Abstract
Elongation factor Tu (EF-Tu), the protein responsible for delivering aminoacyl-tRNAs (aa-tRNAs) to ribosomal A site during translation, belongs to the group of guanosine-nucleotide (GTP/GDP) binding proteins. Its active ‘on’-state corresponds to the GTP-bound form, while the inactive ‘off’-state corresponds to the GDP-bound form. In this work we focus on the chemical step, GTP + H2O → GDP + Pi, of the hydrolysis mechanism. We apply molecular modeling tools including molecular dynamics simulations and the combined quantum mechanical – molecular mechanical calculations for estimates of reaction energy profiles for two possible arrangements of switch II regions of EF-Tu. In the first case we presumably mimic binding of the ternary complex EF-Tu·GTP·aatRNA to the ribosome and allow the histidine (His85) side chain of the protein to approach the reaction active site. In the second case, corresponding to the GTP hydrolysis by EF-Tu alone, the side chain of His85 stays away from the active site, and the chemical reaction GTP + H2O → GDP + Pi proceeds without participation of the histidine but through water molecules. In agreement with the experimental observations which distinguish rate constants for the fast chemical reaction in EF-Tu·GTP·aatRNA·ribosome and the slow spontaneous GTP hydrolysis in EF-Tu, we show that the activation energy barrier for the first scenario is considerably lower compared to that of the second case.
Keywords: EF-Tu protein, GTP hydrolysis, molecular modeling, enzymatic mechanism
Introduction
The hydrolysis cycle of guanosine triphosphate (GTP) provides a universal switch mechanism in controlling numerous cellular functions [1–4]. One of the important guanosine-nucleotide binding proteins, elongation factor EF-Tu, is responsible for delivering all elongator aminoacyl-tRNAs (aa-tRNAs) to the ribosomal A site during translation. The protein consists of a Ras-like domain (the G domain) that binds GTP (or guanosine diphosphate, GDP) and two β-barrel domains which, together with the G-domain, account for interactions with the acceptor domain of aa-tRNA. EF-Tu, GTP and aa-tRNA form a stable ternary complex that binds to the ribosome. Codon–anticodon interaction leads to conformational changes in EF-Tu and to the formation of the activated GTPase state of the ribosome·EF-Tu·aatRNA complex which is followed by the intrinsic chemical reaction of GTP hydrolysis. Subsequent conformational changes in EF-Tu from the GTP- to GDP-bound form lead to the aa-tRNA release while EF-Tu·GDP leaves the ribosome. Elongation factor EF-Ts catalyzes reconstruction of the active form EF-Tu·GTP.
The complete kinetic mechanism of EF-Tu binding of aa-tRNA to the E.coli ribosome was described in Ref.[5]. The chemical reaction of GTP hydrolysis leading to GDP and inorganic phosphate Pi
| (1) |
by EF-Tu itself is known to be slow while its complex with the ribosome performs very efficiently, enhancing the reaction rate about 105 fold [6]. In this work we focus only on this chemical step of the mechanism following our previous studies of GTP hydrolysis by the Ras protein [7,8] and of adenosine triphosphate (ATP) hydrolysis by myosin [9].
The reaction scheme of the hydrolysis of GTP by the protein matrix (M) may be described by the following scheme
| (2) |
It includes the following stages: (1) active site reorganization leading to formation of the enzyme-substrate (ES) complex M*·GTP, (2) formation of the ternary product complex M*·GDP·Pi through the chemical transition state (TS), and (3) product (Pi) release. Rearrangements of the protein structure occur in steps 1 and 3. Our quantum based modeling refers to stage 2 in Eq. (2), while molecular dynamics simulations refer to the protein reorganization in step 1.
By performing molecular modeling we address, in particular, the important question of the role the histidine side chain His85 (or His84 in another numbering) plays in the chemical step of GTP hydrolysis by EF-Tu. This problem was discussed in the literature [10–13] in relevance to site-directed mutational studies. In particular, these studies revealed that replacement of this residue with Ala considerably reduced the rate constant of GTP hydrolysis [13], while replacement with Gln showed only a slightly reduced rate of intrinsic hydrolysis reaction compared to the native protein [11–13]. However, analysis of the available crystal structures from the PDB archive [14] of EF-Tu complexed with GTP analogs 1EXM [15], 1EFT [16], indicates that in these structures His85 points away from the active site of the enzyme. Moreover, the side chains of Val20 and Ile61 forming the so-called ‘hydrophobic gate’ prevent His85 to approach the active site.
According to the working hypotheses discussed in Ref.[13] the ribosome stimulates the intrinsic GTP hydrolysis by 5 orders of magnitude by promoting the conformational reorganization that favors His85 approach to the active site. This suggestion is supported by the observation that His85 is a part of the switch II region which endures the greatest reorganization on the way from the GTP-bound to the GDP-bound form of EF-Tu and, therefore, re-orientation of His85 during conformational changes in EF-Tu is structurally possible. Secondly, the crystal structure (PDBID: 1HA3) of the complex between EF-Tu*·GDP and aurodox is reported [17] in which EF-Tu is believed to be in the GTP-bound form. In this structure, the His85 side chain is oriented toward the nucleotide-binding site and the flexible chain of residues 53–64, including Ile61, does not prevent His85 from approaching the active site. Our simulations described below are in accord with this hypothesis of direct involvement of His85 in the hydrolysis reaction and provide detailed visualization of the reaction mechanism.
More generally, in this work we address the question of the mechanism of the intrinsic chemical reaction in enzymatic hydrolysis of nucleoside triphosphates which is still under active debate [3,18–31]. The key issue is an assignment of the reaction mechanism to a certain type depending on the nature of a transition state. On one extreme, a fully dissociative transition state for the GTP hydrolysis is represented by an intermediate that shows bond cleavage between the γ-phosphate group and GDP. On the other extreme, a fully associative transition state is represented by a penta-coordinated intermediate that shows no β-γ bridge bond cleavage but significant amount of bond formation between the incoming lytic water molecule and the γ-phosphate of GTP. It should be noted that none of the available experiments can determine whether the mechanism is associative or dissociative [32]. Another popular model suggests that the substrate GTP itself, specifically, γ-phosphate group of GTP, serves as the general base in its own hydrolysis [33,34].
Our previous simulations of the intrinsic chemical reaction of the GTP hydrolysis in the Ras protein either by itself [7] or in the Ras-RasGAP complex [8], of the ATP hydrolysis in myosin [9], and of the methyl triphosphate hydrolysis in water [35,36] performed in the combined quantum mechanical – molecular mechanical (QM/MM) approximation allow us to conclude that in all enzymatic reactions the reasonable minimum energy profiles with low activation energies are consistent with the following reaction route. In all cases a low activation energy cleavage of the Pγ−Oβγ bond in nucleotide triphospate and separation of the γ-phosphate from nucleotide diphosphate occurs through a planar arrangement of the PγO3− group in TS with almost unaltered lytic water molecule. Formation of the inorganic phosphate results as a consequence of proton transfers mediated by the neighboring molecular groups of the protein matrix. In Ras and Ras-RasGAP catalyzed hydrolysis of GTP, the Gln61 side chain serves as a crucial mediator. Therefore, when proceeding to simulations of GTP hydrolysis in EF-Tu we expected to find similar features of the reaction mechanism with an active participation of His85 in the case of rapid enzymatic reaction.
