Abstract
To enhance genetic manipulation of the Lyme disease spirochete Borrelia burgdorferi, we assayed the aadA gene for the ability to confer resistance to the antibiotics spectinomycin and streptomycin. Using the previously described pBSV2 as a backbone, a shuttle vector, termed pKFSS1, which carries the aadA open reading frame fused to the B. burgdorferi flgB promoter was constructed. The hybrid flgB promoter-aadA cassette confers resistance to spectinomycin and streptomycin in both B. burgdorferi and Escherichia coli. pKFSS1 has a replication origin derived from the 9-kb circular plasmid and can be comaintained in B. burgdorferi with extant shuttle vector pCE320, which has a replication origin derived from a 32-kb circular plasmid, or pBSV2, despite the fact that pKFSS1 and pBSV2 have the same replication origin. Our results demonstrate the availability of a new selectable marker and shuttle vector for genetically dissecting B. burgdorferi at the molecular level.
Borrelia burgdorferi is the etiologic agent of Lyme disease, the most common arthropod-borne disease in the United States (22, 34). The progressive development of a system to genetically manipulate B. burgdorferi has provided many of the requisite tools to analyze molecular mechanisms in this pathogenic spirochete (4, 9, 10, 26, 29, 31, 33, 35).
Four antibiotic resistance markers have been used for molecular genetic analyses of B. burgdorferi (4, 12, 29, 31). The first marker developed, a mutant gyrB allele that confers coumermycin A1 resistance (26, 29, 30), has been used for targeted gene disruption in B. burgdorferi (5, 11, 17, 38-40). However, caveats regarding the coumermycin A1 resistance marker include the tendency of the gyrB gene to recombine at the chromosomal locus, which necessitates extensive screening of transformants (26), and pleiotropic effects that are likely caused by perturbed levels of DNA supercoiling (1, 2) (D. S. Samuels, C. R. Kuchel, and M. E. Kresge, unpublished data). In response to the need for an exogenous selectable marker, Bono et al. fused the open reading frame of aphI, a kanamycin resistance gene from Tn903, to either the B. burgdorferi flaB or flgB promoters, thereby developing kanamycin resistance cassettes that function in B. burgdorferi (4). These hybrid resistance markers have been used to disrupt genes (7, 12, 18, 20, 23), complement a gene (16, 25), demonstrate transduction by bacteriophage φBB1 (10), and construct Escherichia coli-B. burgdorferi shuttle vectors (9, 35, 36). A third selectable marker, the ermC gene conferring erythromycin resistance, was originally introduced into B. burgdorferi on a broad-host-range plasmid from gram-positive bacteria (31). It has been used to disrupt genes (15, 16, 18) and to conduct complementation studies in B. burgdorferi (32). A gentamicin resistance marker was constructed by fusing the aacC1 open reading frame to the flgB promoter and used to disrupt genes (12, 36). Other antibiotic resistance markers have been identified recently but not yet used to genetically manipulate B. burgdorferi (33).
With the new tools available to study B. burgdorferi at the molecular level, genetic manipulation is poised to become powerful and convenient. Currently, the genetic system needs to be augmented by developing additional selectable markers for complementation studies, multiple gene disruptions, and dual-plasmid experiments. Therefore, we investigated whether the aadA gene, which encodes an aminoglycoside-3′′-adenylyltransferase that confers resistance to spectinomycin and streptomycin (19, 24), can function in B. burgdorferi.
Shuttle vector construction and transformation.
