Abstract
The type IV bundle-forming pili (BFP) of enteropathogenic Escherichia coli (EPEC) are required for virulence in orally challenged human volunteers and for the localized adherence and autoaggregation in vitro phenotypes. BFP filament biogenesis and function are encoded by the 14-gene bfp operon. The BFP assembly complex, containing a BfpB-His6 fusion protein, was chemically cross-linked in situ, and the complex was then purified from BFP-expressing EPEC by a combination of nickel- and BfpB antibody-based affinity chromatography. Characterization of the isolated complex by immunoblotting using BFP protein-specific antibodies showed that at least 10 of the 14 proteins specified by the bfp operon physically interact to form an oligomeric complex. Proteins localized to the outer membrane, inner membrane, and periplasm are within this complex, thus demonstrating that the complex spans the periplasmic space. A combination of immunofluorescence and immuno-gold thin-section transmission electron microscopy studies localized this complex to one pole of the cell.
The bundle-forming pili (BFP) of enteropathogenic Escherichia coli (EPEC), a member of the type IV family of pilus proteins, are required for virulence in orally challenged human volunteers and for the localized adherence (LA) and autoaggregation (AA) in vitro phenotypes (2, 16, 24). The 14-gene bfp operon (located on the 69-kb EPEC adherence factor [EAF] plasmid), together with the genes encoding its transcriptional activator PerABC/BfpTVW, specifies the biogenesis of the pilus filament and the AA phenotype in wild-type EPEC strains and when harbored by E. coli strains that normally do not express BFP (5, 21, 26). Accordingly, the expression of proteins encoded by the bfp operon (in association with certain accessory proteins that are common to both EPEC and lab-adapted E. coli strains) (29) is sufficient for BFP biogenesis and for the BFP-mediated functions that confer the AA phenotype. By contrast, the LA phenotype also requires genes in the chromosomal locus of enterocyte effacement (11, 23).
In-frame disruption of each of the 14 bfp operon genes (designated bfpA to bfpL) (Table 1) and analysis of the resulting mutants show that with the exception of bfpH, each of the remaining 13 genes is required for normal BFP filament production and phenotype expression (1, 13). These findings for the BFP system and similar results from the study of other type IV pili have led to the idea that bfp operon-encoded proteins comprise an oligomeric structural and functional complex (4, 10, 14, 15, 22, 24). The topographical features of this complex have been explored by localizing individual proteins to compartments of the cell through the use of protein-specific antibodies and immunoblot assays to detect their presence in compartment-specific cell fractions. In this manner, BfpB and BfpG have been shown to localize exclusively to the outer membrane (12, 17); BfpU and BfpH mainly localize to the periplasmic space (see references 13 and 18 and unpublished data), and BfpA (as a pool of unassembled pilins), BfpC and BfpE, and BfpI, BfpJ, and BfpK (the last three are stoichiometrically minor pilin-like proteins of the assembly complex) localize to the inner membrane (3, 13). By contrast, BfpL (while predominantly an inner membrane protein) can be consistently detected in small amounts in outer-membrane fractions prepared from French pressure cell-disrupted bacteria and sucrose gradient centrifugation (13). BfpP, which encodes the prepilin peptidase that processes BfpA (the major repeating subunit of the pilus filament) (30) and BfpI, BfpJ, and BfpK (13), is presumed (on the basis of its functional role) to localize to the cytoplasmic face of the inner membrane, but biochemical evidence for this prediction has not been reported (21, 30). BfpD and BfpF, which contain Walker box motifs that are presumed to hydrolyze ATP and thus energize the extrusion and retraction of the pilus filament, respectively, have not been localized (13, 21).
TABLE 1.