Methods
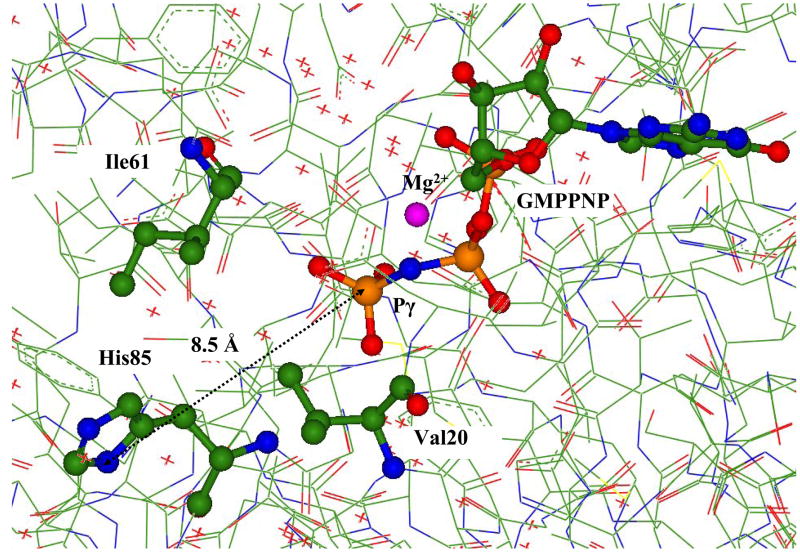
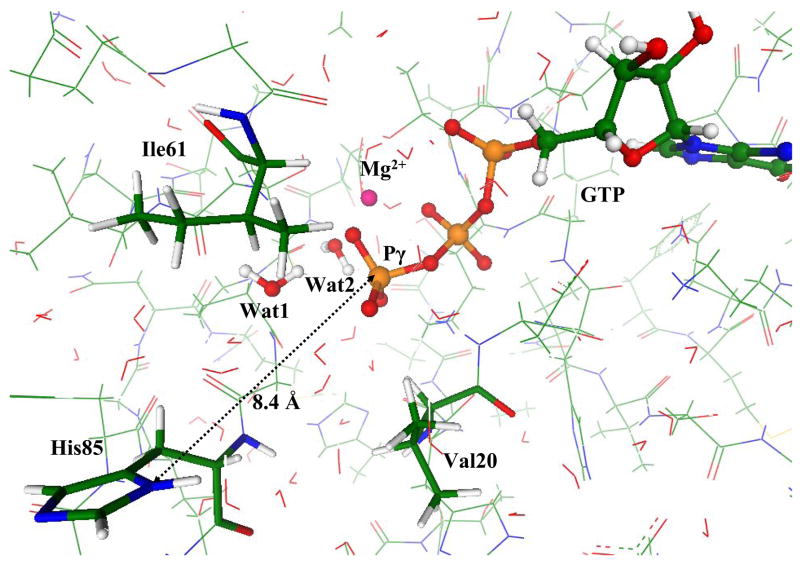
We started simulations from the coordinates of heavy atoms in the crystal structures PDBID:1EFT solved at the resolution 2.50 Å [16] of EF-Tu from Thermus aquaticus and PDBID:1EXM solved at the resolution 1.70 Å [15] of EF-Tu from Thermus thermophilus. These crystals accommodate the slowly hydrolizable GTP analog, 5’-guanylylimidodiphosphate (GMPPNP), containing the NH group instead of the bridging βγ-oxygen atom. Fig. 1 shows the positions of GTP analog, GMPPNP, magnesium cation, His85, and the side chains of the ‘hydrophobic gate’ Val20 and Ile61. The distance between Nδ of His85 and Pγ of GMPPNP constitutes 8.5 Å in PDBID:1EXM, as shown in Fig. 1, and 7.1 Å in PDBID:1EFT (in the latter case, a slightly different orientation of the His85 side chain occurs).
Figure 1.
The fragment of the crystal structure PDBID:1EXM (15) showing the positions of GTP analog, GMPPNP, magnesium cation, His85, and the side chains of the ‘hydrophobic gate’ Val20 and Ile61. Here and below the carbon atoms are distinguished by green, oxygen atoms by red, nitrogen atoms by blue, phosphorus atoms by brown, magnesium by magenta.
At the first stages of the work we used the computer package NAMD2 [37] of molecular dynamics (MD) simulations to model the structures of enzyme-substrate (ES) complexes for subsequent quantum based calculations. The native GTP molecule was reconstructed from its analog GMPPNP, the coordinates of hydrogen atoms were introduced, and the protein was solvated by water molecules. The model for the ES complex included 6348 protein and ligand atoms, 108 crystal water molecules and 4047 solvent water molecules. The CHARMM force field parameters [38] extended with those for GTP from Ref.[39] were used in MD simulations. Initially, we optimized positions of water molecules by the minimum energy criterion with the fixed positions of protein and ligand atoms. Then the constraints were lifted, and positions of all atoms were optimized. The MD runs were performed for the model system within a rectangular box with dimensions 61 × 72.5 × 48 Å3 with the imposed periodic boundary conditions. No constraints were imposed on the bond lengths allowing a 1 fs time step for integration at temperature 300K.
When starting from coordinates of the crystal structures PDBID: 1EFT or PDBID: 1EXM in MD simulations we always arrived at the structure (called Hisout below) with the remote position of the His85 side chain relative to the γ-phosphate group of GTP. In this arrangement, His85 side chain could not approach the active site due to steric hindrance of the hydrophobic gate of Val20 and Ile61. Then we prepared manually another model structure (called Hisin below) by re-orienting the His85 side chain toward the γ-phosphate group of GTP following the motifs of the crystal structure PDBID: 1HA3 [17]. For such re-arrangement we temporarily moved apart the residues of the hydrophobic gate and shifted the residues of switch II. Then we restored the hydrogen bond network and re-optimized atomic coordinates. A fairly long MD trajectory (700 ps) executed for this structure showed the His85 side chain remained close to the active site. Rout mean square fluctuations of Cα protein atoms during the MD trajectory was found to be relatively small, typically less than 1.2 Å indicating the relative stability of the system.