We constructed a hybrid antibiotic resistance marker by fusing the aadA open reading frame to the strong constitutive flgB promoter (13) on the backbone of the extant E. coli-B. burgdorferi shuttle vector pBSV2 (35). The aadA gene from R100, a broad-host-range plasmid originally isolated from Shigella flexneri (21, 37), was generously provided by Joachim Frey (Institute for Veterinary Bacteriology, University of Berne, Berne, Switzerland) on plasmid pHP45Ω (24). Primers aadA F+NdeI (CATATGAGGGAAGCGGTGATC) and aadA R+AatII (GACGTCATTATTTGCCGACTACC), which introduced restriction sites for enzymes NdeI and AatII, respectively, were used to amplify the aadA gene by PCR, essentially as described previously (17, 29). The 802-bp aadA PCR product was then cloned into plasmid pCR2.1-TOPO (Invitrogen) to create pTAaadA1. pBSV2, generously provided by Philip Stewart (Rocky Mountain Laboratories, Hamilton, Mont.), and pTAaadA1 were each digested with AatII and NdeI. The digestion removes the kanamycin resistance open reading frame and part of the zeocin resistance gene from pBSV2. The appropriate restriction fragments were resolved by agarose gel electrophoresis and purified with a QIAEX II gel extraction kit (QIAGEN). pKFSS1 (Fig. 1) was constructed by ligating the open reading frame of aadA (from pTAaadA1) to the flgB promoter (flgBp) carried on the 5.3-kb backbone of AatII- and NdeI-digested pBSV2 (35).
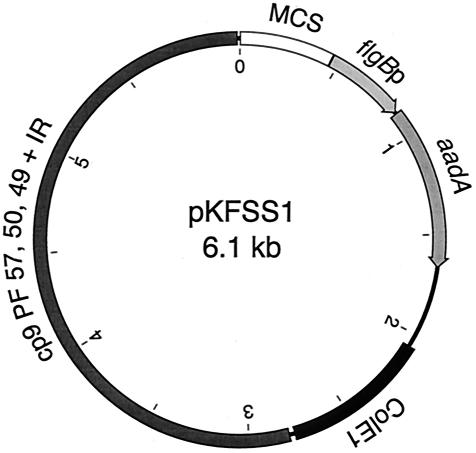
FIG. 1.
Schematic of pKFSS1. pKFSS1 was derived from pBSV2 by replacing the kanamycin resistance open reading frame, under the control of the flgB promoter (flgBp), with a spectinomycin and streptomycin resistance open reading frame (aadA) from pHP45Ω. pBSV2 and pKFSS1 contain the lacZα fragment and multiple cloning site (MCS) and the replication origin (ColE1) of pCR-XL-TOPO. Replication in B. burgdorferi is mediated by a sequence containing genes in paralogous families (PF) 57, 50, and 49 (BBC01, BBC02, and BBC03; open reading frames 1, 2, and 3) and two inverted repeats (IR) from cp9, the 9-kb circular plasmid of B. burgdorferi.
B. burgdorferi sensu stricto strain B31-A, a high-passage avirulent clone of strain B31 (4), was routinely cultivated in modified Barbour-Stoenner-Kelly (BSK-H) complete medium (Sigma) at 34°C with a ∼5% CO2 atmosphere. B. burgdorferi was electroporated with 1 to 10 μg of plasmid DNA and plated in semisolid medium as previously described (27), although 10× medium 199 was fully or partially substituted for 10× CMRL-1066 in some experiments. Transformants were selected in either 160 to 400 μg of kanamycin ml−1, 1.5 μg of spectinomycin ml−1, or 20 to 80 μg of streptomycin ml−1. A mock electrotransformation (no DNA) served as a control.
Selection of transformants.
B. burgdorferi was transformed with either pBSV2, which carries the flgBp::aphI kanamycin resistance cassette, or pKFSS1, which carries the flgBp::aadA spectinomycin and streptomycin resistance cassette. Cells were plated in medium containing kanamycin, spectinomycin, or streptomycin to assess whether these antibiotics can be used to select for B. burgdorferi transformants (Table 1). Transformation of B. burgdorferi with pKFSS1 yielded colonies on medium containing either streptomycin or spectinomycin, but not kanamycin. Electroporation with no DNA, pBSV2, or pKFSS1 yielded high numbers of colonies when plated on medium containing spectinomycin, but only cells transformed with pKFSS1 produced colonies on plates containing streptomycin.
TABLE 1.
