis in tumorigenesis. The Rab proteins are involved in regulating intracellular membrane-trafficking (Pfeffer, 2007). Dysregulation of Rab25 gene expression has been noted in multiple cancers including ovarian and breast cancers (Cheng et al., 2004). On the other hand, RASA4/CAPRI (RAS p21 protein activator 4), a suppressor of epithelial cell transformation (Westbrook et al., 2005), functions as a Ca(2+)-dependent Ras GTPase-activating protein (GAP) to inactivate the Ras-MAPK pathway following a stimulus that elevates intracellular calcium (Lockyer et al., 2001). A common role of RASA4 in regulating both necroptosis and apoptosis may provide a potential mechanism for its proposed role as a tumor suppressor. DAB2IP/AIP1, another Ras-GAP and an ASK1-interacting protein, has been previously shown to bind to RIP1 to mediate the activation of ASK1-JNK/p38 signaling in endothelial cells (EC) (Zhang et al., 2007). We found that knockdown of DAB2IP partially inhibited apoptosis but had no effect on necroptosis (data not shown). Since two GAPs, DAB2IP and RASA4, have been found to mediate the signaling of RIP1, it is possible that multiple Ras-GAP proteins may be involved in mediating downstream signaling of RIP1 in different cell types to regulate cell death. Finally, Rassf7 and Rassf8 are members of the Ras-association domain family (RASSF) that are Ras effectors characterized by a conserved motif (the RalGDS/AF6 Ras association (RA) domain) and are known as putative tumor suppressors that modulate some of the growth inhibitory responses mediated by Ras. Overexpression of some members of RASSF family, such as Rassf6, has been shown to trigger both caspase-dependent and caspase-independent cell death. Rassf family has also been implicated in regulating death receptor mediated cell death (Ikeda et al., 2007).
We propose that necroptosis may be selectively activated when there is a failure of caspase activation, which may occur under pathological conditions such as viral infection or oncogenic mutations. Since deficiency in apoptosis has been noted as one of the hallmarks of cancers (Hanahan and Weinberg, 2000), necroptosis may play a very important role in tumorigenesis as a backup cell death mechanism in apoptosis deficient cancer cells.
Materials and Methods
siRNA screen
siRNA screen was performed using 16,873 siRNA pools covering most of the mouse genome (Dharmacon mouse kinase & phosphatase, G protein-coupled receptor, druggable genome and remaining genome libraries, Thermo Fisher Scientific). Mouse fibrosacroma cell line L929 cells cultured in 384-well white plates (Corning) were robotically transfected with 50nM siRNA by reverse transfection method using HiPerFect transfection reagent according to a protocol from the manufacturer (Qiagen). Forty-eight hrs after the transfection, the cells were treated with 20μM pan-caspase inhibitor zVAD.fmk and cultured for an additional 18 hrs to induce necroptosis. Viability was measured using luminescence-based ATP levels as a surrogate marker in surviving cells using CellTiterGlo ATP assay (Promega). Positive control (ripk1, Dharmacon) and negative controls (non-targeting control siRNA: siCONTROL non-targeting #2, Dharmacon) were present in every plate. The screen was performed in duplicate. siRNAs for each gene were classified as “hits” if their viability showed > 2 standard deviations (SD) above the mean ATP value of the plate on duplicate plates.
In the confirmation screen, 4 siRNAs of each pool were individually transfected into cells and screened using the same procedure as in the primary screen. In the TNFα tertiary screen, L929 cells were transfected with individual siRNAs and 48 hrs later, treated with 40ng/ml TNFα for an additional 72hr. In the apoptosis tertiary screen, NIH3T3 cells were transfected with individual siRNA and 48 hr later, treated with 10ng/ml TNFα+1.0μg/ml cycloheximide (CHX) for an additional 12hr to induce apoptosis.
Enrichment analyses
Sets of hit genes were classified into functional categories such as biological process, molecular function (PANTHER classification system) (Mi et al., 2005), canonical pathways (MSigDB) (Subramanian et al., 2005) and transcription factor (TF)-binding sites (MSigDB and TRANSFAC v7.4 (www.gene-regulation.com)). Enrichment analyses of canonical pathways and TF-binding motifs were performed on gene sets that were mapped to human orthologs (Homologene) (Wheeler et al., 2008). To assess the statistical enrichment or over-representation of these categories for the hit genes relative to their representation in the global set of genes examined in the siRNA screen, P-values were computed using the hypergeometric test (Rivals et al., 2007) as described in the GOHyperGAll module (Horan et al., 2008) implemented in the R programming language. Comprehensive background information about the hypergeometric distribution can be found here: http://mathworld.wolfram.com/HypergeometricDistribution.html.
