Abstract
Background
The C1858T polymorphism in PTPN22 has been associated with risk of systemic lupus erythematosus (SLE), as well as multiple other autoimmune diseases. We have previously shown that high serum interferon alpha (IFN-α) activity is a heritable risk factor for SLE, and we hypothesized that the PTPN22 risk variant may shift serum cytokine profiles to higher IFN-α activity resulting in risk of disease.
Methods
IFN-α was measured in 143 SLE patients using a functional reporter cell assay, and TNF-α was measured with ELISA. The rs2476601 SNP in PTPN22 (C1858T) was genotyped in the same patients. Patients were grouped using a clustering algorithm into four cytokine groups (IFN-α predominant, IFN-α and TNF-α correlated, TNF-α predominant, and IFN-α and TNF-α both low).
Results
SLE patients carrying the risk allele of PTPN22 had higher serum IFN-α activity than patients lacking the risk allele (p=0.027). TNF-α levels were lower in risk allele carriers (p=0.030), and the risk allele was more common in patients with an IFN-α predominant or IFN-α and TNF-α correlated cytokine profile as compared to patients with TNF-α predominance or both cytokines low (p=0.002). 25% of male patients carried the risk allele, compared to 10% of female patients (p=0.02), however cytokine skewing was similar in both sexes.
Conclusions
The autoimmune disease risk allele of PTPN22 is associated with skewing of serum cytokine profiles toward higher IFN-α activity and lower TNF-α in SLE patients in vivo. This serum cytokine pattern may be relevant in other autoimmune diseases associated with the PTPN22 risk allele.
The pathogenesis of SLE is likely driven by a combination of genetic risk factors and environmental events which lead to an irreversible break in immunologic self-tolerance. IFN-α is a pleiotropic type I interferon with the potential to break self-tolerance by activating antigen presenting cells after uptake of self material (1). Serum IFN-α levels are frequently elevated in SLE patients (2). Additionally, a number of patients treated with recombinant human IFN-α for malignancy and chronic viral hepatitis have developed de novo SLE, which typically resolves after the IFN-α is discontinued (3). These data suggest a potential role for IFN-α in SLE susceptibility. In previous work, we have demonstrated that abnormally high serum IFN-α is common in SLE families in both healthy and SLE-affected members, as compared to unrelated individuals (4). These data implicate high serum IFN-α as a heritable SLE risk factor, however the causative genes underlying this risk factor are not known.
The C1858T polymorphism in PTPN22 (rs2476601) has been associated with risk of SLE, as well as multiple other autoimmune diseases including autoimmune thyroid disease, juvenile idiopathic arthritis, rheumatoid arthritis, and type I diabetes (5). One genetic association study found the risk allele of PTPN22 was associated with SLE only in those SLE patients with concomitant autoimmune thyroid disease (AITD) (6). We have recently shown that AITD is associated with high serum IFN-α activity (7), and AITD is a frequent complication of recombinant IFN-α therapy for chronic viral hepatitis (8). IFN-α has also been implicated in the pathogenesis of many of the other diseases associated with the PTPN22 risk allele, including juvenile idiopathic arthritis (9) and type I diabetes (10). Interestingly, the PTPN22 risk allele is not associated with multiple sclerosis (5). Multiple sclerosis is commonly treated with IFN-β, a type I IFN which signals through the same receptor as IFN-α.
The mechanism by which the risk variant of PTPN22 predisposes to autoimmunity is unknown. The polymorphism results in an arginine to tryptophan coding change in the Lyp protein, and work in lymphocytes suggests that the mutation results in decreased T- and B-cell responsiveness, as well as alterations in cytokine production in lymphocytes in vitro (11). While work has thus far focused on lymphocytes, the Lyp protein could presumably alter function in myeloid and dendritic cells. IFN-α has not previously been studied in the context of the PTPN22 risk allele, and any relationship between the risk allele and in vivo serum cytokine profiles is unknown. Given the clustering of high serum IFN-α activity in certain SLE families (4), and the overlapping association of PTPN22 with SLE and a number of other autoimmune diseases in which IFN-α is thought to be important in pathogenesis, we set out to examine serum IFN-α activity in SLE patients as it relates to PTPN22 genotype. We hypothesized that PTPN22 may be associated with high serum IFN-α, which could potentially explain its association with SLE as well as other autoimmune diseases.