Transformation efficiencies of B. burgdorferi with pBSV2 or pKFSS1 in semisolid medium containing different antibioticsa
| Plasmid | No. of colonies/μg of DNA (±SEM)
|
||
|---|---|---|---|
| Streptomycin | Spectinomycin | Kanamycin | |
| pBSV2 (aphI) | 0 (±0) | 327 (±41) | 220 (±134) |
| pKFSS1 (aadA) | 79 (±27) | 695 (±136) | 0 (±0) |
Control transformations with no DNA yielded no colonies when plated on medium containing streptomycin or kanamycin, and the equivalent of 863 (±234) colonies when plated on medium containing spectinomycin.
B. burgdorferi transformants isolated by selection with spectinomycin or streptomycin were analyzed for the presence of pKFSS1 by transforming total genomic DNA from these clones into E. coli. Genomic DNA was extracted from five randomly chosen spectinomycin-resistant and two streptomycin-resistant B. burgdorferi colonies as described previously (28). DNA was transformed into chemically competent E. coli JM109, and the cells were plated on semisolid Luria-Bertani medium containing 60 to 100 μg of spectinomycin ml−1. flgBp::aadA confers resistance to both streptomycin and spectinomycin in E. coli, but many common cloning strains are resistant to streptomycin. Plasmid DNA was recovered from E. coli transformants by alkaline lysis (3). Only those E. coli cells transformed with DNA from streptomycin-resistant pKFSS1 transformants of B. burgdorferi acquired resistance to spectinomycin; pKFSS1 was recovered from these E. coli transformants. pKFSS1 was not recovered from any B. burgdorferi colonies isolated on plates containing spectinomycin, suggesting that they were background mutants.
Plasmid stability.
A randomly chosen B. burgdorferi transformant carrying pKFSS1 was serially passaged in liquid BSK-H medium without streptomycin selection at 34°C. After about 50 generations, the culture was plated on semisolid medium without selection, and 10 colonies were isolated. Total genomic DNA was extracted from B. burgdorferi clones and used to transform E. coli DH5α to screen for pKFSS1, as described above. Six of the 10 colonies isolated in the absence of streptomycin selection contained pKFSS1. Therefore, B. burgdorferi transformants maintained pKFSS1 for 50 generations. However, pKFSS1 is not as stable as the parental plasmid, pBSV2, which is maintained in all 20 colonies following ∼90 passages in the absence of selection, as assayed by PCR (36). This difference in stability was unexpected, because the two plasmids have the same replication origin and are nearly identical except for the selectable marker; pKFSS1 also has a truncated zeocin resistance gene downstream of the aadA gene.
Antibiotic resistance in B. burgdorferi.
The ability of pKFSS1 to confer resistance to selected aminoglycosides and spectinomycin was measured to assess whether the flgBp::aadA marker could be used in conjunction with the extant selectable markers that confer resistance to similar antibiotics. Resistance was assayed by inoculating media containing various concentrations of streptomycin, spectinomycin, kanamycin, and gentamicin antibiotics. Cultures of wild-type and transformed B. burgdorferi were inoculated into 4-ml cultures of BSK-H at approximately 106 cells ml−1 and grown at 34°C for about 72 h in the presence of the antibiotics. Cultures were assayed for growth by spectrophotometry as previously described (28). Assays were replicated three to five times. The antibiotic concentration that inhibits 50% of bacterial growth was determined for wild-type B. burgdorferi and pBSV2 and pKFSS1 transformants (Table 2). The flgBp::aadA cassette in B. burgdorferi transformants confers 10-fold resistance to spectinomycin and approximately 100-fold resistance to streptomycin. No cross-resistance to kanamycin or gentamicin was observed with the flgBp::aadA cassette. Additionally, the flgBp::aphI cassette carried on pBSV2 does not confer resistance to either spectinomycin or streptomycin.
TABLE 2.