bfp operon-encoded gene products: molecular masses and cellular localization
| Protein | Molecular mass (kDa)a | Localizationb |
|---|---|---|
| BfpA | 20.4 | Inner membrane (nonpolymerized monomers) |
| BfpG | 13.3 | Outer membrane |
| BfpB | 58.2 | Outer membrane |
| BfpC | 45.4 | Inner membrane |
| BfpU | 17.7 | Periplasm |
| BfpD | 60.5 | Unknown |
| BfpE | 39.8 | Inner membrane |
| BfpF | 36.7 | Unknown |
| BfpP | 28 | Inner membranec |
| BfpH | 16.5 | Periplasmd |
| BfpI | 20.3 | Inner membrane |
| BfpJ | 20.3 | Inner membrane |
| BfpK | 18.1 | Inner membrane |
| BfpL | 16.9 | Inner membrane |
Several reviews of type IV pilus biogenesis have postulated the existence of a hetero-oligomeric assembly complex that spans the periplasmic space (4, 10, 14, 15, 22, 24). Although this is a plausible structural model of the type IV pilus assembly complex, biochemical evidence in support of a complex of this kind has not been reported. With respect to the putative BFP assembly complex, only BfpB and BfpG, which compose a secretin-like complex that is believed to form a transpilus pore in the outer membrane, have been shown by chemical cross-linking and coimmunoprecipitation methods to physically interact (17). Consequently, in the absence of further information of this kind, the idea that bfp operon-encoded proteins physically interact as a macromolecular assembly complex is at present an attractive but unproven hypothesis. The studies reported here address this issue by providing evidence that (i) at least 10 of the bfp operon-encoded proteins can be extracted from the cell as a chemically cross-linked complex; (ii) the complex contains proteins from the inner and outer membranes and from the periplasmic space and thus most likely spans the periplasmic space; and (iii) BFP operon-encoded proteins preferentially localize to one of the poles of the cell.
Construction and functional studies of a BfpB-His6 fusion protein.
To determine whether proteins encoded by the BFP operon physically associate in situ as a complex, BFP-expressing cells were incubated with a membrane-permeable, disulfide-cleavable cross-linker and the cross-linked complex was isolated by affinity chromatography as described below. The cross-linked eluted complex was treated with 2-mercaptoethanol (to cleave the cross-linker), and proteins in the complex were then identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and by immunoblotting with antisera to 10 of the 14 bfp operon-encoded proteins. This experimental strategy required that we replace one of the 14 wild-type bfp operon proteins with a hexahistidine-tagged version of the same protein by complementing the in-frame mutant of the corresponding bfp operon gene with a plasmid encoding the His-tagged variant. Proof that functional complementation had occurred was sought by determining whether the complemented strain exhibited normal LA and AA phenotypes. To efficiently express His-tagged bfp operon proteins, we constructed a plasmid containing the bfp operon promoter and the coding sequence for a His6 tag located at the C terminus of the expressed BFP protein. Inclusion of the bfp operon promoter allowed the His-tagged protein to be expressed under the control of the normal BFP transcription factor, PerABC/BfpTVW (5, 26). The multicloning site (MCS) and polyhistidine (His6) region of pBAD/Myc-HisA (Invitrogen, Carlsbad, Calif.) was obtained by PCR and used to replace the MCS of pTrc99A (Pharmacia, Peapack, N.J.) to produce pTrc99-His. Then, the upstream region of bfpA, which contains the bfp operon promoter, was obtained from the EAF plasmid by PCR and used to replace the promoter region of pTrc99-His. The resulting plasmid, pTrcAp-His, has the bfp operon promoter and C-terminal His6-tag coding sequence flanked by the MCS of pBAD/Myc-HisA. The strains and plasmids used in this study are listed in Table 2.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| B171-8 (wt)a | Wild-type EPEC O111:NM; carries the 69-kb EAF plasmid | 2 |
| B171-8ΔAcm (ΔA) | B171-8 EAF plasmid mutant specifically lacking the bfpA gene product, BfpA; 133 of 193 amino acids removed and replaced by the Cmr geneb | 2 |
| B171-8dbfp (dbfp) | B171-8 EAF plasmid bfp operon mutant; expresses none of the Bfp proteins | 12 |
| B171-8ΔB (ΔB) | B171-8 EAF plasmid mutant specifically lacking the bfpB gene product, BfpB; 9 of 453 amino acids deleted; two stop codons introduced into each reading frame | 12 |
| B171-8ΔF (ΔF) | B171-8 EAF plasmid mutant specifically lacking the bfpF gene product, BfpF; 90 of 331 amino acids deleted | 2 |
| Plasmids | ||
| pBAD/Myc-HisA | Expression vector containing C-terminal His6 tag | Invitrogen |
| pTrc99A | Expression vector | Pharmacia |
| pTrc99-His | Subclone MCS and C-terminal His6 tag of pBAD/Myc-HisA into MCS region of pTrc99A | This work |
| pTrcAp-His | Replacement of pTrc99-His promoter with bfp operon promoter | This work |
| pApB-His | Subclone bfpB into MCS of pTrcAp-His; produces BfpB-His6 fusion protein | This work |
| pApF-His | Subclone bfpF into MCS of pTrcAp-His; produces BfpF-His6 fusion protein | This work |
wt, wild type.