Methods
Patients and samples
Serum and genomic DNA samples were obtained from the Hospital for Special Surgery (HSS) Lupus Family Registry, HSS Lupus Registry, and the Translational Research Initiative in the Department of Medicine (TRIDOM) at the University of Chicago. 143 SLE patients of European-American and Hispanic ancestry were studied. 141 healthy donor serum samples were used to standardize the IFN-α assay, as previously described (4). The study was approved by the institutional review boards at all institutions, and informed consent was obtained from all subjects in the study.
Reporter cell assay for IFN-α
The reporter cell assay for IFN-α has been described in detail elsewhere (4, 12). In this assay, reporter cells were used to measure the ability of patient sera to cause IFN-induced gene expression. The reporter cells (WISH cells, ATCC #CCL-25) were cultured with 50% patient sera for 6 hours, and then lysed. mRNA was purified from cell lysates, and cDNA was made from total cellular mRNA. cDNA was then quantified using real-time PCR with the Biorad SYBR Green fluorophore system. Forward and reverse primers for the genes MX1, PKR, and IFIT1, which are known to be highly and specifically induced by IFN-α, were used in the reaction (4). GAPDH was amplified in the same samples to control for background gene expression.
Real Time PCR Data Analysis
The amount of PCR product of the IFN-α-induced gene was normalized to the amount of product for the housekeeping gene GAPDH in the same sample. The relative expression of each of the three tested IFN-induced genes was calculated as a fold increase compared to its expression in WISH cells cultured with media alone. Healthy unrelated donor sera (n=141) were tested in the WISH assay to establish a normal value for IFN-α activity, and the mean and standard deviation (SD) of IFN-α-induced gene relative expression induced by healthy donor sera were calculated. The ability of patient serum samples to cause IFN-induced gene expression in the reporter cells was then compared to the mean and SD induced by healthy unrelated donor serum. The number of SD above healthy donors for each gene was calculated as described in (4).
TNF-alpha ELISA
TNF alpha was measured in serum samples using the Pierce Human Monoclonal TNF-α ELISA per manufacturer instructions. Healthy unrelated donor samples were tested (n=18) and performed as expected (mean =1.50 pg/mL, SD = 2.43 pg/mL).
Genotyping
Individuals in the HSS registries were genotyped at the rs2476601 SNP using ABI Taqman Assays-by-Design primers and probes on an ABI 7900HT PCR machine. SNP genotyping was performed with >99% completeness among registry samples.
Statistical Analysis
Two-sided Fisher's exact test (sum of small p's method for observed ≥ expected) was used to analyze categorical data, and Mann-Whitney non-parametric t-test was used to compare quantitative data. K-median clustering of SLE patients by IFN-α and TNF-α levels was performed using Cluster software by Eisen MB, et al (http://rana.lbl.gov/EisenSoftware.htm). Parameters were set to three clusters and 10,000 iterations. This resulted in three groups of patients, one in which IFN-α levels were much higher than TNF-α levels, one group in which IFN-α and TNF-α levels were correlated, and one group in which TNF-α levels were much higher than IFN-α levels. Patients who did not have any significant elevation in TNF-α or IFN-α (not more than 1 SD above the mean of healthy unrelated donors) were separated into a 4th group following the clustering algorithm. Thus, the group of patients designated as IFN-α and TNF-α correlated consists of patients with significant elevations in the levels of both cytokines, and those with low levels of both were considered separately.
Results
PTPN22 genotyping
18 of the 143 SLE patients studied carried the risk allele (17 C/T genotypes and one T/T genotype). Interestingly, 6 of the 24 (25%) male SLE patients studied carried the risk allele, compared to 12 of 119 (10%) of the female SLE patients. The one patient with T/T genotype was male, so comparing allelic frequencies by sex, 7 of 48 (15%) alleles in the male patients were risk alleles, as compared to 12 of 238 (5%) in females (p=0.02).