Antibiotic susceptibility of B. burgdorferi clones containing pBSV2 or pKFSS1
| Plasmid | IC50 (μg ml−1)a
|
|||
|---|---|---|---|---|
| Streptomycin | Spectinomycin | Kanamycin | Gentamicin | |
| None | 7 | 0.1 | 7 | 2 |
| pBSV2 (aphI) | 3 | 0.2 | >1,400 | 6 |
| pKFSS1 (aadA) | 650 | 1.4 | 10 | 3 |
The IC50 is the concentration of antibiotic that inhibits growth by 50%.
Plasmid compatibility.
The ability to transform B. burgdorferi with two different shuttle vectors would expand experimental opportunities. Therefore, we transformed cells carrying pKFSS1 with the shuttle vector pCE320 (9), which uses a compatible replication origin from a 32-kb circular plasmid and the flgBp::aphI kanamycin resistance cassette. Plasmids carrying the same replication origin typically are not maintained together in the same cell (14), but plasmid incompatibility has not been extensively studied in B. burgdorferi (9, 35, 36). Therefore, pKFSS1 was also cotransformed with its parental plasmid pBSV2. Plasmids were electroporated into B. burgdorferi, and cells were plated in semisolid medium containing both kanamycin and streptomycin, as described above. As expected, transforming cells carrying pKFSS1 with pCE320 yielded colonies on plates with both kanamycin and streptomycin, indicating that the cells contained both resistance cassettes. Surprisingly, colonies resistant to both kanamycin and streptomycin were also obtained from cells carrying pKFSS1 transformed with pBSV2, although both plasmids carry the same replication origin derived from the 9-kb circular plasmid cp9 (35).
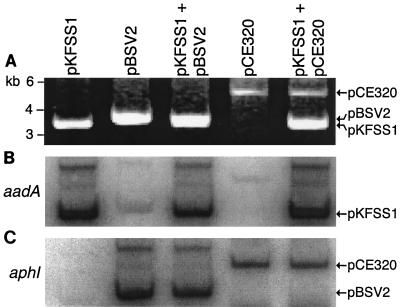
To test that these plasmids autonomously replicated in the cotransformant clones, DNA was isolated using a Wizard Plus Midipreps DNA purification system (Promega) and ∼7 μg was loaded on a 1% agarose gel in 1× TAE (40 mM Tris-20 mM acetate-1 mM EDTA) buffer. Electrophoresis and Southern blotting to an Immobilon-Ny+ membrane (Millipore) were performed essentially as described previously (1, 10), except the hybridization signal was detected using a FLA3000G radioisotope imaging system (Fuji film). The probes were amplified by two rounds of PCR using oligonucleotides aadA F+NdeI and aadA R+AatII or oligonucleotides kanR 88F (AATGTCGGGCAATCAGGTG) and kanR 488R (TCACTCGCATCAACCAAACC), and labeled using a Prime-It II random primer labeling kit (Stratagene). pKFSS1 and pCE320 differ in size, so they are readily resolved and detected in the cloned transformants by ethidium bromide staining (Fig. 2A). pKFSS1 and pBSV2 are similar in size, but they can be distinguished using either an aadA probe specific for pKFSS1 (Fig. 2B) or an aphI probe specific for pBSV2 and pCE320 (Fig. 2C).
FIG. 2.
Plasmid compatibility of pKFSS1 with pBSV2 and pCE320. A B. burgdorferi clone carrying pKFSS1 (6.1 kb; streptomycin resistance) was cotransformed with either pBSV2 (6.4 kb; kanamycin resistance) or pCE320 (8.1 kb; kanamycin resistance) and grown with both kanamycin and streptomycin in semisolid medium. Plasmid DNA, isolated from B. burgdorferi clones carrying pKFSS1 alone, pBSV2 alone, pKFSS1 and pBSV2, pCE320 alone, or pKFSS1 and pCE320, was resolved by 1% agarose gel electrophoresis and visualized by ethidium bromide staining (A). The positions of molecular mass markers (in kilobase pairs [kb]) are indicated to the left of the gel. The gel was transferred to a nylon membrane and probed with either the spectinomycin and streptomycin resistance (aadA) open reading frame (B) or the kanamycin resistance (aphI) open reading frame (C). The aadA probe cross-reacts to a small extent with pBSV2 and pCE320.