The coordinates obtained in preliminary MD simulations for both model systems, Hisout and Hisin, were used as an initial guess for more rigorous calculations of equilibrium geometry parameters by using the combined quantum mechanical – molecular mechanical (QM/MM) method [40]. To ensure that the QM/MM optimization resulted in minimum energy points corresponding to the stable ES complexes of Hisout and Hisin structures we numerously varied the starting sets of coordinates around those obtained in preliminary MD simulations and repeated QM/MM minimization calculations until the lowest energy structures were reached.
In QM/MM calculations we used the same version of the theory as in previous studies of enzymatic GTP and ATP hydrolysis [7–9], i.e., the flexible effective fragment variant [41,42] of the effective fragment potential QM/MM [43] method. This is an approach which allows one to perform calculations close to an ab initio QM treatment of the entire molecular system. Molecular groups assigned to the MM part are represented by effective fragments which introduce their electrostatic potentials expanded up to octupoles as one-electron contributions to the quantum Hamiltonian. These potentials, as well as contributions from interactions of effective fragments with the QM region, are obtained in preliminary quantum chemical calculations for separated fragments by using ab initio electron densities. The exchange-repulsion potentials to be combined with the electrostatic terms are also created in preliminary ab initio calculations. Thus, all empirical parameters are entirely within the MM subsystem. In the flexible effective fragment version [41,42] the fragment-fragment interactions are modeled by the force field parameters from the AMBER library [44]. The computer program used in the simulations is based on the GAMESS(US) [45] (more specifically, its Intel-specific version, PC GAMESS [46]) quantum chemistry package and on the TINKER [47] molecular modeling system.
In this application we performed partitioning of the model system into QM and MM parts as follows. All phosphate groups of fully unprotonated GTP molecule, the lytic water molecule, the magnesium cation, and the side chain of His85 were assigned to the QM subsystem in the case of the Hisin structure, while for calculations for the Hisout structure, an additional water molecule was introduced to the QM subsystem instead of the His85 side chain. In total, 33 atoms constituted the quantum subsystem and 1556 atoms combined to 473 effective fragments formed the MM subsystem for the Hisin structure. Correspondingly, 24 atoms in the QM part and 1481 MM atoms combined to 448 effective fragments were considered for the Hisout structure.
The simulations included scans of the composite multidimensional QM/MM potential energy surface in the regions where chemical bonds or hydrogen bonds could be cleaved or formed. As a result, the basins around presumable stationary points were specified for more careful calculations of the local minima or saddle points. The stationary points were located by unconstrained minimizations (for local minima) or by constrained minimizations (for saddle points) of the QM/MM energy. The location of a transition states (TS) was determined based on the criterion that the gradient of the constrained internal coordinate along an assumed reaction path must change its sign at the presumed TS. The internal coordinates of all atoms in the QM subsystem and positions of effective fragments in the MM subsystem including all water molecules were optimized. Positions of remote effective fragments at distance greater ~ 10 Å from the reaction center were kept fixed as in the crystal structure.
As in our previous simulations of enzymatic GTP and ATP hydrolysis [7–9], quantum calculations were carried out by using the Hartree-Fock approach in the QM part. The polarized “LANL2DZdp ECP” basis set (and the corresponding pseudopotential for phosphorus [48]) was used for all atoms except magnesium. For magnesium, the standard 6-31G basis was employed. It should be noted that multiple minimum energy points could be located in geometry optimizations. We attempted to overcome this difficulty by performing in each case numerous selections of the starting sets of coordinates for minimization until the lowest energy was reached under the condition that the hydrogen bond network in the immediate vicinity of the active site. Below we shall use the notations Hisin and Hisout for the QM/MM optimized model structures.
Results
(i) The reaction path for the Hisin model structure
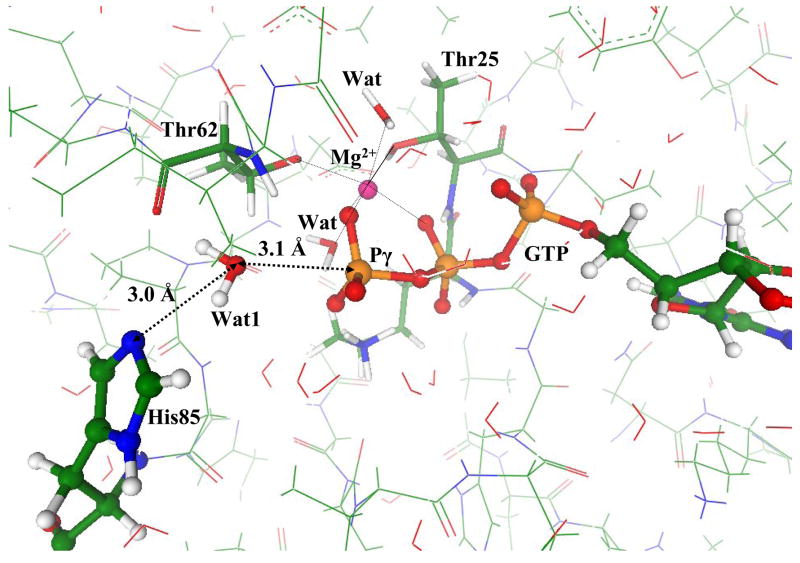
Fig. 2 illustrates the geometry configuration of the ES complex as optimized in QM/MM calculations. We show the immediate participants of the chemical reaction, the phosphate groups of GTP and the lytic water molecule, Wat1. The latter is oriented in a perfect position for the in-line attack on γ-phosphate by the hydrogen bond network which includes the side chain of His85. The equilibrium distance between oxygen from the lytic water oxygen and γ-phosphorus, 3.1 Å, is typical for ES complexes for the enzymatic hydrolysis of nucleotide triphosphates [7–9,31]. The characteristic 6-fold coordination shell of magnesium cation is formed by oxygen atoms: Oγ and Oβ from GTP, two from water molecules denoted as Wat in Fig. 2, and two from the side chains of Thr25 and Thr65.
Figure 2.
Geometry configuration of the ES complex for the Hisin model structure.