PTPN22 T/- genotype is associated with high serum IFN-α activity in SLE patients
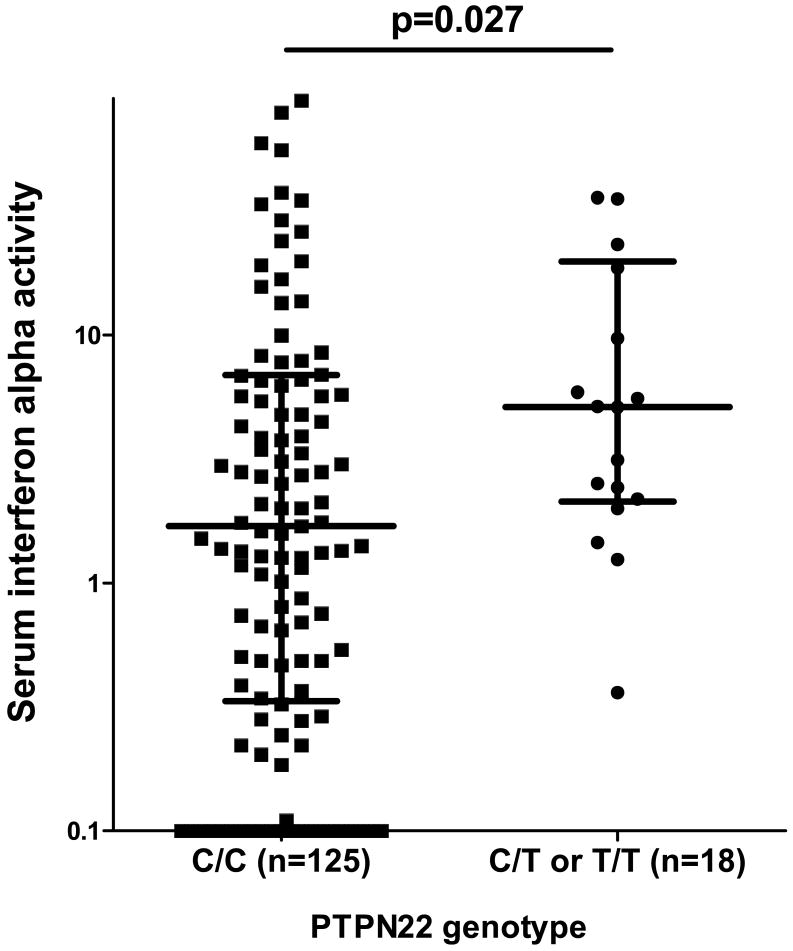
When SLE patients are stratified by PTPN22 genotype, the patients with the SLE-risk T allele had higher serum IFN-α than those lacking the risk allele (p=0.03), as shown in Figure 1. We have previously shown that anti-RNA-binding protein (anti-RBP) antibodies such as Ro, La, Sm, and RNP, as well as anti-dsDNA antibodies are associated with high serum IFN-α in SLE patients (4), however there were no significant differences in the proportion of patients positive for these antibodies in the PTPN22 risk allele vs. non-risk allele carriers (p=0.9 and p=0.8 for anti-dsDNA and anti-RBP respectively).
Figure 1.
Serum interferon alpha activity in SLE patients stratified by PTPN22 genotype (please see methods for a description of the IFN-α activity measurement). Line represents the median, error bars show the inter-quartile range. p value by Mann-Whitney t-test.
PTPN22 T/- genotype is associated with skewing of serum cytokine profile away from high TNF-α and toward high IFN-α in SLE patients
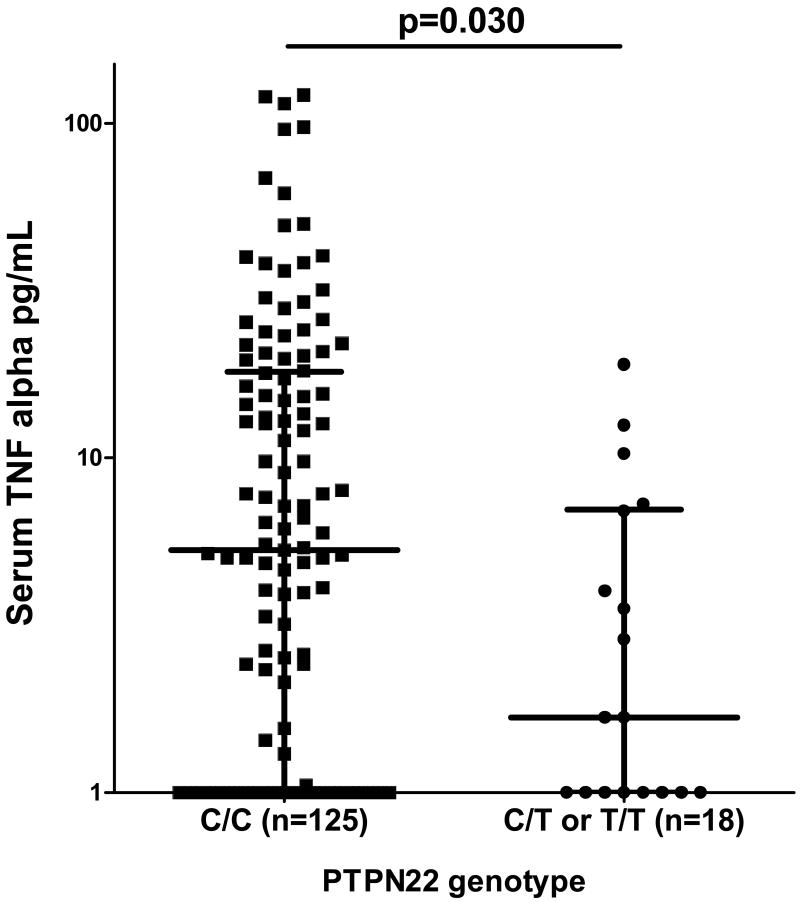
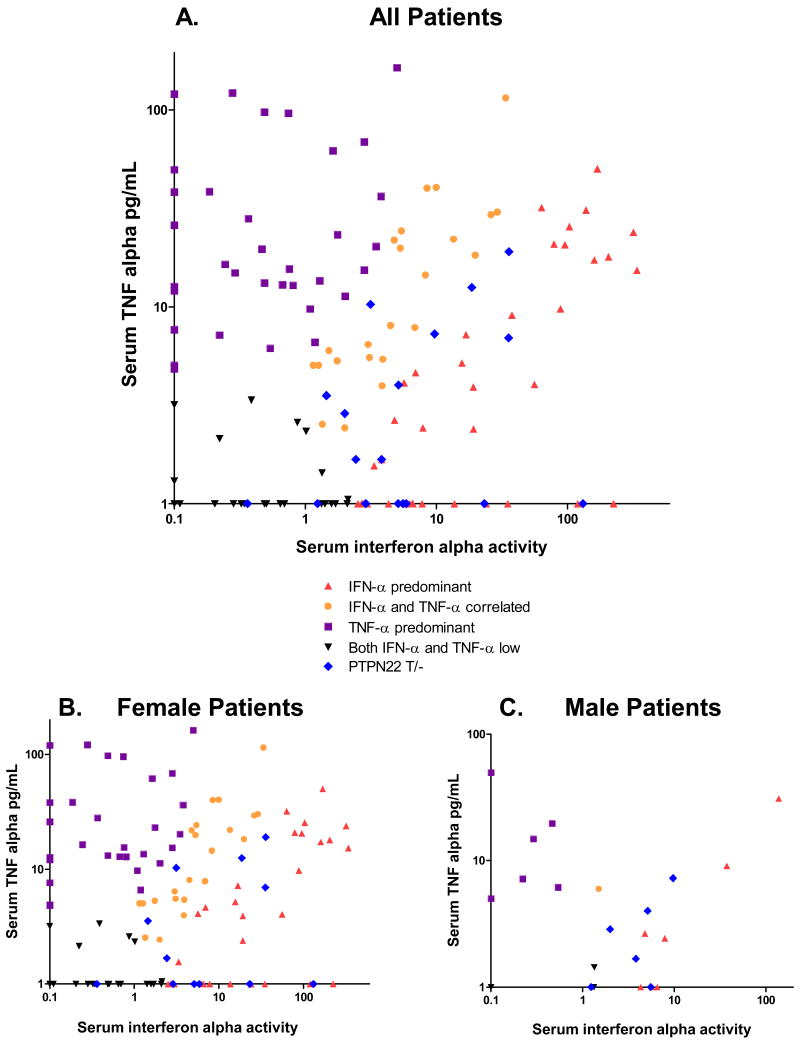
When TNF-α levels were examined in the same patients, patients with the SLE-risk T allele of PTPN22 showed a strong trend toward lower serum TNF-α levels as compared to those lacking the risk allele (p=0.03). Quantitative TNF-α data is shown in Figure 2 stratified by PTPN22 genotype. When patients were grouped according to levels of both cytokines, the PTPN22 risk allele were largely represented in the IFN-α predominant (10 of 18) or in the IFN-α and TNF-α correlated categories (6 of 18), as shown in Figure 3. None of the risk allele carriers were TNF-α predominant, and two were IFN-α and TNF-α low. Thus, 16 of the 76 (21%) patients in the IFN-α predominant or IFN-α and TNF-α correlated groups carried the PTPN22 risk allele, as compared to 2 of 67 patients (3%) in the TNF-α predominant or IFN-α and TNF-α both low groups (p=0.001). Males and females showed similar skewing of cytokine profiles (Figure 3), and cytokine skewing in female risk allele carriers as compared to female non-risk allele carriers was independently significant (p=0.01), demonstrating that an increased proportion of males in the risk allele group was not driving the association between skewed serum cytokine patterns and PTPN22 risk allele carriage.