Because pKFSS1 and pBSV2 carry the same origin of replication, we assayed plasmid compatibility by growing the cotransformant in the presence or absence of both kanamycin and streptomycin for 40 generations, as described above. Cells were then plated in the absence of antibiotic or in the presence of kanamycin, streptomycin, or both kanamycin and streptomycin to assess the stability of pKFSS1 and pBSV2 (Table 3). We expected that the same number of colonies would be obtained on all four plates from cells grown in the presence of antibiotics, which would maintain selective pressure on those cells containing both plasmids. Interestingly, cells on plates with streptomycin (either alone or with kanamycin) formed about 30% fewer colonies than cells on plates with kanamycin. We speculate that cells carrying pKFSS1 have a lower plating efficiency than cells carrying pBSV2, which may explain the observed lower transformation efficiency of pKFSS1 (Table 1). The mechanism for this lower plating efficiency is unknown and unanticipated, considering the similarity of pKFSS1 and pBSV2. Most cells grown in the absence of antibiotic maintained pBSV2. However, only about half the colonies were obtained on plates containing streptomycin (selecting for pKFSS1) from the cells grown without antibiotic compared to cells grown with an antibiotic. This is consistent with the above observation that pKFSS1 is not as stable as pBSV2. Several colonies from the B. burgdorferi cotransformant grown without antibiotic were isolated and assayed for plasmid DNA by transformation of E. coli, as described above. All colonies contained the expected plasmid(s) selected with the antibiotic(s) used for plating (data not shown).
TABLE 3.
Plating efficiencies in the presence of two antibiotics of a B. burgdorferi clone carrying both pBSV2 (kanamycin resistance) and pKFSS1 (streptomycin resistance) after 40 generations with or without selection
| Treatmenta | % of colonies compared to no antibioticb (±SEM)
|
||
|---|---|---|---|
| Kanamycin | Streptomycin | Kanamycin and streptomycin | |
| With selection | 95 (±1) | 70 (±5) | 79 (±8) |
| Without selection | 93 (±12) | 36 (±9) | 27 (±11) |
Selection with kanamycin and streptomycin.
Plates without antibiotics yielded a mean of 448 colonies (= 100%) from the cultures grown for 40 generations with selection (both kanamycin and streptomycin) and 379 colonies (= 100%) from the cultures grown for 40 generations without selection.
Conclusions.
Molecular genetic manipulation in B. burgdorferi, though relatively immature, has finally advanced to the stage where complex genetic studies can be conducted. Several methods to introduce foreign DNA into B. burgdorferi have been described, including electroporation (27, 29), transduction (10), and chemical transformation (12). Gene disruption and complementation experiments, made possible by the development of selectable markers (4, 12, 29, 31, 33) and shuttle vectors (9, 31, 35), are now available for probing gene function in B. burgdorferi.
We examined the ability of the aadA gene to confer spectinomycin and streptomycin resistance in B. burgdorferi. This resistance gene functions in several bacteria, including Myxococcus xanthus, another genetically challenging organism (19). We demonstrated that a hybrid flgBp::aadA cassette provides approximately 100-fold resistance to streptomycin in B. burgdorferi (Table 2) and allows for selection of streptomycin-resistant transformants in semisolid medium. The aadA gene with its native promoter confers low-level antibiotic resistance (B. J. Kimmel, M. E. Kresge, and D. S. Samuels, unpublished data). Spectinomycin-resistant B. burgdorferi arose at a high frequency, regardless of whether they had been electroporated with pKFSS1, pBSV2, or no DNA (Table 1). We were unable to recover plasmid DNA from these spectinomycin-resistant colonies, which suggests that they were naturally occurring background mutants.