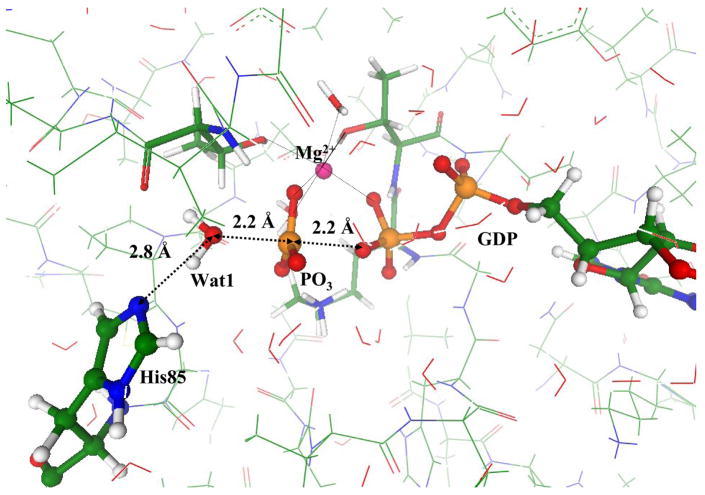
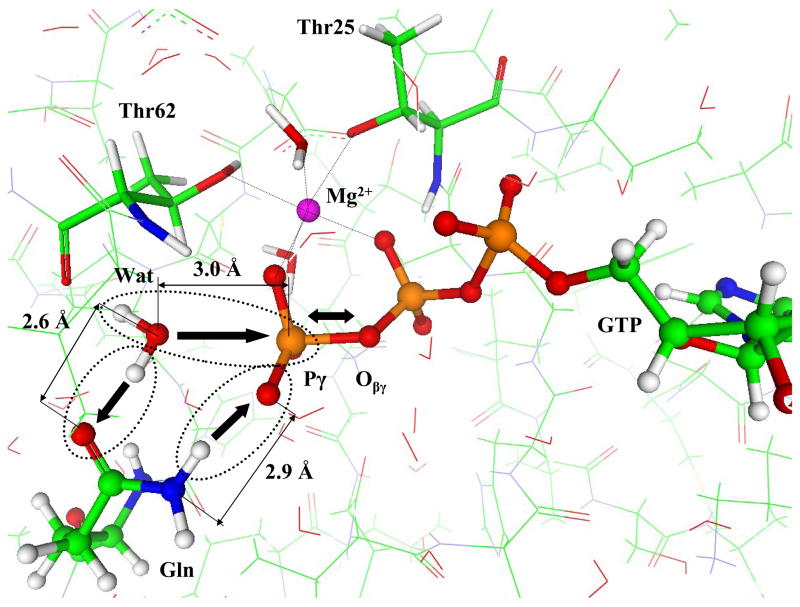
Fig. 3 illustrates the geometry configuration of the saddle point on the potential energy surface or, in other words, of a transition state (TS) for the only stage of the intrinsic chemical reaction occurring in hydrolysis of GTP by EF-Tu. The Pγ − Oβγ bond is cleaved, but the lytic water molecule, Wat1, remains essentially unaltered. We recognize a characteristic planar arrangement of the PγO3 group with the typical distances, 2.2 Å between Pγ and Oβγ and between Pγ oxygen of Wat1. The QM/MM energy of TS is 32 kJ/mol higher than the level of ES.
Figure 3.
Geometry configuration of TS for the Hisin model structure.
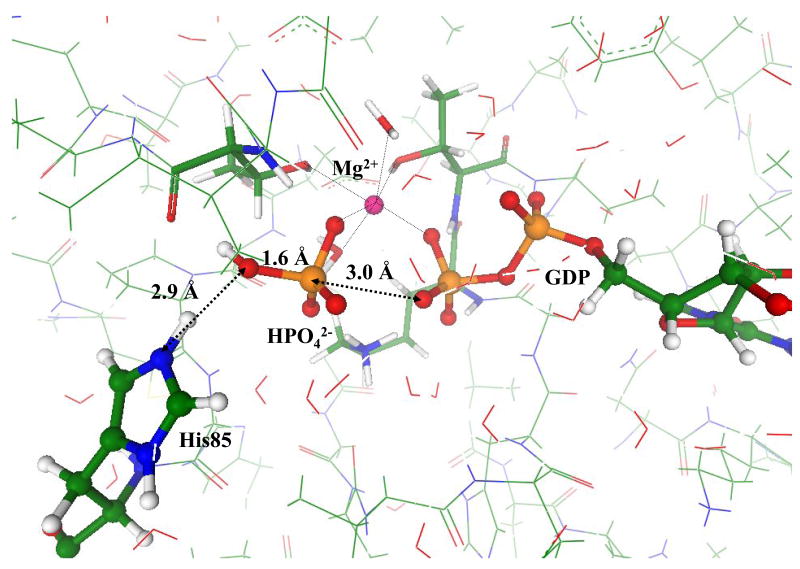
The steepest descent from TS toward products led the system to the stationary point on the potential energy surface, corresponding to the enzyme-product (EP) complex, geometry of which is shown in Fig. 4. The inorganic phosphate, HPO42− is formed and spatially separated from GDP (the Pγ − Oβγ distance is 3.0 Å). The protonated side chain of His85 is a part of the hydrogen bond network. However, it is reasonable to assume that after release of Pi, i.e. the stage which is not modeled in this work, His85 returns to the conventional neutral status by donating an extra proton to the environmental molecular groups. The QM/MM energy of the EP complex is estimated to be 34 kJ/mol lower than the level of ES.
Figure 4.
Geometry configuration of the EP complex for the Hisin model structure.
Calculations of vibrational frequencies for the geometry configurations of ES, TS and EP allowed us to estimate entropic contributions to the energies in the conventional rigid rotor – harmonic oscillator approximation. We obtained the following free energy differences at 300K: 45 kJ/mol between TS and ES levels and 21 kJ/mol between ES and EP levels.
(ii) The reaction path for the Hisout model structure
In Fig. 5 we show a view of the computed equilibrium geometry configuration of the Hisout model structure indicating positions of the side chain of His85 and of the hydrophobic gate Val20 and Ile61. This picture may be compared to the view of heavy atoms in the crystal structure 1EXM as illustrated in Fig. 1. Apparently, His85 in this position cannot help to hydrolyze GDP in EF-Tu. Instead, the reaction can proceed with the help of water molecules like GTP hydrolysis in free Ras [8,21], ATP hydrolysis in myosin [9] or triphosphate hydrolysis in water clusters [24,35,36]. The GTP molecule in EF-Tu resides fairly close to the surface and, therefore, the protein cavity around the phosphate groups of GTP can accommodate several water molecules. We show in Fig. 5 positions of the lytic molecule, Wat1, and the water molecule, Wat2, which assists the reaction as clarified below.
Figure 5.
Geometry configuration of the ES complex for the Hisout model structure showing the side chain of His85.