Figure 2.
Serum TNF-α (pg/mL) in SLE patients stratified by PTPN22 genotype. Line represents the median, error bars show the inter-quartile range. p value by Mann-Whitney t-test.
Figure 3.
Serum IFN-α activity plotted against serum TNF-α levels from the same sample in SLE patients. Colors represent different groupings as designated by the clustering algorithm run with Cluster software. PTPN22 risk allele carriers are indicated separately in blue to show their location on the X-Y plot. A. shows all patients in the study, B. and C. show female and male patients separately.
Discussion
The C1858T variant of PTPN22 has been associated with susceptibility to multiple autoimmune diseases, including SLE (5). We demonstrate skewing of serum cytokine profiles in SLE patients carrying the PTPN22 risk allele toward high serum IFN-α and low serum TNF-α. IFN-α has been implicated as a heritable risk factor for human SLE (4), and the present study suggests that variation at PTPN22 contributes to this heritable risk factor. While the PTPN22 risk allele is rare and cannot account for a large proportion of the high IFN-α seen in SLE patients, this study demonstrates that the subgroup of SLE patients carrying the risk allele of PTPN22 have a distinct serum cytokine phenotype including high IFN-α and low TNF-α as compared to SLE patients lacking this risk allele. This study is cross-sectional in nature, and potential variation in cytokine profiles due to temporal variables such as disease activity are not assessed, although in general large fluctuations in cytokine profiles over time would tend to abolish the patterns we have observed, unless these fluctuations are themselves related to genotype. For example, if PTPN22 risk allele carriage conferred a more stable presence of high serum IFN-α activity and less stable serum TNF-α activity over time, this could result in findings similar to those observed in our study.
In vitro experiments have shown that TNF-α can inhibit the release of IFN-α from plasmacytoid dendritic cells, suggesting that IFN-α and TNF-α can cross-regulate each other (9). Our group and others have shown that anti-TNF-α treatment in Sjogren's syndrome (12) and systemic onset juvenile arthritis (9) results in increased serum IFN-α activity, suggesting that such cross-regulation could be present in vivo in patients with autoimmune disease. A previous study reported that the PTPN22 risk allele was associated with invasive bacterial infection (13), and bacterial infection is a situation in which TNF-α may be a more important defensive cytokine than IFN-α. One of the known complications of therapy with anti-TNF-α agents is an increased risk of serious bacterial infection. If otherwise healthy subjects carrying the PTPN22 risk allele demonstrate a similar skewing of serum cytokine profile away from TNF-α when challenged with a bacterial pathogen, this may explain the seemingly paradoxical finding in which an autoimmune risk allele is also associated with susceptibility to bacterial infection.
Finding an increased proportion of males in the risk allele group is interesting, and to our knowledge has not been reported in previous PTPN22 genetic association studies in SLE. Studies suggest that the PTPN22 risk allele exerts a greater influence on risk of RA in men than in women (14). While high serum IFN-α seems to be equally common and of similar magnitude in females and males with SLE (15), the risk factors underlying the high serum IFN-α trait may differ between the sexes. The cohort presented in this study is small, and replication of PTPN22 sex-skewing in SLE will be important.
Skewing of serum cytokine profiles toward high IFN-α in SLE patients suggests that PTPN22 may exert some of risk of autoimmunity via the IFN-α pathway in other autoimmune diseases. Further study of the IFN-α system in the other PTPN22-associated diseases and better understanding of the underlying cell biology will likely improve our understanding of a diverse range of autoimmune phenomena.
Footnotes
Disclosures and Support: SN Kariuki – none; MK Crow – patent pending for interferon assay, research grants from NIAID R01 AI059893, Alliance for Lupus Research, Mary Kirkland Center for Lupus Research, and Lupus Research Institute; TB Niewold – NIH T32 AR07517, NIAID Clinical Research Loan Repayment AI071651, Arthritis Foundation Post-Doctoral Fellowship Award; support for HSS Family Lupus Registry – Toys ‘R Us Foundation and the S.L.E. Foundation, Inc.