The ability to use streptomycin resistance as a selectable marker adds significantly to the existing antibiotic-resistant genes. Streptomycin is not clinically used to treat B. burgdorferi infections in humans, and aadA does not have the potential for homologous recombination into the B. burgdorferi genome. Limitations imposed by the coumermycin A1 marker have resulted in recent genetic studies relying on only the erythromycin resistance and kanamycin resistance markers (10, 12, 15, 16, 18, 20, 23, 25, 32). flgBp::aadA does not confer cross-resistance to either kanamycin or gentamicin (Table 2), allowing this resistance cassette to be used in conjunction with flgBp::aphI and flgBp::aacC1.
pKFSS1 is compatible with either pCE320 or pBSV2, even though pKFSS1 and pBSV2 have the same cp9 origin of replication. The comaintenance of these two plasmids is surprising because pBSV2 appears to displace the parental cp9 plasmid (35). In addition, other B. burgdorferi plasmids with the same replication origin and compatibility locus demonstrate plasmid incompatibility (9, 35, 36). However, unlike the other larger B. burgdorferi plasmids, cp9 does not encode a gene for a paralogous family 32 (PF32) protein (6, 8, 35). PF32 proteins are homologous to the ParA proteins that play a critical role in the stable maintenance of several well-characterized plasmids, including P1 and F (14). A number of factors, including copy number and the absence of the ParA homolog, may contribute to the maintenance of pBSV2 and pKFFS1 in the same B. burgdorferi cell. Whatever the reason, compatibility of pKFSS1 with either pCE320 or pBSV2 enables experimental approaches that require introducing genes or regulatory sequences on two different vectors. flgBp::aadA and pKFSS1 add an essential new capability to the genetic study of B. burgdorferi by furthering techniques to complement mutants, simultaneously disrupt multiple genes, monitor several promoters, or assay gene interactions.
Acknowledgments
K. L. Frank and S. F. Bundle contributed equally to this work.
We thank Meghan Lybecker, Mike Minnick, and Justin Radolf for thoughtful and critical reading of the manuscript, Philip Stewart for providing pBSV2, Joachim Frey for providing pHP45Ω, Betsy Kimmel and Jill Dion for assistance, and Phil Youderian, Philip Stewart, Chuck Sohaskey, Jon Skare, Nyles Charon, Henry Krisch, Jim Bono, Lori Lubke, and Noel Keen for useful discussions or providing other materials.
This work was supported in part by grants from the National Science Foundation (MCB-9722408), National Institutes of Health (AI39695 and AI53195), and the University Grant Program. K.L.F. is the recipient of an IBS-CORE Undergraduate Research Fellowship through a grant from the Howard Hughes Medical Institute, a Watkins Scholarship from The University of Montana, and an Undergraduate Research Assistantship from UM NSF EPSCoR. K.L.F. was also supported by a NSF Research Experience for Undergraduates supplement. C.H.E. was supported by a grant from the National Institutes of Health (AI29735) to Justin D. Radolf and Melissa J. Caimano. D.S.S. was supported in part by a sabbatical award from The University of Montana.
REFERENCES
- 1.Alverson, J., S. F. Bundle, C. D. Sohaskey, M. C. Lybecker, and D. S. Samuels. 2003. Transcriptional regulation of the ospAB and ospC promoters from Borrelia burgdorferi. Mol. Microbiol. 48:1665-1677. [DOI] [PubMed] [Google Scholar]
- 2.Alverson, J., and D. S. Samuels. 2002. groEL expression in gyrB mutants of Borrelia burgdorferi. J. Bacteriol. 184:6069-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1999. Short protocols in molecular biology, 4th ed. Wiley, New York, N.Y.