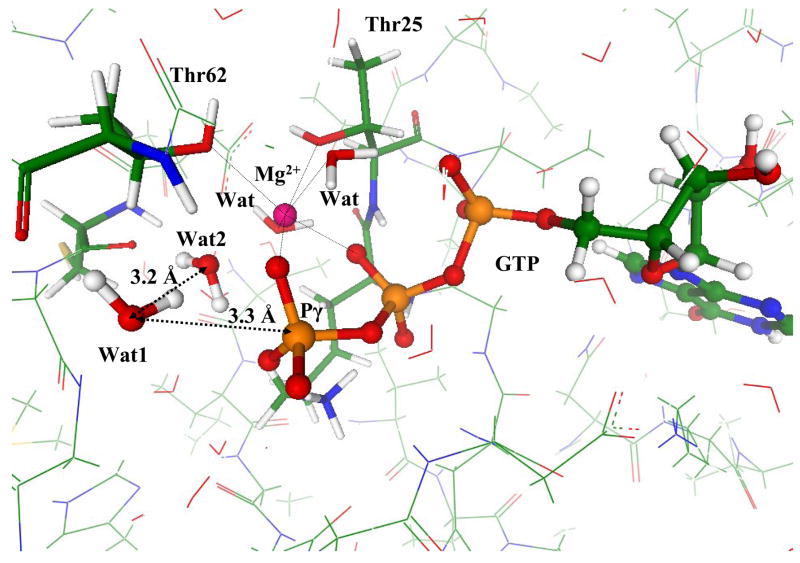
Fig. 6 presents another perspective of the computed geometry configuration of the ES complex showing GTP, two active water molecules, Wat1 and Wat2, and the magnesium coordination shell. The lytic water Wat1 is oriented in favorable position for an in-line attack on γ-phosphate; however, it stays a little farther (3.3 Å) from Pγ compared to the case of the Hisin model system (3.1 Å) as shown in Fig. 2.
Figure 6.
Geometry configuration of the ES complex for the Hisout model structure.
By selecting the distance between oxygen of Wat1 and Pγ as a reaction coordinate we computed the minimum energy profile from ES to TS for the Hisout model structure. The geometry configuration of TS (or the saddle point) is illustrated in Fig. 7. Again we recognize the characteristic motif of TS for the cleavage of the Pγ − Oβγ bond in triphosphates with a planar arrangement of the PO3 group and almost unaltered lytic water molecule Wat1. Compared to the case of the Hisin structure (Fig. 3) the Pγ atom stays closer to the lytic water oxygen (2.0 Å) than to Oβγ (2.6 Å) along the line O(Wat1) – Pγ − Oβγ. The estimated QM/MM energy of this TS is much higher (80 kJ/mol above the corresponding level of ES) than that for the Hisin catalyzed reaction (32 kJ/mol). Therefore, the results of modeling are consistent with the working hypothesis [13] on a substantial enhancement of the reaction if the His85 residue directly participates in hydrolysis.
Figure 7.
Geometry configuration of TS for the Hisout model structure.
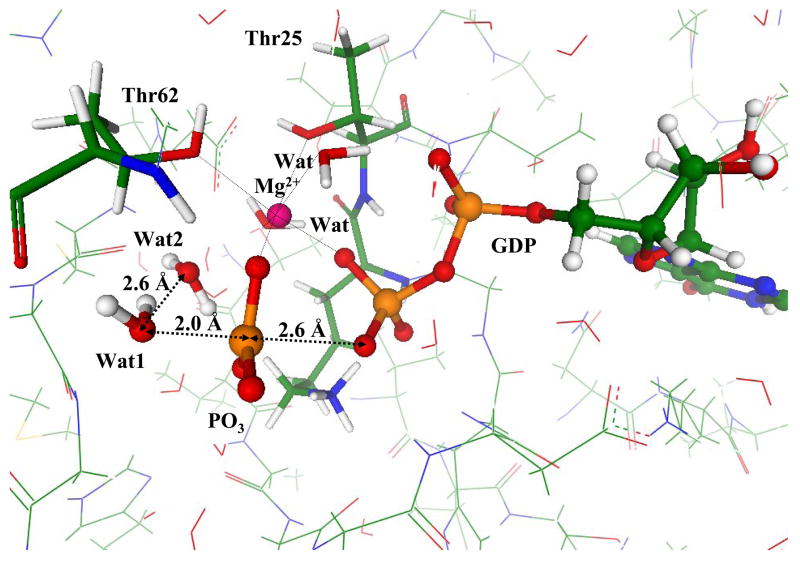
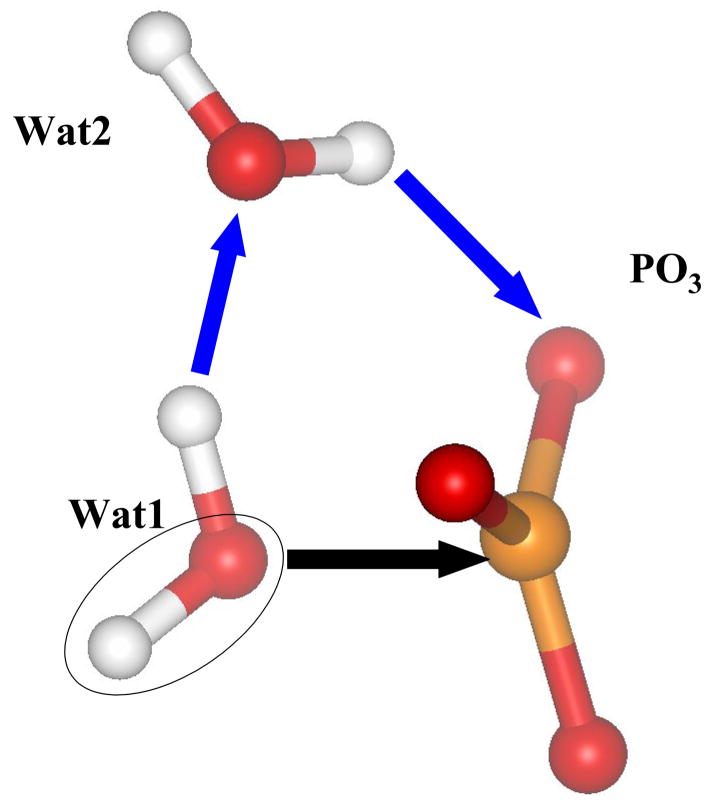
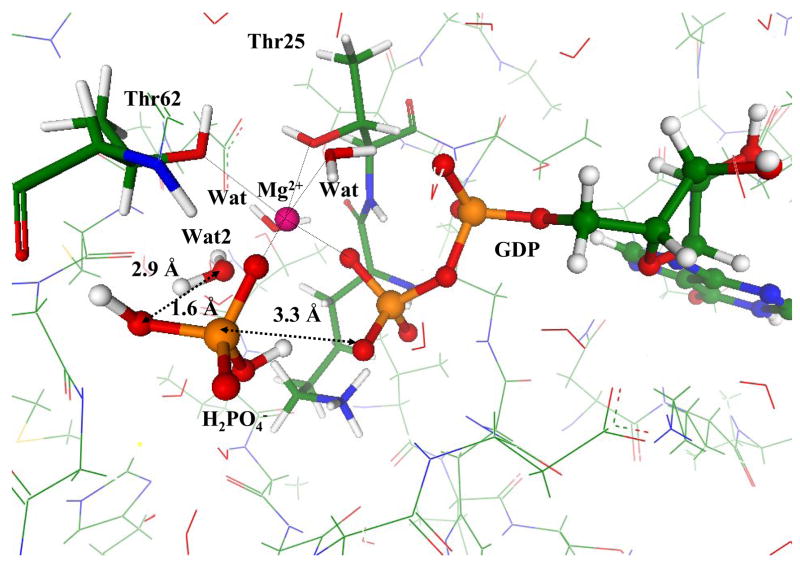
The steepest descent from the computed TS toward products allowed us to arrive to the stationary point on the potential energy surface corresponding to the enzyme-product (EP) complex. The inorganic phosphate, in this case, H2PO4− is formed as a result of the chain of proton transfers as illustrated in Fig. 8. This includes the nucleophilic attack of hydroxyl from the breaking lytic water molecule Wat1 on the γ-phosphorus atom as shown by black arrow and two proton transfers shown by blue arrows in Fig. 8. All these transformations are barrier-less in this particular environment.