Cmr, chloramphenicol resistance.
Since the putative BFP assembly complex is hypothesized to be a heteropolymer composed of bfp operon-encoded proteins, the introduction of a His-tagged variant of one of these proteins might disrupt the quaternary structure and therefore the biogenic function of the assembly complex. To identify His-tagged fusion proteins that could replace the native molecule without disrupting assembly complex function, we subcloned six individual bfp operon genes (bfpB, bfpC, bfpD, bfpE, bfpF, or bfpJ) into pTrcAp-His. When each of the resulting plasmids was used to complement the corresponding in-frame deletion mutant (B171-8 ΔB, ΔC, ΔD, ΔE, ΔF, or ΔJ, respectively) (13), only B171-8 ΔB/pApB-His and B171-8 ΔF/pApF-His cells retained the characteristic BFP-mediated LA and AA phenotypes (data not shown). B171-8 ΔB/pApB-His was selected for the cross-linking studies because BfpB forms a multimer in the outer membrane, thus affording the possibility of multiplicative binding of the C-terminal His6-tagged BfpB fusion protein to the Ni affinity column (12, 17).
To prove that the BfpB-His tag fusion protein is expressed in B171-8 ΔB/pApB-His, this recombinant strain was grown under BFP-expressing conditions and the bacteria were collected, washed, boiled in sample buffer, and analyzed by immunoblotting with a rabbit polyclonal BfpB antiserum or with a murine monoclonal anti-penta-His antibody (Qiagen, Inc., Valencia, Calif.). As the fusion protein is expressed and detectable by both antibodies (Fig. 1), it was judged suitable for Ni and/or BfpB antibody-based affinity purification of the cross-linked BFP assembly complex.
FIG. 1.
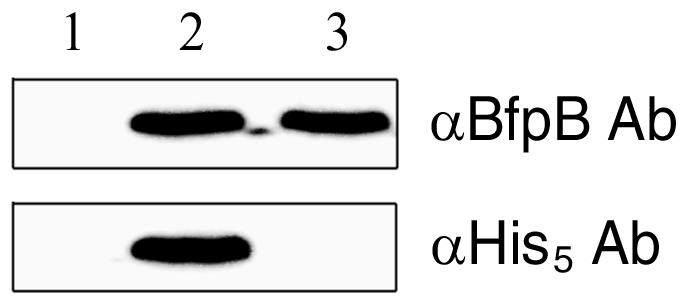
Immunoblot analysis of bacteria expressing either BfpB or the His5-tagged BfpB fusion protein. Lanes: 1, B171-8ΔB; 2, B171-8ΔB/pApB-His; 3, wild-type B171-8. The primary antibodies (Ab) are indicated in the right margin.
Affinity purification of the chemically cross-linked BFP assembly complex.
Chemical cross-linking was carried out using either the BFP-expressing recombinant B171-8ΔB/pApB-His or, as a negative control, the non-BfpB-expressing recombinant B171-8ΔB/pTrcAp-His (which lacks the BfpB-His-tagged fusion protein coding sequence but expresses each of the other Bfp operon proteins, with the exception of BfpB) and viable, exponential-phase bacteria. The negative control was used to identify proteins that interact nonspecifically with the affinity matrices and thus would be erroneously identified as being components of the BFP assembly complex. Cells grown overnight from B171-8ΔB/pTrcAp-His or B171-8ΔB/pApB-His in Luria-Bertani broth were diluted to an optical density at 600 nm of 0.02 in Dulbecco's modified Eagle's medium containing 0.45% glucose and 100 μg of ampicillin/ml. After incubation with shaking at 37°C for 3.5 h, at which time expression of all the BFP proteins and the AA phenotype are maximal (13), the cells were harvested by centrifugation, washed, and resuspended in phosphate-buffered saline (PBS). Disulfide-cleavable DSP (dithiobis[succinimidyl propionate]; Pierce, Rockford, Ill.) (final concentration, 0.2 mg/ml) was added for 30 min at room temperature (6). Following the addition of 0.1 M Tris-HCl (pH 7.4) to quench the reaction, the cross-linked cells were harvested, washed with PBS, and resuspended in phosphate buffer containing Triton X-100. The bacterial suspension was passed through a French press at 14,000 lb/in2. Unbroken cells were removed by centrifugation at 3,000 × g for 15 min; then, the supernatant was applied to the Ni affinity column according to the manufacturer's recommendations (Qiagen, Inc.). To promote exposure of the His6 tag at the C terminus of the BfpB fusion protein and the dissociation of non-cross-linked proteins from the assembly complex, 1% (vol/vol) Triton X-100 was added to each of the Ni affinity steps. The imidazole-eluted material from the Ni affinity column was then dialyzed against PBS and further purified by immunoaffinity chromatography using BfpB antibodies conjugated to a protein A column (8, 9).