Briefly, the hypergeometric distribution describes the probability of finding at least s genes associated with a particular category, in a set of g genes involved in zVAD.fmk-induced necroptosis, given that there are S genes associated with that same category in the global set of G genes examined in the genome-wide siRNA screen. For each category, c and the list of genes, l, the P value was calculated as:
Here represents the binomial coefficient. An unadjusted P-value <0.05 was considered significant. Categories assigned with at least 10 genes are displayed in the Figure 5A and B.
Human protein interaction network
The network was constructed by iteratively connecting interacting proteins, with data extracted from a collection of genome-wide interactome screens and curated literature entries in HPRD (Mishra et al., 2006). For the network analysis, mouse components were mapped to human orthologs (Homologene) (Wheeler et al., 2008). The network uses graph theoretic representations, which abstract components (gene products) as nodes and relationships (e.g. interactions) between components as edges, implemented in the Perl programming language.
Supplementary Material
L929 cells were treated with 20μM zVAD.fmk (left panel) for 20hr, or with 10ng/ml TNFα for 48rh (right panel) with or without co-treatment of 1.0μg/ml cycloheximide (CHX) and 30μM nec-1. The cellular viability was measured as described in Figure 1.
Acknowledgments
We thank Dodzie Sogah, Marta Lipinski and Caroline Yi for helpful comments on this manuscript. We are grateful for the support of Caroline Shamu, Stewart Rudnicki, Sean Johnston and David Wrobel at the ICCB Longwood Facility during screening. We thank SC Sun (UT MD Anderson) and J Blenis (HMS) for CYLD antibody and FADD def Jurkat cells, respectively. This work was supported in parts by grants from the National Institute of Health (DP1OD000580), the National Institute on Aging (R37 AG012859-14), the National Institute on Neurological Disorders and Stroke (U01NS050560) and NIH Director's Pioneer Award (to JY), the National Institute of Allergy and Infectious Diseases (AI062773) and DK43351 (to RJX). JH was supported in part by fellowships from the Mochida Memorial Foundation for Medical and Pharmaceutical Research, the Uehara Memorial Foundation, the Japan Society for Promotion of Science. AN is a recipient of a Fellowship Award from the Crohn's and Colitis Foundation of America.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cheng KW, Lahad JP, Kuo WL, Lapuk A, Yamada K, Auersperg N, Liu J, Smith-McCune K, Lu KH, Fishman D, et al. The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat Med. 2004;10:1251–1256. doi: 10.1038/nm1125. [DOI] [PubMed] [Google Scholar]
- Clarke PG. Developmental cell death: morphological diversity and multiple mechanisms. Anat Embryol (Berl) 1990;181:195–213. doi: 10.1007/BF00174615. [DOI] [PubMed] [Google Scholar]
- Degterev A, Hitomi J, Germscheid M, Ch'en IL, Korkina O, Teng X, Abbott D, Cuny GD, Yuan C, Wagner G, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N, Cuny GD, Mitchison TJ, Moskowitz MA, Yuan J. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- Fiers W, Beyaert R, Boone E, Cornelis S, Declercq W, Decoster E, Denecker G, Depuydt B, De Valck D, De Wilde G, et al. TNF-induced intracellular signaling leading to gene induction or to cytotoxicity by necrosis or by apoptosis. J Inflamm. 1995;47:67–75. [PubMed] [Google Scholar]
- Fukushima K, Ishiyama C, Yamashita K. Recognition by TNF-alpha of the GPI-anchor glycan induces apoptosis of U937 cells. Arch Biochem Biophys. 2004;426:298–305. doi: 10.1016/j.abb.2004.02.028. [DOI] [PubMed] [Google Scholar]
- Grimm S, Stanger BZ, Leder P. RIP and FADD: two “death domain”-containing proteins can induce apoptosis by convergent, but dissociable, pathways. Proc Natl Acad Sci U S A. 1996;93:10923–10927. doi: 10.1073/pnas.93.20.10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Li L, Qiu S, Lu Q, Pan Q, Gu Y, Luo J, Hu X. Shikonin circumvents cancer drug resistance by induction of a necroptotic death. Mol Cancer Ther. 2007;6:1641–1649. doi: 10.1158/1535-7163.MCT-06-0511. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hijikata A, Kitamura H, Kimura Y, Yokoyama R, Aiba Y, Bao Y, Fujita S, Hase K, Hori S, Ishii Y, et al. Construction of an open-access database that integrates cross-reference information from the transcriptome and proteome of immune cells. Bioinformatics. 2007;23:2934–2941. doi: 10.1093/bioinformatics/btm430. [DOI] [PubMed] [Google Scholar]