- 4.Bono, J. L., A. F. Elias, J. J. Kupko III, B. Stevenson, K. Tilly, and P. Rosa. 2000. Efficient targeted mutagenesis in Borrelia burgdorferi. J. Bacteriol. 182:2445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bono, J. L., K. Tilly, B. Stevenson, D. Hogan, and P. Rosa. 1998. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology 144:1033-1044. [DOI] [PubMed] [Google Scholar]
- 6.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 7.Coburn, J., and C. Cugini. 2003. Targeted mutation of the outer membrane protein P66 disrupts attachment of the Lyme disease agent, Borrelia burgdorferi, to integrin αvβ3. Proc. Natl. Acad. Sci. USA 100:7301-7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn, J. J., S. R. Buchstein, L.-L. Butler, S. Fisenne, D. S. Polin, B. N. Lade, and B. J. Luft. 1994. Complete nucleotide sequence of a circular plasmid from the Lyme disease spirochete, Borrelia burgdorferi. J. Bacteriol. 176:2706-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eggers, C. H., M. J. Caimano, M. L. Clawson, W. G. Miller, D. S. Samuels, and J. D. Radolf. 2002. Identification of loci critical for replication and compatibility of a Borrelia burgdorferi cp32 plasmid and use of a cp32-based shuttle vector for expression of fluorescent reporters in the Lyme disease spirochaete. Mol. Microbiol. 43:281-296. [DOI] [PubMed] [Google Scholar]
- 10.Eggers, C. H., B. J. Kimmel, J. L. Bono, A. Elias, P. Rosa, and D. S. Samuels. 2001. Transduction by φBB-1, a bacteriophage of Borrelia burgdorferi. J. Bacteriol. 183:4771-4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elias, A. F., J. L. Bono, J. A. Carroll, P. Stewart, K. Tilly, and P. Rosa. 2000. Altered stationary-phase response in a Borrelia burgdorferi rpoS mutant. J. Bacteriol. 182:2909-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elias, A. F., P. E. Stewart, D. Grimm, M. J. Caimano, C. H. Eggers, K. Tilly, J. L. Bono, D. R. Akins, J. D. Radolf, T. G. Schwan, and P. Rosa. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 70:2139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ge, Y., I. G. Old, I. Saint Girons, and N. W. Charon. 1997. Molecular characterization of a large Borrelia burgdorferi motility operon which is initiated by a consensus σ70 promoter. J. Bacteriol. 179:2289-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerdes, K., J. Møller-Jensen, and R. B. Jensen. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37:455-466. [DOI] [PubMed] [Google Scholar]
- 15.Hübner, A., A. T. Revel, D. M. Nolen, K. E. Hagman, and M. V. Norgard. 2003. Expression of a luxS gene is not required for Borrelia burgdorferi infection of mice via needle inoculation. Infect. Immun. 71:2892-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hübner, A., X. Yang, D. M. Nolen, T. G. Popova, F. C. Cabello, and M. V. Norgard. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. USA 98:12724-12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knight, S. W., B. J. Kimmel, C. H. Eggers, and D. S. Samuels. 2000. Disruption of the Borrelia burgdorferi gac gene, encoding the naturally synthesized GyrA C-terminal domain. J. Bacteriol. 182:2048-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, C., R. G. Bakker, M. A. Motaleb, M. L. Sartakova, F. C. Cabello, and N. W. Charon. 2002. Asymmetrical flagellar rotation in Borrelia burgdorferi nonchemotactic mutants. Proc. Natl. Acad. Sci. USA 99:6169-6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magrini, V., C. Creighton, D. White, P. L. Hartzell, and P. Youderian. 1998. The aadA gene of plasmid R100 confers resistance to spectinomycin and streptomycin in Myxococcus xanthus. J. Bacteriol. 180:6757-6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Motaleb, M. A., L. Corum, J. L. Bono, A. F. Elias, P. Rosa, D. S. Samuels, and N. W. Charon. 2000. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc. Natl. Acad. Sci. USA 97:10899-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakaya, R., A. Nakamura, and Y. Murata. 1960. Resistance transfer agents in Shigella. Biochem. Biophys. Res. Commun. 3:654-659. [DOI] [PubMed] [Google Scholar]