The sequential two-step Ni and BfpB antibody affinity chromatography purification procedure began with equal cell numbers and used the same elution volumes for the His-tagged BfpB fusion protein-expressing strain (B171-8ΔB/pApB-His) and the non-BfpB-expressing negative-control strain (B171-8ΔB/pTrcAp-His). Figure 2A shows the SDS-PAGE protein profiles for these strains during each purification step. Bound and unbound samples from the Ni and immunoaffinity columns were boiled in sample buffer containing 5% (vol/vol) 2-mercaptoethanol to cleave the cross-linker and subjected to SDS-12% PAGE, and the proteins were detected by silver staining. Lanes 7 and 8 compare the protein profiles from the BfpB negative-control and the BfpB-His-tagged fusion protein-expressing strains, respectively, after combined, sequential Ni and immunoaffinity chromatography. A prominent ∼58-kDa band is evident in both lanes. Though this appears to have comigrated with BfpB, the use of the BfpB-specific antibody in immunoblotting experiments did not detect the BfpB protein in lane 7, which contains proteins from the BfpB negative-control strain. Accordingly, this protein likely interacts nonspecifically with both matrices and thus is not a component of the BFP assembly complex. Moreover, none of the other proteins identified in lane 7 were detected by antisera to 10 proteins encoded by the bfp operon and therefore are not likely to be components of the BFP assembly complex.
FIG. 2.
Identification of bfp operon-encoded proteins in chemically cross-linked, affinity-purified complexes. (A) Silver-stained SDS-12% PAGE analysis of cross-linked preparations boiled in 2-mercaptoethanol-containing sample buffer during sequential steps of Ni and BfpB antibody-based affinity chromatography. The results for B171-8ΔB/pTrcAp-His are shown in odd-numbered lanes, and those for B171-8ΔB/pApB-His are shown in even-numbered lanes. Lanes 1 and 2, unbound fractions from the Ni affinity column; lanes 3 and 4, bound and eluted fractionsfrom the Ni affinity column. The fractions in lanes 3 and 4 were then subjected to BfpB antibody affinity chromatography. Lanes 5 and 6, unbound fractions from the BfpB antibody affinity column; lanes 7 and 8, bound and eluted fractions from the BfpB antibody affinity column. Non-bfp operon-encoded proteins that copurify with the assembly complex are denoted by an asterisk. A prominent ∼58-kDa protein represented in lanes 7 and 8 that nonspecifically interacted with the affinity column matrices is denoted by an arrow. bfp operon-encoded proteins that are grouped by estimated molecular masses are denoted by brackets. (B) bfp operon-encoded protein-specific immunoblot analysis of samples sequentially purified by Ni and BfpB antibody affinity chromatography (lanes 7 and 8 of panel A) from B171-8ΔB/pTrcAp-His cells (lanes 1 and 2) and B171-8ΔB/pApB-His cells (lanes 3 and 4). The samples were treated in either the absence (lanes 1 and 3) or the presence (lanes 2 and 4) of 5% (vol/vol) 2-mercaptoethanol to retain or cleave the cross-linking, respectively. Arrows indicate expected proteins.