- Hildt E, Oess S. Identification of Grb2 as a novel binding partner of tumor necrosis factor (TNF) receptor I. J Exp Med. 1999;189:1707–1714. doi: 10.1084/jem.189.11.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler N, Zaru R, Micheau O, Thome M, Attinger A, Valitutti S, Bodmer JL, Schneider P, Seed B, Tschopp J. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat Immunol. 2000;1:489–495. doi: 10.1038/82732. [DOI] [PubMed] [Google Scholar]
- Holmes SE, O'Hearn E, Rosenblatt A, Callahan C, Hwang HS, Ingersoll-Ashworth RG, Fleisher A, Stevanin G, Brice A, Potter NT, et al. A repeat expansion in the gene encoding junctophilin-3 is associated with Huntington disease-like 2. Nat Genet. 2001;29:377–378. doi: 10.1038/ng760. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Hirabayashi S, Fujiwara N, Mori H, Kawata A, Iida J, Bao Y, Sato Y, Iida T, Sugimura H, et al. Ras-association domain family protein 6 induces apoptosis via both caspase-dependent and caspase-independent pathways. Exp Cell Res. 2007;313:1484–1495. doi: 10.1016/j.yexcr.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Kalai M, Van Loo G, Vanden Berghe T, Meeus A, Burm W, Saelens X, Vandenabeele P. Tipping the balance between necrosis and apoptosis in human and murine cells treated with interferon and dsRNA. Cell Death Differ. 2002;9:981–994. doi: 10.1038/sj.cdd.4401051. [DOI] [PubMed] [Google Scholar]
- Kawahara A, Ohsawa Y, Matsumura H, Uchiyama Y, Nagata S. Caspase-independent cell killing by Fas-associated protein with death domain. J Cell Biol. 1998;143:1353–1360. doi: 10.1083/jcb.143.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Won J, Sohn S, Joe CO. DNA-binding activity of the N-terminal cleavage product of poly(ADP-ribose) polymerase is required for UV mediated apoptosis. J Cell Sci. 2000;113(Pt 6):955–961. doi: 10.1242/jcs.113.6.955. [DOI] [PubMed] [Google Scholar]
- Kurth I, Hentschke M, Hentschke S, Borgmeyer U, Gal A, Hubner CA. The forkhead transcription factor Foxi1 directly activates the AE4 promoter. Biochem J. 2006;393:277–283. doi: 10.1042/BJ20051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labi V, Erlacher M, Kiessling S, Manzl C, Frenzel A, O'Reilly L, Strasser A, Villunger A. Loss of the BH3-only protein Bmf impairs B cell homeostasis and accelerates gamma irradiation-induced thymic lymphoma development. J Exp Med. 2008;205:641–655. doi: 10.1084/jem.20071658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockyer PJ, Kupzig S, Cullen PJ. CAPRI regulates Ca(2+)-dependent inactivation of the Ras-MAPK pathway. Curr Biol. 2001;11:981–986. doi: 10.1016/s0960-9822(01)00261-5. [DOI] [PubMed] [Google Scholar]
- Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, Schreiber V, Ame JC, Dierich A, LeMeur M, et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. Embo J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Burns K, Hofmann K, Blancheteau V, Martinon F, Kelliher M, Tschopp J. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat Immunol. 2004;5:503–507. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan TM, et al. Human protein reference database--2006 update. Nucleic Acids Res. 2006;34:D411–414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR. Unsolved mysteries in membrane traffic. Annu Rev Biochem. 2007;76:629–645. doi: 10.1146/annurev.biochem.76.061705.130002. [DOI] [PubMed] [Google Scholar]
- Puthalakath H, Villunger A, O'Reilly LA, Beaumont JG, Coultas L, Cheney RE, Huang DC, Strasser A. Bmf: a proapoptotic BH3-only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis. Science. 2001;293:1829–1832. doi: 10.1126/science.1062257. [DOI] [PubMed] [Google Scholar]