Figure 8.
The transformations that lead to the formation of inorganic phosphate from the TS shown in Fig.7: the attack of hydroxyl from Wat1 and proton transfers.
The resulting enzyme-product complex is illustrated in Fig. 9. Its QM/MM energy is estimated to be 45 kJ/mol lower than the level of the corresponding ES complex.
Figure 9.
Geometry configuration of the EP complex for the Hisout model structure.
The estimates of entropic contributions resulted in the following free energy differences at 300K: 89 kJ/mol between TS and ES levels and 39 kJ/mol between ES and EP levels.
Discussion
The results of molecular modeling GTP hydrolysis reaction for two possible arrangements of switch II regions of EF-Tu are completely consistent with the previously considered mechanisms of enzymatic or solution reactions of triphosphates [7–9]. In both cases of protein arrangements denoted here as Hisin and Hisout the minimum energy reaction profiles correspond to the fairly low activation barriers with the planar structure of the of the PγO3− group in the transition state (TS) and almost unchanged lytic water molecule. The character of TS is closer to the dissociative type reaction mechanism than to the associative type, however, no metaphosphate intermediate is found in the present simulations. Formation of the inorganic phosphate (Pi) is responsible for irreversibility of the chemical reaction since the energy levels of the enzyme-product (EP) complexes are lower by 34 or 45 kJ/mol for the Hisin and Hisout models, respectively, than those of ES complexes. Creation of Pi proceeds differently for the Hisin and Hisout protein matrices. In the Hisin case His85 serves as a general base abstracting the proton from the breaking lytic water molecule. In the case of the Hisout conformation formation of Pi results as a consequence of proton transfers mediated by the water molecules inside the EF-Tu protein cavity.
The results are consistent with the experimental observations according to which GTP hydrolysis in EF-Tu, with the His85 side chain far apart the active site, is very slow, about 5 × 10−5 s, while the hydrolysis rate in the activated by the ribosome ternary complex EF-Tu·GTP·aatRNA, and hence with the His85 side chain staying close to the active site, is considerably higher. Our QM/MM calculations result in the substantial decrease of the activation barrier in case of active participation of His85 in the reaction compared to the case of GTP hydrolysis by water molecules inside the protein cavity.
The reduction of the rate constant upon replacement of His85 by Ala observed experimentally [13] is also consistent with our mechanism, since Ala cannot serve as a proton acceptor from the lytic water molecule. Another experimental result which suggests that replacement of His85 with Gln shows only a slightly reduced rate of intrinsic hydrolysis reaction compared to the native protein [11–13] can be also understood within the present model. To clarify the latter statement we performed additional series of QM/MM calculations by substituting the side chain of His85 in the Hisin model structure by the Gln side chain. Unconstrained optimization of geometry parameters resulted in the structure of the ES complex shown in Fig. 10.
Fig. 10.
QM/MM optimized geometry configuration of the ES complex in the Hisin protein conformation with the His side chain replaced by Gln.
The relative position of the key participants of the reaction is here almost precisely the same as in the ES complex for the Ras-RasGAP catalyzed hydrolysis of GTP [8]. The lytic water molecule (Wat in Fig. 10) is oriented toward the γ-phosphate of GTP by the carbonyl groups of Gln and Thr62. In the Ras-RasGAP case the lytic water is oriented by the Gln61 and Thr35 side chains (see Fig. 3 of Ref.[8]). As illustrated in Fig. 4 of Ref.[8] for such an arrangement of the reagents the reaction should proceed as follows. Upon decreasing the distance between oxygen of Wat and Pγof GTP, the Pγ−Oβγ bond is cleaved (as illustrated by the red thick arrow in Fig. 10), the system overcomes the first transition state and occurs in the configuration of a reaction intermediate (Fig. 3 of Ref.[8]). The barrier height at this stage was estimated as 36 kJ/mol for the Ras-RasGAP system [8]. The separated from GDP γ-phosphate group, the side chain of Gln and lytic water are in perfect position for a nucleophilic attack of the hydroxyl accompanied by concerted proton transfers as illustrated by black arrows in Fig. 10 of this paper (or by the curl arrows in Fig. 3 of Ref.[8]). This stage of the reaction from the intermediate to the products (inorganic phosphate H2PO4−, GDP and tautomerized Gln) should proceed through the barrier of the height less than 40 kJ/mol (Fig. 5 of Ref.[8]). Restoring the amino form of Gln can be easily achieved upon leakage of water molecules after release of inorganic phosphate. Such mechanism of the GTP hydrolysis by G-proteins, according to which Gln abstracts a proton from the attacking water molecule while simultaneously donates a proton to a γ-phosphate oxygen, has been also discussed by Sondek et al. [49]. For the purposes of this paper it is important to understand that mutation of His85 by Gln should not lead to dramatic increase in activation barriers.
The only experimentally motivated conclusion not supported by the present simulations is the proposal that His85 does not act as a general base in the hydrolysis reaction [13]. The authors of Ref.[13] expressed no doubts on the catalytic role of His85, but excluded its efficiency as a general base following the results of studies of pH dependence of the hydrolysis reaction rate for the slowly hydrolizable GTP analog, mant-GTPγS. Within the range of 6.5 – 8.5 pH units compared to the usual experimental conditions at pH=7.5 the reaction rate showed no dependence on pH which was at variance with the expected behavior if a general base with a pKa of about 7, presumably assigned to His85, were involved in catalysis. Instead, the authors of Ref.[13] proposed that the catalytic role of His85 is to stabilize reaction TS by hydrogen bonding to the lytic water molecule or the γ-phosphate group of GTP. We may argue, firstly, that mant-GTPγS is not a full substitute of natural GTP and, secondly, that the pKa value of His85 buried inside the protein cavity for the case of the Hisin structure may lie outside the studied range of 6.5 – 8.5 pH units. It seems difficult to specify another role of His85 in stabilizing the reaction TS at the mechanistic level. Apparently, His85 is a part of hydrogen bond networks in geometry configurations of all stationary points: ES (Fig. 2), TS (Fig. 3) and EP (Fig. 4). It clearly stabilizes the TS (Fig. 3) by fixing the lytic water molecule Wat1 by hydrogen bonds, but not the γ-phosphate group of GTP. However in order to complete the reaction by formation of inorganic phosphate and to arrive to the configuration of the enzyme-product complex His85 accepts the proton. The calculation results indicate that the conformation of the EP complex (Fig. 4) with the protonated His85 corresponds to the true minimum energy point with the energy level lower than that of the ES complex. Therefore, molecular modeling provides a direct evidence of such a development compared to indirect evaluations based on solution pKa values for His.