The proteins represented in lane 8, with the exception of the nonspecifically bound proteins that are shown in lane 7, are considered to be physically associated with a functional BFP assembly complex for the following reasons: (i) the proteins originated from a recombinant strain in which BfpB was replaced with a BfpB-His-tagged fusion protein; (ii) this strain exhibits the AA and LA phenotypes that require the expression of functional BFP; (iii) the proteins were chemically cross-linked and isolated by sequential Ni and BfpB antibody affinity chromatography; and (iv) the proteins appear in lane 8 but not in the adjacent BfpB negative-control lane. Inspection of the silver-stained proteins in lane 8 shows bands with estimated molecular masses that coincide approximately with the predicted masses of 12 of the 14 proteins encoded by the BFP operon. However, we were not able to unambiguously correlate any of the bands with a particular BFP operon-encoded protein because several BFP proteins have similar predicted molecular masses (BfpA, BfpG, BfpI, BfpJ, BfpK, BfpL, and BfpU, 15 to 19 kDa; BfpC, BfpE, and BfpF, 38 to 45 kDa; and BfpB and BfpD, ∼60 kDa) (Table 1) (21, 23). Because BFP assembly complex proteins might have been associated with proteins that were not encoded by the BFP operon, lane 8 was also inspected for bands that are not present in lane 7 and do not correspond in size with any of the BFP proteins. Two such proteins were identified (Fig. 2A), including one represented by a prominent and well-resolved band of ∼100 kDa. The identities and possible functional significance of these non-BFP operon-encoded proteins were not further investigated.
Characterization of the affinity-purified BFP assembly complex by immunoblot analysis.
To determine whether the cross-linked protein complex isolated by the combination of sequential Ni and immunoaffinity chromatography contains proteins encoded by the BFP operon, immunoblot assays of the eluted complex were performed with antibodies to each of the 14 BFP proteins, excepting BfpP (13). Antibodies to BfpI, BfpK, and BfpH failed to detect the corresponding protein in the isolated cross-linked complex. Results obtained with antibodies to the 10 remaining bfp operon-encoded proteins are shown in Fig. 2B, in which lanes 1 and 2 represent proteins isolated from the BfpB negative-control strain and lanes 3 and 4 represent proteins from the strain that expresses the BfpB-His-tagged fusion protein and produces functional BFP. The protein complexes represented in lanes 2 and 4 were treated with 5% (vol/vol) 2-mercaptoethanol to reduce the cross-linker, thus dissociating the complex and allowing the proteins to migrate as monomers. By contrast, the proteins represented in lanes 1 and 3 were not treated with 2-mercaptoethanol; thus, these lanes depict nondissociated complexes.
Treatment of the cross-linked complex that was isolated by sequential Ni and BfpB antibody affinity chromatography with 2-mercaptoethanol allowed each of the 10 detected proteins to migrate in accordance with its predicted molecular mass. By contrast, none of these proteins was detected in lanes 1 and 2, indicating that in this bfpB deletion strain the remaining 13 bfp operon-encoded proteins that are expressed by this mutant do not nonspecifically bind and elute from the affinity columns. These Western blot results and the small number of proteins detected in lane 8 of Fig. 2A provide compelling evidence that at least 10 of the 14 proteins encoded by the bfp operon form a macromolecular complex. Prior localization studies show that two of these proteins (BfpB and BfpG) reside exclusively in the outer membrane and that one (BfpE) resides exclusively in the inner membrane (3, 13). Thus, this complex appears to span the periplasmic space, a conclusion that is supported by the finding that BfpU, a soluble protein that is principally located in the periplasmic space, is also a component of the cross-linked complex (13, 18). Not surprisingly, BfpA, the principal repeating subunit of the pilus fiber, was also identified in the complex, as was the pilin-like protein BfpJ. Likewise, BfpD and BfpF were found in the complex, thus demonstrating a close physical association between proteins required for the kinetic properties (extrusion and retraction, respectively) of the pilus with structural features of the assembly complex (2).
To investigate the role of cross-linking per se in the results depicted in Fig. 2, non-cross-linked B171-8ΔB/pApB-His was subjected to all steps in the affinity purification protocol and the proteins that were eluted from the Ni and BfpB antibody affinity columns were characterized by immunoblot analysis using the same antisera. Only BfpB and BfpG were copurified in the absence of chemical cross-linking under the conditions used in this study (data not shown). Because both of these proteins reside in the outer membrane, this control experiment shows that cross-linking is required to copurify components of the assembly complex that reside in the periplasmic space and inner membrane.