- Simonson SJ, Wu ZH, Miyamoto S. CYLD: a DUB with many talents. Dev Cell. 2007;13:601–603. doi: 10.1016/j.devcel.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Smith CC, Davidson SM, Lim SY, Simpkin JC, Hothersall JS, Yellon DM. Necrostatin: a potentially novel cardioprotective agent? Cardiovasc Drugs Ther. 2007;21:227–233. doi: 10.1007/s10557-007-6035-1. [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Gong S, Carmody RJ, Hilliard A, Li L, Sun J, Kong L, Xu L, Hilliard B, Hu S, et al. TIPE2, a negative regulator of innate and adaptive immunity that maintains immune homeostasis. Cell. 2008;133:415–426. doi: 10.1016/j.cell.2008.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temkin V, Huang Q, Liu H, Osada H, Pope RM. Inhibition of ADP/ATP exchange in receptor-interacting protein-mediated necrosis. Mol Cell Biol. 2006;26:2215–2225. doi: 10.1128/MCB.26.6.2215-2225.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting AT, Pimentel-Muinos FX, Seed B. RIP mediates tumor necrosis factor receptor 1 activation of NF-kappaB but not Fas/APO-1-initiated apoptosis. Embo J. 1996;15:6189–6196. [PMC free article] [PubMed] [Google Scholar]
- Vercammen D, Beyaert R, Denecker G, Goossens V, Van Loo G, Declercq W, Grooten J, Fiers W, Vandenabeele P. Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J Exp Med. 1998;187:1477–1485. doi: 10.1084/jem.187.9.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercammen D, Vandenabeele P, Beyaert R, Declercq W, Fiers W. Tumour necrosis factor-induced necrosis versus anti-Fas-induced apoptosis in L929 cells. Cytokine. 1997;9:801–808. doi: 10.1006/cyto.1997.0252. [DOI] [PubMed] [Google Scholar]
- Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, Zhao JJ, Roberts TM, Mandel G, Hannon GJ, et al. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A, Reiley WW, Chang M, Jin W, Lee AJ, Zhang M, Sun SC. Regulation of early wave of germ cell apoptosis and spermatogenesis by deubiquitinating enzyme CYLD. Dev Cell. 2007;13:705–716. doi: 10.1016/j.devcel.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Chua CC, Kong J, Kostrzewa RM, Kumaraguru U, Hamdy RC, Chua BH. Necrostatin-1 protects against glutamate-induced glutathione depletion and caspase-independent cell death in HT-22 cells. J Neurochem. 2007;103:2004–2014. doi: 10.1111/j.1471-4159.2007.04884.x. [DOI] [PubMed] [Google Scholar]
- Xu Y, Kim SO, Li Y, Han J. Autophagy contributes to caspase-independent macrophage cell death. J Biol Chem. 2006;281:19179–19187. doi: 10.1074/jbc.M513377200. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Jono H, Kai H, Li JD. The tumor suppressor cylindromatosis (CYLD) acts as a negative regulator for toll-like receptor 2 signaling via negative cross-talk with TRAF6 AND TRAF7. J Biol Chem. 2005;280:41111–41121. doi: 10.1074/jbc.M509526200. [DOI] [PubMed] [Google Scholar]
- Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S, Baehrecke EH, Lenardo MJ. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science. 2004;304:1500–1502. doi: 10.1126/science.1096645. [DOI] [PubMed] [Google Scholar]
- Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, Baehrecke EH, Lenardo M. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci U S A. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang H, Lin Y, Li J, Pober JS, Min W. RIP1-mediated AIP1 phosphorylation at a 14-3-3-binding site is critical for tumor necrosis factor-induced ASK1-JNK/p38 activation. J Biol Chem. 2007;282:14788–14796. doi: 10.1074/jbc.M701148200. [DOI] [PubMed] [Google Scholar]
- Zheng L, Bidere N, Staudt D, Cubre A, Orenstein J, Chan FK, Lenardo M. Competitive control of independent programs of tumor necrosis factor receptor-induced cell death by TRADD and RIP1. Mol Cell Biol. 2006;26:3505–3513. doi: 10.1128/MCB.26.9.3505-3513.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004;18:1272–1282. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
L929 cells were treated with 20μM zVAD.fmk (left panel) for 20hr, or with 10ng/ml TNFα for 48rh (right panel) with or without co-treatment of 1.0μg/ml cycloheximide (CHX) and 30μM nec-1. The cellular viability was measured as described in Figure 1.