Conclusion
The results of molecular modeling provide support to the working hypotheses [13] that the ribosome stimulates the intrinsic GTP hydrolysis by the EF-Tu protein. This is achieved by promoting the conformational reorganization that favors the side chain of His85 approach to the active site. The simulated reaction routes for the Hisin and Hisout protein conformations are characterized by distinctive energy profiles with the considerably lower activation barrier for the Hisin scenario.
Acknowledgments
This work was partly supported by the grant from the Russian Foundation for Basic Research (project #07-03-00060). We thank the staff and administration of the Advanced Biomedical Computing Center for their support of this project. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organization imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–131. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 2.Sprang SR. G protein mechanisms: Insights from structural analysis. Annu Rev Biochem. 1997;66:639–672. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 3.Li G, Zhang XC. GTP Hydrolysis Mechanism of Ras-like GTPases. J Mol Biol. 2004;340:921–932. doi: 10.1016/j.jmb.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Sprang SR, Chen Z, Du X. Structural basis of effector regulation and signal termination in heterotrimeric Gα proteins. Advances in Protein Chemistry. 2007;74:1–65. doi: 10.1016/S0065-3233(07)74001-9. [DOI] [PubMed] [Google Scholar]
- 5.Pape T, Wintermeyer W, Rodnina M. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminiacyl-tRNA to the A site of the E.coli ribosome. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodnina MV, Pape T, Fricke R, Wintermeyer W. Elongation factor Tu, a GTPase triggered by codon recognition on the ribosome: mechanism and GTP consumption. Biochem, Cell Biol. 1995;73:1221–1227. doi: 10.1139/o95-132. [DOI] [PubMed] [Google Scholar]
- 7.Grigorenko BL, Nemukhin AV, Topol IA, Cachau RE, Burt SK. QM/MM modeling the Ras–GAP catalyzed hydrolysis of guanosine triphosphate. Proteins: Struct Funct Bioinf. 2005;60:495–503. doi: 10.1002/prot.20472. [DOI] [PubMed] [Google Scholar]
- 8.Grigorenko BL, Shadrina MS, Topol IA, Burt SK, Nemukhin AV. Mechanisms of guanosine triphosphate hydrolysis by Ras and Ras-GAP proteins as rationalized by ab initio QM/MM simulations. Proteins: Struct Funct Bioinf. 2007;66:456–466. doi: 10.1002/prot.21228. [DOI] [PubMed] [Google Scholar]
- 9.Grigorenko BL, Rogov AV, Topol IA, Burt SK, Martinez HM, Nemukhin AV. Mechanism of the myosin catalyzed hydrolysis of ATP as rationalized by molecular modeling. Proc Natl Acad Sci USA. 2007;104:7057–7061. doi: 10.1073/pnas.0701727104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cool RH, Parmeggiani A. Substitution of Histidine-84 and the GTPase Mechanism of Elongation Factor Tu. Biochemistry. 1991;30:362–366. doi: 10.1021/bi00216a008. [DOI] [PubMed] [Google Scholar]
- 11.Scarano G, Krab IM, Bocchini V, Parmeggiani A. Relevance of histidine-84 in the elongation factor Tu GTPase activity and in poly(Phe) synthesis: its substitution by glutamine and alanine. FEBS Letters. 1995;365:214–218. doi: 10.1016/0014-5793(95)00469-p. [DOI] [PubMed] [Google Scholar]
- 12.Zeidler W, Egle C, Ribeiro S, Wagner A, Katunin V, Kreutzer R, Rodnina M, Wintermeyer W, Sprinzl M. Site-directed mutagenesis of Thermus thermophilus elongation factor Tu. Replacement of His85, Asp81 and Arg300. Eur J Biochem. 1995;229:596–604. doi: 10.1111/j.1432-1033.1995.tb20503.x. [DOI] [PubMed] [Google Scholar]
- 13.Daviter T, Wieden HJ, Rodnina MV. Essential role of Histidine 84 in elogation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J Mol Biol. 2003;332:689–699. doi: 10.1016/s0022-2836(03)00947-1. [DOI] [PubMed] [Google Scholar]
- 14.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. (2000) The Protein Data Bank. Nucleic Acids Research. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hilgenfeld R, Mesters JR, Hogg T. Insights into the GTPase Mechanism of EF-Tu from Structural Studies. The Ribosome: Struct, Funct, Antibiotics, and Cellular Interactions. 2000;28:347–357. [Google Scholar]
- 16.Kjeldgaard M, Nissen P, Thirup S, Nyborg J. The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure. 1993;1:35–50. doi: 10.1016/0969-2126(93)90007-4. [DOI] [PubMed] [Google Scholar]
- 17.Vogeley L, Palm GJ, Mesters JR, Hilgenfeld R. Conformational change of elongation factor Tu (EF-Tu) induced by antibiotic binding. Crystal structure of the complex between EF-Tu*·GDP and aurodox. J Biol Chem. 2001;276:17149–17155. doi: 10.1074/jbc.M100017200. [DOI] [PubMed] [Google Scholar]
- 18.Langen R, Schweins T, Warshel A. On the mechanism of guanosine triphosphate hydrolysis in ras p21 proteins. Biochemistry. 1992;31:8691–8696. doi: 10.1021/bi00152a002. [DOI] [PubMed] [Google Scholar]
- 19.Schweins T, Langen R, Warshel A. Why have mutagenesis studies not located the general base in ras p21? Nat Struct Biol. 1994;1:476–484. doi: 10.1038/nsb0794-476. [DOI] [PubMed] [Google Scholar]
- 20.Maegley KA, Admiraal SJ, Herschlag D. Ras-catalyzed hydrolysis of GTP: A new perspective from model studies. Proc Natl Acad Sci USA. 1996;93:8160–8166. doi: 10.1073/pnas.93.16.8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheidig AJ, Burmester C, Goody RG. The pre-hydrolysis state of p21ras in complex with GTP: New insights into the role of water molecules in the GTP hydrolysis reaction of ras-like proteins. Structure. 1999;7:1311–1324. doi: 10.1016/s0969-2126(00)80021-0. [DOI] [PubMed] [Google Scholar]
- 22.Glennon TM, Villa J, Warshel A. How does GAP catalyze the GTP reaction of Ras: A computer simulation study. Biochemistry. 2000;39:9641–9651. doi: 10.1021/bi000640e. [DOI] [PubMed] [Google Scholar]