More-slowly migrating bands are evident in lane 4 of the BfpE, BfpF, and BfpJ immunoblots, indicating the presence of a molecular species that is larger than the expected molecular mass of the assayed protein. The identification of these higher-molecular-mass bands was not investigated, but they could be oligomeric forms of the protein or complexes that contain the protein together with other components of the assembly complex.
BfpH was not detected in the complex with these antibodies. This was an expected finding, since the BfpH anti-peptide antibody used here failed to detect this protein in the wild-type parent strain (13). We were able to use an antibody to the His6 epitope to localize a His-tagged variant of this protein to the periplasmic space only recently (data not shown). Failure to detect BfpI and BfpK in the complex requires the consideration of other explanations, since both were detected in the wild-type parent by anti-peptide antibodies (13). One possibility is that the stoichiometric ratios of these pilin-like proteins to other components of the complex are too low for detection by this method. A second possibility is that they are not sufficiently near other components of the complex to be cross-linked by the reagent (Pierce) used here, which has an estimated effective length of 12 angstroms. Finally, it is possible that the use of the His-tagged BfpF fusion protein (which also yielded functional BFP), rather than that of the His-tagged BfpB fusion protein, would have provided a different portrait of complex membership.
The BFP assembly complex localizes to the poles of the cell.
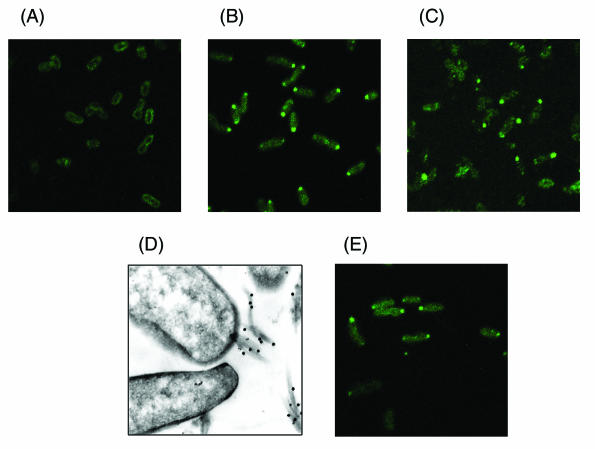
The well-studied type IV pili of Caulobacter crescentus, Myxococcus xanthus, and Pseudomonas aeruginosa are located at the poles of these cells, a topographical feature that is likely critical for the functions they confer in these species (19, 20, 28). By contrast, the type IV pili of Neisseria gonorrhoeae are believed to be peritrichous (25, 27). The polar or peritrichous location of BFP pili is unresolved: electron micrographs of BFP-expressing EPEC have failed to disclose where they emanate from the cell surface, because BFP from adjacent cells tend to aggregate, forming a meshwork that obscures their site of origination. To address this question, the BFP assembly complex was localized on the cell surface by immunofluorescence and immuno-gold electron microscopy through the use of antibodies to the proteins shown in Fig. 2B to be part of the assembly complex.
Immunofluorescence microscopy (7) was conducted by incubating an anti-penta-His murine monoclonal antibody of the immunoglobulin G class with the BfpF deletion strain B171-8ΔF/pApF-His that had been functionally complemented for the AA and LA phenotypes with a plasmid expressing the His6-tagged BfpF fusion protein. B171-8dbfp (Table 2), which lacks part of the principal promoter of the bfp operon and thus produces none of the BFP proteins, served as a negative control for these studies. Detection of the anti-penta-His antibody with a fluorescein-conjugated anti-mouse immunoglobulin G antibody showed distinctive polar staining of this recombinant strain, which produces functional BFP assembly complexes containing the His-tagged BfpF fusion protein (Fig. 3B). By contrast, B171-8dbfp showed only faint circumferential staining (Fig. 3A).
FIG. 3.
Localization of BFP filaments and the BfpF and BfpL protein components of the BFP assembly complex. (A) B171-8dbfp, a negative-control strain that does not express bfp-operon-encoded proteins, stained with a penta-His murine monoclonal antibody and a fluorescein-conjugated anti-mouse secondary antibody. (B) B171-8ΔF/pApF-His, expressing the BfpF-His-tagged fusion protein and functional BFP phenotypes stained with the same antibodies as described for panel A. (C) Wild-type EPEC strain B171-8 stained with a BfpL anti-peptide rabbit antiserum and a fluorescein-conjugated anti-rabbit antibody. (D) Thin-sectioned wild-type EPEC strain B171-8 stained with a rabbit BfpA antibody and a gold-labeled anti-rabbit secondary antibody. (E) B171-8ΔA/pApF-His, which does not produce BfpA, stained with the same antibodies as described for panel A.