- 22.Nordstrand, A., A. G. Barbour, and S. Bergström. 2000. Borrelia pathogenesis research in the post-genomic and post-vaccine era. Curr. Opin. Microbiol. 3:86-92. [DOI] [PubMed] [Google Scholar]
- 23.Östberg, Y., M. Pinne, R. Benz, P. Rosa, and S. Bergström. 2002. Elimination of channel-forming activity by insertional inactivation of the p13 gene in Borrelia burgdorferi. J. Bacteriol. 184:6811-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 25.Purser, J. E., M. B. Lawrenz, M. J. Caimano, J. K. Howell, J. D. Radolf, and S. J. Norris. 2003. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol. Microbiol. 48:753-764. [DOI] [PubMed] [Google Scholar]
- 26.Rosa, P., D. S. Samuels, D. Hogan, B. Stevenson, S. Casjens, and K. Tilly. 1996. Directed insertion of a selectable marker into a circular plasmid of Borrelia burgdorferi. J. Bacteriol. 178:5946-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samuels, D. S. 1995. Electrotransformation of the spirochete Borrelia burgdorferi, p. 253-259. In J. A. Nickoloff (ed.), Electroporation protocols for microorganisms, vol. 47. Humana Press, Totowa, N.J. [DOI] [PMC free article] [PubMed]
- 28.Samuels, D. S., and C. F. Garon. 1993. Coumermycin A1 inhibits growth and induces relaxation of supercoiled plasmids in Borrelia burgdorferi, the Lyme disease agent. Antimicrob. Agents Chemother. 37:46-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samuels, D. S., K. E. Mach, and C. F. Garon. 1994. Genetic transformation of the Lyme disease agent Borrelia burgdorferi with coumarin-resistant gyrB. J. Bacteriol. 176:6045-6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samuels, D. S., R. T. Marconi, W. M. Huang, and C. F. Garon. 1994. gyrB mutations in coumermycin A1-resistant Borrelia burgdorferi. J. Bacteriol. 176:3072-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sartakova, M., E. Dobrikova, and F. C. Cabello. 2000. Development of an extrachromosomal cloning vector system for use in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 97:4850-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sartakova, M. L., E. Y. Dobrikova, M. A. Motaleb, H. P. Godfrey, N. W. Charon, and F. C. Cabello. 2001. Complementation of a nonmotile flaB mutant of Borrelia burgdorferi by chromosomal integration of a plasmid containing a wild-type flaB allele. J. Bacteriol. 183:6558-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sartakova, M. L., E. Y. Dobrikova, D. A. Terekhova, R. Devis, J. V. Bugrysheva, O. V. Morozova, H. P. Godfrey, and F. C. Cabello. 2003. Novel antibiotic-resistance markers in pGK12-derived vectors for Borrelia burgdorferi. Gene 303:131-137. [DOI] [PubMed] [Google Scholar]
- 34.Steere, A. C. 2001. Lyme disease. N. Engl. J. Med. 345:115-125. [DOI] [PubMed] [Google Scholar]
- 35.Stewart, P., R. Thalken, J. Bono, and P. Rosa. 2001. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol. Microbiol. 39:714-721. [DOI] [PubMed] [Google Scholar]
- 36.Stewart, P. E., G. Chaconas, and P. Rosa. 2003. Conservation of plasmid maintenance functions between linear and circular plasmids in Borrelia burgdorferi. J. Bacteriol. 185:3202-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugino, Y., and Y. Hirota. 1962. Conjugal fertility associated with resistance factor R in Escherichia coli. J. Bacteriol. 84:902-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tilly, K., S. Casjens, B. Stevenson, M. Bono, D. S. Samuels, D. Hogan, and P. Rosa. 1997. The Borrelia burgdorferi circular plasmid cp26: conservation of plasmid structure and targeted inactivation of the ospC gene. Mol. Microbiol. 23:361-373. [DOI] [PubMed] [Google Scholar]
- 39.Tilly, K., A. F. Elias, J. Errett, E. Fischer, R. Iyer, I. Schwartz, J. L. Bono, and P. Rosa. 2001. Genetics and regulation of chitobiose utilization in Borrelia burgdorferi. J. Bacteriol. 183:5544-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tilly, K., L. Lubke, and P. Rosa. 1998. Characterization of circular plasmid dimers in Borrelia burgdorferi. J. Bacteriol. 180:5676-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]