References
- 1.Blanco P, Palucka AK, Gill M, Pascual V, Banchereau J. Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science. 2001;294(5546):1540–3. doi: 10.1126/science.1064890. [DOI] [PubMed] [Google Scholar]
- 2.Hooks JJ, Moutsopoulos HM, Geis SA, Stahl NI, Decker JL, Notkins AL. Immune interferon in the circulation of patients with autoimmune disease. N Engl J Med. 1979;301(1):5–8. doi: 10.1056/NEJM197907053010102. [DOI] [PubMed] [Google Scholar]
- 3.Niewold TB, Swedler WI. Systemic lupus erythematosus arising during interferon-alpha therapy for cryoglobulinemic vasculitis associated with hepatitis C. Clin Rheumatol. 2005;24(2):178–81. doi: 10.1007/s10067-004-1024-2. [DOI] [PubMed] [Google Scholar]
- 4.Niewold TB, Hua J, Lehman TJ, Harley JB, Crow MK. High serum IFN-alpha activity is a heritable risk factor for systemic lupus erythematosus. Genes Immun. 2007;8:492–502. doi: 10.1038/sj.gene.6364408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee YH, Rho YH, Choi SJ, Ji JD, Song GG, Nath SK, et al. The PTPN22 C1858T functional polymorphism and autoimmune diseases--a meta-analysis. Rheumatology (Oxford) 2007;46(1):49–56. doi: 10.1093/rheumatology/kel170. [DOI] [PubMed] [Google Scholar]
- 6.Wu H, Cantor RM, Graham DS, Lingren CM, Farwell L, Jager PL, et al. Association analysis of the R620W polymorphism of protein tyrosine phosphatase PTPN22 in systemic lupus erythematosus families: increased T allele frequency in systemic lupus erythematosus patients with autoimmune thyroid disease. Arthritis Rheum. 2005;52(8):2396–402. doi: 10.1002/art.21223. [DOI] [PubMed] [Google Scholar]
- 7.Mavragani CP, Danielides S, Niewold TB, Kirou KA, Moutsopoulos HM, Crow MK. Activation of the type I interferon pathway in autoimmune thyroid disease. Arthritis Rheum. 2007;56(suppl):S229. [Google Scholar]
- 8.Ioannou Y, Isenberg DA. Current evidence for the induction of autoimmune rheumatic manifestations by cytokine therapy. Arthritis Rheum. 2000;43(7):1431–42. doi: 10.1002/1529-0131(200007)43:7<1431::AID-ANR3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 9.Palucka AK, Blanck JP, Bennett L, Pascual V, Banchereau J. Cross-regulation of TNF and IFN-alpha in autoimmune diseases. Proc Natl Acad Sci U S A. 2005;102(9):3372–7. doi: 10.1073/pnas.0408506102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devendra D, Eisenbarth GS. Interferon alpha--a potential link in the pathogenesis of viral-induced type 1 diabetes and autoimmunity. Clin Immunol. 2004;111(3):225–33. doi: 10.1016/j.clim.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 11.Rieck M, Arechiga A, Onengut-Gumuscu S, Greenbaum C, Concannon P, Buckner JH. Genetic variation in PTPN22 corresponds to altered function of T and B lymphocytes. J Immunol. 2007;179(7):4704–10. doi: 10.4049/jimmunol.179.7.4704. [DOI] [PubMed] [Google Scholar]
- 12.Mavragani CP, Niewold TB, Moutsopoulos NM, Pillemer SR, Wahl SM, Crow MK. Augmented interferon-alpha pathway activation in patients with Sjogren's syndrome treated with etanercept. Arthritis Rheum. 2007;56(12):3995–4004. doi: 10.1002/art.23062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman SJ, Khor CC, Vannberg FO, Maskell NA, Davies CW, Hedley EL, et al. PTPN22 and invasive bacterial disease. Nat Genet. 2006;38(5):499–500. doi: 10.1038/ng0506-499. [DOI] [PubMed] [Google Scholar]
- 14.Pierer M, Kaltenhauser S, Arnold S, Wahle M, Baerwald C, Hantzschel H, et al. Association of PTPN22 1858 single-nucleotide polymorphism with rheumatoid arthritis in a German cohort: higher frequency of the risk allele in male compared to female patients. Arthritis Res Ther. 2006;8(3):R75. doi: 10.1186/ar1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niewold TB, Adler JE, Glenn SB, Lehman TJA, Harley JB, Crow MK. Lupus families reveal age- and sex- related patterns of interferon alpha expression. Arthritis Rheum. 2008 doi: 10.1002/art.23619. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]