The use of the BfpF epitope-tagged fusion protein allowed its detection by a high-avidity murine monoclonal antibody that recognizes the C-terminal hexahistidine. However, if overexpressed, the BfpF-His-tagged fusion protein encoded by the recombinant plasmid might deposit in an inclusion body. If so, the apparent polar staining depicted in Fig. 3B might have been due to staining of a BfpF-containing inclusion body located at one end of the cell. To address this possibility, an affinity-purified anti-peptide rabbit antibody to BfpL (13), a protein shown in Fig. 2B to be part of the cross-linked assembly complex, was used with a fluorescein-labeled anti-rabbit antibody to stain wild-type B171-8 grown under BFP-inducing conditions. Polar staining with this BfpL antibody was evident (Fig. 3C), suggesting that the polar staining shown in Fig. 3B was not due to the accumulation of the BfpF-His-tagged fusion protein in an inclusion body. The polar localization of the BFP assembly complex was also examined by immuno-gold transmission electron microscopy of thin-sectioned wild-type EPEC strain B171-8 stained with a rabbit polyclonal antibody to BfpA and a goat-anti-rabbit antibody labeled with 10-nm-diameter gold particles. The non-BFP-expressing strain B171-8dbfp served as a negative control (data not shown). One of the bacteria shown in Fig. 3D was sectioned through a site at which a cluster of BFP filaments emanate from a polar site on the cell surface. Since sections through BFP assembly complexes would be an infrequent event, too few images of pili protruding from the bacterial cell surface were identified for statistical analysis. However, these immuno-gold electron microscopy findings and the polar staining shown in Fig. 3B (for BfpF) and Fig. 3C (for BfpL) provide strong evidence for the polar localization of the BFP assembly complex.
The ability to detect the BfpF-His6-tagged fusion protein within BFP assembly complexes at the cell pole led us to initiate studies aimed at identifying determinants of this organelle's polar localization. To learn whether BfpA is required for the polar localization of BfpF, a derivative of the wild-type strain B171-8 was prepared that produced all of the bfp operon proteins except BfpA (2). Immunofluorescence microscopy (using the anti-penta-His antibody) of this strain (B171-8ΔA/pApF-His) showed clear polar localization of the His-tagged BfpF fusion protein (Fig. 3E). Thus, the polar localization of BfpF does not require BfpA or the production of pilus filaments per se, since such cells are nonpiliated.
Conclusion.
Chemical cross-linking experiments and immunofluorescence and immuno-gold transmission electron microscopy have shown that proteins encoded by the bfp operon form a hetero-oligomeric complex that spans the periplasmic space and is preferentially localized to one pole of the cell. These results provide the basis for ongoing studies that seek to determine how the individual proteins are organized in the complex, how this organizational plan contributes to their biogenic roles, and by what mechanism they are targeted to the pole. From a pathogenic perspective, future studies will examine whether the polar location of the organelle favors the formation of infectious units and epithelial cell colonization.
Acknowledgments
This work was supported by grant AI39521 from the National Institutes of Health.
We thank Jon Mulholland for assistance with fluorescence microscopy.