- 23.Katagiri D, Hata M, Itoh T, Neya S, Hoshino T. Atomic-scale mechanism of the GTP → GDP hydrolysis reaction by the Giα1 protein. J Phys Chem B. 2003;107:3278–3283. [Google Scholar]
- 24.Wang YN, Topol IA, Collins JR, Burt SK. Theoretical studies on the hydrolysis of mono-phosphate in gas phase and aqueous solution. J Am Chem Soc. 2003;125:13265–13273. doi: 10.1021/ja0279794. [DOI] [PubMed] [Google Scholar]
- 25.Topol IA, Cachau RE, Nemukhin AV, Grigorenko BL, Burt SK. Quantum chemical modeling of the GTP hydrolysis by the RAS–GAP protein complex. Biochim Biophys Acta. 2004;1700:125–136. doi: 10.1016/j.bbapap.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 26.Shurki A, Warshel A. Why does the Ras switch “break” by oncogenic mutations? Proteins: Struct Funct Bioinf. 2004;55:1–10. doi: 10.1002/prot.20004. [DOI] [PubMed] [Google Scholar]
- 27.Li G, Cui Q. Mechanochemical coupling in myosin: A theoretical analysis with molecular dynamics and combined QM/MM reaction path calculations. J Phys Chem B. 2004;108:3342–3357. [Google Scholar]
- 28.Schwarzl S, Smith JC, Fisher S. Insights into the chemomechanical coupling of the myosin motor from simulation of its ATP hydrolysis mechanism. Biochemistry. 2006;45:5830–5847. doi: 10.1021/bi052433q. [DOI] [PubMed] [Google Scholar]
- 29.Kötting C, Blessonohl M, Suveydis Y, Goody RC, Wittinghofer A, Gerwert K. (2006) A phosphoryl transfer intermediate in the GTPase reaction of Ras in complex with its GTPase-activating protein. Proc Natl Acad Sci USA. 2006;103:13911–13916. doi: 10.1073/pnas.0604128103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wittinghofer A. Phosphoryl transfer in Ras proteins, conclusive or elusive? Trends Biochem Sci. 2006;31:20–23. doi: 10.1016/j.tibs.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 31.Te Heesen H, Gerwert K, Schlitter J. Role of the arginine finger in Ras·RasGAP revealed by QM/MM calculations. FEBS Lett. 2007;581:5677–5684. doi: 10.1016/j.febslet.2007.11.026. [DOI] [PubMed] [Google Scholar]
- 32.Åqvist J, Kolmodin K, Florian J, Warshel A. Mechanistic alternatives in phosphate monoester hydrolysis: what conclusions can be drawn from available experimental data? Chemistry & Biology. 1999;6:R71–R80. doi: 10.1016/S1074-5521(99)89003-6. [DOI] [PubMed] [Google Scholar]
- 33.Kosloff M, Selinger Z. Substrate assisted catalysis – application to G proteins. Trends Biochem Sci 26. 2001:257–262. doi: 10.1016/s0968-0004(00)01748-5. [DOI] [PubMed] [Google Scholar]
- 34.Pasqualato S, Cherfils J. Crystallographic evidence for substrate-assisted GTP hydrolysis by a small GTP binding protein. Structure. 2005;13:533–540. doi: 10.1016/j.str.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 35.Grigorenko BL, Rogov AV, Nemukhin AV. On the mechanism of triphosphate hydrolysis in aqueous Solution: QM/MM simulations in water clusters. J Phys Chem B. 2006;110:4407–4412. doi: 10.1021/jp056395w. [DOI] [PubMed] [Google Scholar]
- 36.Rogov AV, Grigorenko BL, Bochenkova AV, Granovsky AA, Nemukhin AV. The role of magnesium in hydrolysis reaction of triphosphates in water: Modeling by the quantum mechanical – molecular mechanical method. Moscow Univ Chem Bull. 2007;62:123–127. [Google Scholar]
- 37.Kale L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. NAMD2: Greater scalability for parallel molecular dynamics. J Comp Phys. 1999;151:283–312. [Google Scholar]
- 38.Brooks BR, Bruccoleri RE, Olafson BD, States DS, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 39.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fisher S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 40.Warshel A, Levitt M. Theoretical studies of enzymatic reactions: Dielectric, electrostatic and steric stabilization of the carbonium ion in the reaction of lyzozyme. J Molec Biol. 1976;103:227–249. doi: 10.1016/0022-2836(76)90311-9. [DOI] [PubMed] [Google Scholar]
- 41.Grigorenko BL, Nemukhin AV, Topol IA, Burt SK. Modeling of biomolecular systems with the quantum mechanical and molecular mechanical method based on the effective fragment potential technique: Proposal of flexible fragments. J Phys Chem A. 2002;106:10663–10672. [Google Scholar]
- 42.Nemukhin AV, Grigorenko BL, Topol IA, Burt SK. Flexible effective fragment QM/MM method: Validation through the challenging tests. J Comput Chem. 2003;24:1410–1420. doi: 10.1002/jcc.10309. [DOI] [PubMed] [Google Scholar]
- 43.Gordon MS, Freitag MA, Bandyopadhyay P, Jensen JH, Kairys V, Stevens WJ. The effective fragment potential method: A QM-based MM approach to modeling environmental effects in chemistry. J Phys Chem A. 2001;105:293–307. [Google Scholar]
- 44.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nuclear acids, and organic molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 45.Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su SJ, Windus TL, Dupuis M, Montgomery JA. General atomic and molecular electronic structure system. J Comp Chem. 1993;14:1347–1363. [Google Scholar]
- 46.Nemukhin AV, Grigorenko BL, Granovsky AA. Molecular modeling with the PC GAMESS package: From diatomic molecules to enzymes. Moscow Univ Chem Bull. 2004;45:75–102. [Google Scholar]
- 47.Ren P, Ponder JW. Polarizable Atomic Multipole Water Model for Molecular Mechanics Simulation. J Phys Chem B. 2003;107:5933–5947. [Google Scholar]
- 48.Hay PJ, Wadt WR. Ab initio effective core potentials for molecular calculations. Potentials for main group elements Na to Bi. J Chem Phys. 1985;82:284–298. [Google Scholar]
- 49.Sondek J, Lambright DG, Noel JP, Hamm HE, Sigler PB. GTPase mechanism of G proteins from the 1.7-Å crystal structure of transducin α GDP AlF4−. Nature. 1994;372:276–279. doi: 10.1038/372276a0. [DOI] [PubMed] [Google Scholar]