REFERENCES
- 1.Anantha, R. P., K. D. Stone, and M. S. Donnenberg. 2000. Effects of bfp mutations on biogenesis of functional enteropathogenic Escherichia coli type IV pili. J. Bacteriol. 182:2498-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. [DOI] [PubMed] [Google Scholar]
- 3.Blank, T. E., and M. S. Donnenberg. 2001. Novel topology of BfpE, a cytoplasmic membrane protein required for type IV fimbrial biogenesis in enteropathogenic Escherichia coli. J. Bacteriol. 183:4435-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnenberg, M. S. 2000. Pathogenic strategies of enteric bacteria. Nature 406:768-774. [DOI] [PubMed] [Google Scholar]
- 5.Gomez-Duarte, O. G., and J. B. Kaper. 1995. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect. Immun. 63:1767-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hardie, K. R., S. Lory, and A. P. Pugsley. 1996. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 15:978-988. [PMC free article] [PubMed] [Google Scholar]
- 7.Harry, E. J., K. Pogliano, and R. Losick. 1995. Use of immunofluorescence to visualize cell-specific gene expression during sporulation in Bacillus subtilis. J. Bacteriol. 177:3386-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lane, D., and E. Harlow. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 9.Layh-Schmitt, G., A. Podtelejnikov, and M. Mann. 2000. Proteins complexed to the P1 adhesin of Mycoplasma pneumoniae. Microbiology 146:741-747. [DOI] [PubMed] [Google Scholar]
- 10.Mattick, J. S., B. J. Anderson, P. T. Cox, B. P. Dalrymple, M. M. Bills, M. Hobbs, and J. R. Egerton. 1991. Gene sequences and comparison of the fimbrial subunits representative of Bacteroides nodosus serotypes A to I: class I and class II strains. Mol. Microbiol. 5:561-573. [DOI] [PubMed] [Google Scholar]
- 11.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 12.Ramer, S. W., D. Bieber, and G. K. Schoolnik. 1996. BfpB, an outer membrane lipoprotein required for the biogenesis of bundle-forming pili in enteropathogenic Escherichia coli. J. Bacteriol. 178:6555-6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramer, S. W., G. K. Schoolnik, C. Y. Wu, J. Hwang, S. A. Schmidt, and D. Bieber. 2002. The type IV pilus assembly complex: biogenic interactions among the bundle-forming pilus proteins of enteropathogenic Escherichia coli. J. Bacteriol. 184:3457-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakai, D., and T. Komano. 2002. Genes required for plasmid R64 thin-pilus biogenesis: identification and localization of products of the pilK, pilM, pilO, pilP, pilR, and pilT genes. J. Bacteriol. 184:444-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271-283. [DOI] [PubMed] [Google Scholar]
- 16.Scaletsky, I. C., M. L. Silva, and L. R. Trabulsi. 1984. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect. Immun. 45:534-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt, S. A., D. Bieber, S. W. Ramer, J. Hwang, C. Y. Wu, and G. Schoolnik. 2001. Structure-function analysis of BfpB, a secretin-like protein encoded by the bundle-forming-pilus operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:4848-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schreiber, W., K. D. Stone, M. A. Strong, L. J. DeTolla, Jr., M. Hoppert, and M. S. Donnenberg. 2002. BfpU, a soluble protein essential for type IV pilus biogenesis in enteropathogenic Escherichia coli. Microbiology 148:2507-2518. [DOI] [PubMed] [Google Scholar]
- 19.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skerker, J. M., and L. Shapiro. 2000. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 19:3223-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sohel, I., J. L. Puente, S. W. Ramer, D. Bieber, C. Y. Wu, and G. K. Schoolnik. 1996. Enteropathogenic Escherichia coli: identification of a gene cluster coding for bundle-forming pilus morphogenesis. J. Bacteriol. 178:2613-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soto, G. E., and S. J. Hultgren. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J. Bacteriol. 181:1059-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stone, K. D., H. Z. Zhang, L. K. Carlson, and M. S. Donnenberg. 1996. A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for the biogenesis of a type IV pilus. Mol. Microbiol. 20:325-337. [DOI] [PubMed] [Google Scholar]
- 24.Strom, M. S., and S. Lory. 1993. Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47:565-596. [DOI] [PubMed] [Google Scholar]
- 25.Swanson, J., S. J. Kraus, and E. C. Gotschlich. 1971. Studies on gonococcus infection. I. Pili and zones of adhesion: their relation to gonococcal growth patterns. J. Exp. Med. 134:886-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tobe, T., G. K. Schoolnik, I. Sohel, V. H. Bustamante, and J. L. Puente. 1996. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 21:963-975. [DOI] [PubMed] [Google Scholar]
- 27.Todd, W. J., G. P. Wray, and P. J. Hitchcock. 1984. Arrangement of pili in colonies of Neisseria gonorrhoeae. J. Bacteriol. 159:312-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wall, D., and D. Kaiser. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1-10. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, H. Z., and M. S. Donnenberg. 1996. DsbA is required for stability of the type IV pilin of enteropathogenic Escherichia coli. Mol. Microbiol. 21:787-797. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, H. Z., S. Lory, and M. S. Donnenberg. 1994. A plasmid-encoded prepilin peptidase gene from enteropathogenic Escherichia coli. J. Bacteriol. 176:6885-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]